Abstract
Pancreatic ductal adenocarcinoma (PDAC) remains a common and deadly cancer. Despite numerous efforts, no reliable biomarker is available for daily clinical practice. Circular RNAs (circRNAs) are an abundant, stable and conserved class of RNA molecules that exhibit tissue/developmental-stage-specific expression (Salzman et al., 2012; Jeck et al., 2013; Memczak et al., 2013). CircRNAs play a crucial role in disease, especially in cancer, and provide new potential diagnostic and therapeutic targets for disease (Hansen et al., 2013; Qu et al., 2015).This research was designed to explore the expression profile of circRNAs in PDAC to serve as new diagnosis and treatment strategies for PDAC. Microarray and sample annotation data were deposited in Gene Expression Omnibus (GEO) under accession number GSE69362.
Keywords: Pancreatic cancer, Microarray, Noncoding RNAs, Circular RNAs
| Specifications | |
|---|---|
| Organism/cell line/tissue | Homo sapiens/pancreatic ductal adenocarcinoma and adjacent normal tissues |
| Sex | Male and female |
| Sequencer or array type | Arraystar Human CircRNA Array (8 × 15K, Arraystar) |
| Data format | Raw and analyzed |
| Experimental factors | Tumor vs. corresponding normal tissues from 4 pancreatic ductal adenocarcinoma patients |
| Experimental features | Microarray expression profile analysis of circular RNAs in human pancreatic ductal adenocarcinoma |
| Consent | All patients provided written informed consent |
| Sample source location | Xi'an, China |
1. Direct link to deposited data
2. Experimental design, materials and methods
2.1. Tissue samples
The Ethics Review Board of Xijing Hospital (No: XJYYLL-2015075) approved the study. Tissue samples were prospectively collected from patients undergoing operation at the Department of Hepatobiliary Surgery at the Xijing Hospital. Tumor and adjacent normal pancreatic tissue samples were snap-frozen in liquid nitrogen immediately after resection and stored at − 130 °C until use. Histology of the tissue specimen was confirmed by two uropathologists.
2.2. RNA preparation
Total RNA was isolated from 4 PDAC samples and paired adjacent normal tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's protocol. Total RNA from each specimen was quantified and quality was verified using NanoDrop ND-1000 (NanoDrop, Wilmington, DE, USA). Additionally, RNA integrity was assessed by electrophoresis on a denaturing agarose gel.
2.3. Labeling and hybridization
Sample labeling and array hybridization were performed according to the manufacturer's protocol (Arraystar, Rockville, Maryland, USA). In short, circRNAs were treated with Ribonuclease R (RNase R) to remove linear RNAs according to the manufacturer's protocol (Epicenter, Madison, WI, USA). Then, each sample was amplified and transcribed into fluorescent cRNA utilizing a random priming method with a Super RNA Labeling Kit (Arraystar). The labeled cRNAs were purified using an RNeasy Mini Kit (Qiagen, Hilden, Germany). The concentration and specific activity of the labeled cRNAs (pmol Cy3/μg cRNA) were measured using NanoDrop ND-1000. Then, 1 μg of each labeled cRNA was fragmented by adding 5 μl 10 × Blocking Agent and 1 μl of 25 × Fragmentation Buffer. The mixture was heated at 60 °C for 30 min, and 25 μl 2 × Hybridization buffer was added to dilute the labeled cRNA. Then, 50 μl of the hybridization solution was dispensed into the gasket slide, which was assembled with Human Circular RNA Array slides. The slides were incubated for 17 h at 65 °C in an Agilent Hybridization Oven. The hybridized arrays were washed, fixed and scanned using an Agilent G2505C Scanner.
2.4. Microarray and quality control
Scanned images were imported into Agilent Feature Extraction software (version 11.0.1.1) for raw data extraction. Quantile normalization of raw data and subsequent data processing were performed using the R software package (R version 3.1.2). After quantile normalization of the raw data, low intensity filtering was performed, and circRNAs with at least 2 out of 8 samples that had flags in “P” or “M” (“All Targets Value”) were retained for further analyses. The log2-ratio was used for quantile normalization. CircRNAs differentially expressed between the two groups were conveniently estimated by fold-change filtering and Student's t-test. CircRNAs exhibiting fold changes ≥ 2.0 and p-values ≤ 0.05 were selected as significantly differentially expressed circRNAs.
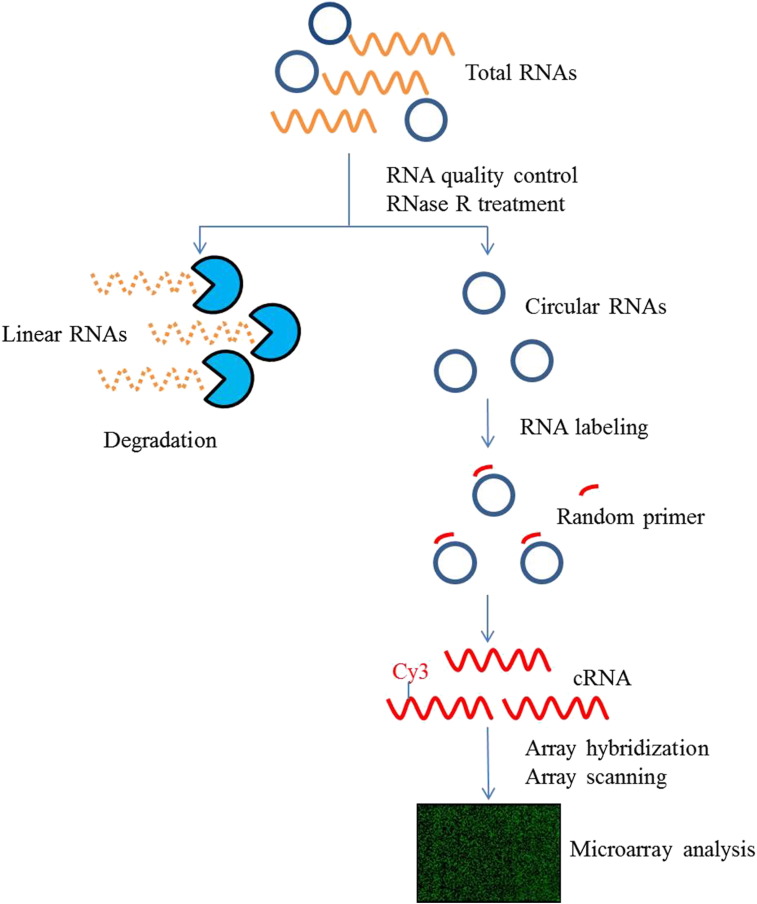
The experiment workflow is listed in Fig. 1.
Fig. 1.
Experiment workflow of microarray expression profile of circular RNAs.
3. Discussion
Large amounts of circRNAs are recently discovered and represent a new special class of endogenous noncoding RNA. Recent researches have revealed that circRNAs are an abundant, stable, diverse and conserved class of RNA molecules [1], [2], [3]. Moreover, circRNAs can function as miRNA sponges or regulate parent gene expression to affect disease initiation and progression [4], [5], [6], [7]. These studies indicate that circRNAs have great potential to become diagnostic or predictive biomarkers of disease and provide new insights into the treatment of diseases. In this study, we have explored the expression profile of circRNAs in 4 PDAC samples and paired adjacent normal tissues. We have also revealed that the circRNA expression signatures of PDAC are dysregulated. The identification of novel differentially expressed circRNAs is a crucial step towards better understanding of PDAC. These findings indicate that circRNAs can be involved in the initiation and progression of PDAC.
Conflict of interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 81772061) and Health Public Welfare Industry Special Scientific Research Projects of China (No. 201202007).
We thank KangChen Bio-tech, Shanghai, China for their technical support for the microarray works.
References
- 1.Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 4.Hansen T.B., Kjems J., Damgaard C.K. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 5.Qu S., Yang X., Li X., Wang J., Gao Y., Shang R. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 7.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X. Exon–intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]