Abstract
RNA sequencing (RNAseq) technology recently allowed the identification of thousands of small RNAs (sRNAs) within the prokaryotic kingdom. However, drawing the comprehensive interaction map of a sRNA remains a challenging task. To address this problem, we recently developed a method called MAPS (MS2 affinity purification coupled with RNA sequencing) to characterize the full targetome of specific sRNAs. This method enabled the identification of target RNAs interacting with sRNAs, regardless of the type of regulation (positive or negative), type of targets (mRNA, tRNA, sRNA) or their abundance. We also demonstrated that we can use this technology to perform a reverse MAPS experiment, where an RNA fragment of interest is used as bait to identify interacting sRNAs. Here, we demonstrated that RybB and MicF sRNAs co-purified with internal transcribed spacers (ITS) of metZ–metW–metV tRNA transcript, confirming results obtained with MS2-RybB MAPS.
Both raw and analyzed RNAseq data are available in GEO database (GSE66517).
Keywords: MS2 affinity purification coupled with RNA sequencing (MAPS), Small RNA (sRNA), Target RNA, RNA sequencing (RNAseq), Internal transcribed spacers (ITS)
| Specifications | |
|---|---|
| Organism/cell line/tissue | Escherichia coli K12 substrain MG1655 |
| Sex | |
| Sequencer or array type | Illumina Miseq |
| Data format | Raw and analyzed |
| Experimental factors | MS2 tagged RNA vs. untagged RNA control |
| Experimental features | MS2-affinity purification coupled with RNA sequencing |
| Consent | |
| Sample source location | |
1. Direct link to deposited data
2. Experimental design, materials and methods
2.1. Constructs
We used MAPS technology to identify sRNA(s) interacting with both internal transcribed spacers (ITS) of metZ–metW–metV tRNA transcript [5]. Because ITSmetZ–metW and ITSmetW–metV have similar but imperfectly repeated sequences (4 mismatches/33 nucleotides), we cloned both sequences individually, under the control of a pBAD promoter (arabinose-inducible promoter). A T7 terminator was added at the end of both ITS to interrupt transcription.
We fused the 5′-end of ITSmetZ–metW and ITSmetW–metV with bacteriophage MS2 RNA stemloops, which are bound by the MS2 coat protein with high specificity. This highly specific interaction was previously shown to allow affinity co-purification of sRNA-bound proteins from bacterial extracts [2].
As controls, we used untagged ITSmetZ–metW and ITSmetW–metV expressed under the same conditions.
We used Northern blot analysis to verify that the MS2-ITSmetZ–metW construct is expressed at a level similar to ITSmetZ–metW and at the expected molecular weight. Similar results were obtained with MS2-ITSmetW–metV and ITSmetW–metV.
2.2. Growth conditions
Cells from an overnight culture grown in rich medium (LB) supplemented with 50 μg/ml ampicillin were diluted 1/1000 in the same fresh medium. For each construct, cells were harvested in exponential (OD600 nm = 0.5; 50 mL) and stationary (OD600 nm = 1; 50 mL) phases.
2.3. Affinity purification
Affinity purification assays were performed as described in Desnoyers and Masse [3] with some modifications (see Fig. S1 in Lalaouna et al. [5] for a schematic representation of MAPS technology). As indicated above, the bacterial strains were grown to an OD600 nm of 0.5 or 1, at which point 0.1% arabinose was added to induce expression from pBAD promoter during 10 min. Cells were then chilled for 10 min on ice. Total RNA was extracted from 600 μL of culture (input) using the hot-phenol procedure [1]. The remaining cells were then centrifuged, resuspended in 1 mL of buffer A (20 mM Tris–HCl pH 8.0, 150 mM KCl, 1 mM MgCl2, 1 mM DTT), and centrifuged again.
At this point, cells carrying MS2-ITSmetZ–metW and MS2-ITSmetW–metV were mixed together. The same was done with cells carrying ITSmetZ–metW and ITSmetW–metV. Cells were resuspended in 3 mL of buffer A and lysed using a French Press (430 psi, four times). Lysate was then cleared by centrifugation (17,000 g, 30 min, 4 °C). The soluble fraction was subjected to affinity chromatography (all steps performed at 4 °C). The column was prepared by adding 100 μL of amylose resin (New England Biolabs) to Bio-Spin disposable chromatography columns (Bio-Rad). The column was then washed with 3 mL of buffer A. Next, 200 pmol of MS2-MBP protein was immobilized on the amylose resin, and the column was washed with 2 mL of buffer A. The cleared lysate was then loaded onto the column, which was washed with 8 mL of buffer A. RNA was eluted from the column with 1 mL of buffer A containing 15 mM maltose. Eluted RNA was extracted with phenol–chloroform, followed by ethanol (3 vol) precipitation of the aqueous phase in the presence of 20 mg of glycogen. RNA samples were then analyzed by Northern blot as described in Lalaouna et al. [5].
2.4. RNA sequencing
After MS2 affinity purification, samples were treated with TURBO™DNase (Ambion). Again, RNA was extracted using phenol–chloroform, followed by ethanol (3 vol) precipitation of the aqueous phase in the presence of 20 mg of glycogen.
RNA quality and quantity assessments were performed on Agilent Nano Chip on the bioanalyzer 2100. The RNAseq library was then built using NEBNext Small RNA Library Prep set E7330S kit from 300 ng total RNA. Size selection was performed with Agencourt AMPure XP beads. Library quality was assessed using Agilent DNA HS Chip. Library quantification was performed by qPCR following Illumina Kappa library quantification protocol. Pooled libraries were sequenced 2 × 50 bp paired-end reads using Illumina MiSeq with 150v3 reagent kit.
2.5. Data processing
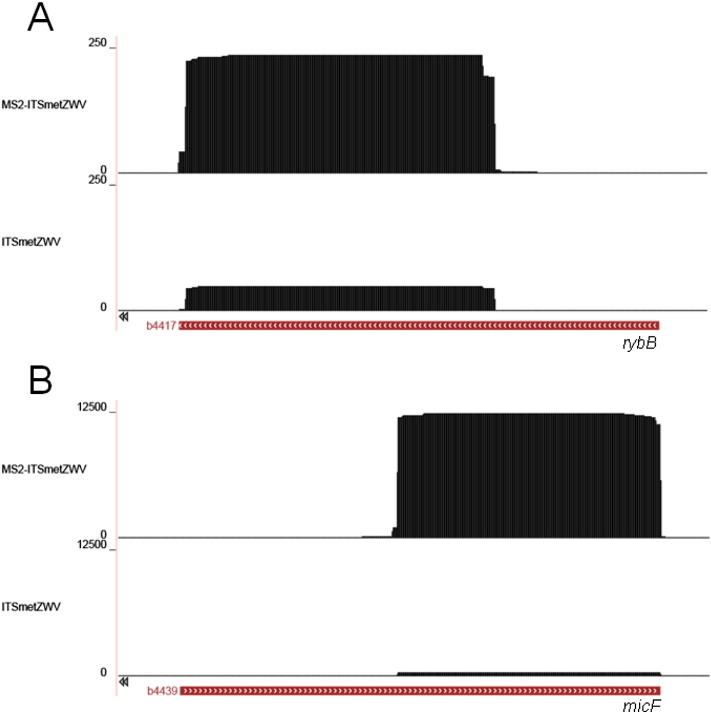
We used Galaxy Project [4] and UCSC Microbial GenomeBrowser [7] to analyze and visualize data. We used FASTQ Groomer (version 1.0.4) to verify and convert FASTQ files. The quality of raw sequence data was controlled with FastQC:Read QC (version 0.52). Then, reads were aligned to the Escherichia coli K12 genome assembly using Map with Bowtie for Illumina (version 1.1.2). We used Create a BedGraph of genome coverage (version 0.1.0) to visualize reads on E. coli MG1655 genome with UCSC Microbial GenomeBrowser. Results obtained for RybB and MicF sRNA are represented in Fig. 1.
Fig. 1.
RNAseq data visualized using GenomeBrowser software. (A) RybB (b4417) and (B) MicF (b4439) sRNAs are highly enriched in MS2-ITSmetZWV pull-down.
Finally, we assigned reads to gene by comparing mapped regions of reads with E. coli gene positions (extracted from GenBank). The processed data files is a tab-delimited text file which includes normalized reads for each sample and MS2-ITSmetZWV/ITSmetZWV ratio. Here, Read counts were normalized by coverage according to Oshlack et al. [6].
Conflict of interest
The authors declare that there are no conflicts of interest.
Acknowledgments
We are grateful to Michelle Scott, Elvy Lapointe and Karine Prévost for excellent bioinformatical and/or technical assistance. This work was supported by an operating grant from the Canadian Institutes of Health Research to E.M. E.M. is a senior scholar from the Fonds de la Recherche en santé du Québec.
References
- 1.Aiba H., Adhya S., de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 2.Corcoran C.P., Rieder R., Podkaminski D., Hofmann B., Vogel J. Use of aptamer tagging to identify in vivo protein binding partners of small regulatory RNAs. Methods Mol. Biol. 2012;905:177–200. doi: 10.1007/978-1-61779-949-5_11. [DOI] [PubMed] [Google Scholar]
- 3.Desnoyers G., Masse E. Noncanonical repression of translation initiation through small RNA recruitment of the RNA chaperone Hfq. Genes Dev. 2012;26:726–739. doi: 10.1101/gad.182493.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goecks J., Nekrutenko A., Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lalaouna D., Carrier M.C., Semsey S., Brouard J.S., Wang J., Wade J.T., Masse E. A 3′ external transcribed spacer in a tRNA transcript acts as a sponge for small RNAs to prevent transcriptional noise. Mol. Cell. 2015;58:393–405. doi: 10.1016/j.molcel.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 6.Oshlack A., Robinson M.D., Young M.D. From RNA-seq reads to differential expression results. Genome Biol. 2010;11:220. doi: 10.1186/gb-2010-11-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneider K.L., Pollard K.S., Baertsch R., Pohl A., Lowe T.M. The UCSC archaeal genome browser. Nucleic Acids Res. 2006;34:D407–D410. doi: 10.1093/nar/gkj134. [DOI] [PMC free article] [PubMed] [Google Scholar]