Abstract
Accumulating evidence indicates that noncoding RNAs play important roles in a multitude of biological processes. The striking findings of miRNAs (microRNAs) and lncRNAs (long noncoding RNAs) as members of noncoding RNAs open up an exciting era in the studies of gene regulation. More recently, the reports of circRNAs (circular RNAs) add fuel to the noncoding RNAs research. Human intervertebral disc degeneration (IDD) is a main cause of low back pain as a disabling spinal disease. We have addressed the expression profiles if miRNAs, lncRNAs and mRNAs in IDD (Wang et al., J Pathology, 2011 and Wan et al., Arthritis Res Ther, 2014). Furthermore, we thoroughly analysed noncoding RNAs, including miRNAs, lncRNAs and circRNAs in IDD using the very same samples. Here we delineate in detail the contents of the aforementioned microarray analyses. Microarray and sample annotation data were deposited in GEO under accession number GSE67567 as SuperSeries. The integrated analyses of these noncoding RNAs will shed a novel light on coding-noncoding regulatory machinery.
Keywords: Intervertebral disc degeneration, Noncoding RNAs, MiRNAs, LncRNAs, CircRNAs
| Specifications: Where applicable, please follow the Ontology for Biomedical Investigations: http://obi-ontology.org/page/Main_Page | |
|---|---|
| Organism/cell line/tissue | Homo sapiens/Nucleus pulposus |
| Sex | Control samples: 4 females, 1 male; IDD samples: 2 females, 3 males |
| Sequencer or array type | miRNA microarray: miRCURYTM LNA Array (v.18.0, Exiqon) lncRNA microarray: Arraystar Human LncRNA Array (v2.0, Arraystar) circRNA microarray: Arraystar Human circRNA Array (8x15K, Arraystar) |
| Data format | Raw and analysed |
| Experimental factors | Degeneration vs. normal (cadavers), nucleus pulposus samples were dissected under a stereoscopic microscope |
| Experimental features | Total RNA was extracted with quality control and subsequently subjected to various microarray analyses |
| Consent | All patients and relatives of cadaveric donors gave their written informed consent before study entry. |
| Sample source location | Xi'an, P.R. China |
1. Direct link to deposited data
2. Experimental design, materials and methods
2.1. Sample collections
The Ethics Review Board of Xijing Hospital (No: 20,111,103–7) approved the study. We collected five normal discs from cadaveric donors and five degenerative discs for patients with IDD as control and IDD samples. The details of sample information have been described in our previous studies [1], [2].
The details of the ten samples are available in samples section of GSE67567.
2.2. Noncoding RNAs microarray data
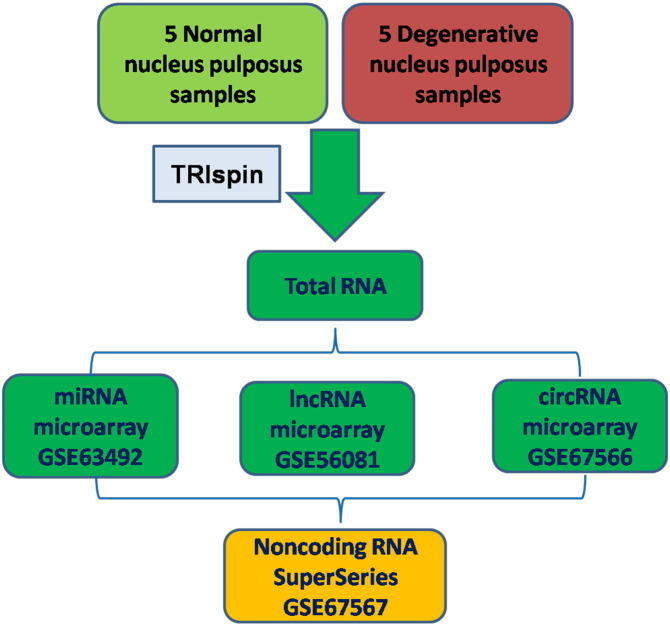
Following total RNA extraction and quality control assay using the TRIspin method [3], we performed miRNA, lncRNA and circRNA microarray on corresponding platforms according to standard procedures. Microarray and sample annotation data were deposited in GEO under accession number GSE67567 as SuperSeries. The SuperSeries is composed of three SubSeries as GSE56081 (lncRNA microarray), GSE63492 (miRNA microarray) and GSE67566 (circRNA microarray). Each of the SubSeries contained in this SuperSeries represents identical RNA samples used for hybridization to different array platforms.
2.3. Quality control
We used Agilent Feature Extraction software (version 11.0.1.1) to analyse acquired array images.
2.4. Normalization and data analysis
Quantile normalization and subsequent data processing were performed with the Gene Spring GX v11.5.1 software package (Agilent Technologies).
After quantile normalization of the raw data, lncRNAs and mRNAs that at least 5 out of 10 samples have flags in Present or Marginal (All Targets Value) were chosen for further data analysis. The normalized intensity for each lncRNA, mRNA, miRNA, circRNA was presented as a log2-transformed pattern. Differentially expressed noncoding RNAs and mRNAs between two groups were identified through fold-change filtering and Student's t-test. Multiple testing corrections were performed by calculating the Benjamini-Hochberg false discovery rate (FDR). Fold-change greater than two, a P-value < 0.05 (two-tailed) and an FDR < 0.05 were considered the criteria for differential expression.
The details of the SuperSeries and SubSeries are listed in Fig. 1.
Fig. 1.
SuperSeries and subseries of noncoding RNAs in human intervertebral disc degeneration.
Acknowledgements
The study was supported by National Natural Science Foundation Grant of China (No. 81270028).
We thank KangChen Bio-tech, Shanghai, P.R. China for their technical support for the microarray works.
References
- 1.Wang H.Q., Yu X.D., Liu Z.H., Cheng X., Samartzis D., Jia L.T., Wu S.X., Huang J., Chen J., Luo Z.J. Deregulated miR-155 promotes Fas-mediated apoptosis in human intervertebral disc degeneration by targeting FADD and caspase-3. J. Pathol. 2011;225:232–242. doi: 10.1002/path.2931. [DOI] [PubMed] [Google Scholar]
- 2.Wan Z.Y., Song F., Sun Z., Chen Y.F., Zhang W.L., Samartzis D., Ma C.J., Che L., Liu X., Ali M., Wang H.Q., Luo Z.J. Aberrantly expressed long noncoding RNAs in human intervertebral disc degeneration: a microarray related study. Arthritis Res. Ther. 2014;16:465. doi: 10.1186/s13075-014-0465-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reno C., Marchuk L., Sciore P., Frank C.B., Hart D.A. Rapid isolation of total RNA from small samples of hypocellular, dense connective tissues. Biotechniques. 1997;22:1082–1086. doi: 10.2144/97226bm16. [DOI] [PubMed] [Google Scholar]