Abstract
Few studies so far examined alternative splicing alterations in blood cells of neurodegenerative disease patients, particularly Parkinson's disease (PD). Prototype junction microarrays interrogate known human genome junctions and enable characterization of alternative splicing events; however, the analysis is not straightforward and different methods can be used to estimate junction-specific alternative splicing events (some of which can also be applied for analyzing RNA sequencing junction-level data). In this study, we characterized alternative splicing changes in blood leukocyte samples from Parkinson's patients prior to, and following deep brain stimulation (DBS) treatment; both on stimulation and following 1 h off electrical stimulation. Here, we describe in detail analysis approaches for junction microarrays and provide suggestions for further analyses to delineate transcript level effects of the observed alterations as well as detection of microRNA binding sites and protein domains in the alternatively spliced target regions spanning across both untranslated and the coding regions of the targets. The raw expression data files are publically available in the Gene Expression Omnibus (GEO) database (accession number: GSE37591) and in Synapse, and can be re-analyzed. The results may be useful for designing of future experiments and cross correlations with other datasets from PD or patients having other neurodegenerative diseases.
Keywords: Alternative splicing, Blood, Parkinson's disease, Deep brain stimulation, Junction microarrays
| Specifications | |
|---|---|
| Organism/cell line/tissue | Human/blood leukocytes |
| Sex | Males |
| Sequencer or array type | Splice junction microarrays |
| Data format | Raw (CELL files) and processed |
| Experimental factors | Parkinson's disease, Deep Brain Stimulation (pre- and post-treatment, on and following 1 h off electrical stimulation) |
| Experimental features | RNA was isolated from blood leukocyte samples of three Parkinson's disease patients prior to deep brain stimulation (DBS) treatment and post-DBS, both on and following 1 h off electrical stimulation, and age- and gender-matched healthy control volunteers. Samples were prepared for hybridization on Affymetrix human junction prototype microarrays (HJAY) and served to characterize alternative splicing changes pre- and post-treatment and as compared with healthy controls. |
| Consent | All of the study participants signed informed consent forms prior to inclusion in the study. |
| Sample source location | Jerusalem, Israel, Department of Neurosurgery, Hadassah University Hospital, The Hadassah-Hebrew University Faculty of Medicine |
1. Direct link to deposited data
2. Experimental design, materials and methods
2.1. Experimental design
Blood leukocyte samples were isolated from Parkinson's disease (PD) male patients one day prior to undergoing deep brain stimulation treatment to electrically bi-directionally stimulate the sub-thalamic nucleus (STN-DBS) [1]. Blood leukocyte samples were also collected from age- and gender-matched male healthy control volunteers.
2.2. Materials and methods
2.2.1. Subject recruitment
Blood samples were collected from three PD patients' pre- and post-STN-DBS neurosurgery while being on stimulation and following a 1-hour of stimulation cessation. All of the volunteers that passed the study exclusion criteria (clinical parameters of the recruited volunteers are given under [2]) signed informed consent forms. Patients went through bilateral STN-DBS electrode implantation (Medtronics, USA) and were under dopamine replacement therapy (DRT) both pre- and post-DBS (on significantly reduced dosage post-DBS, with t-test P < 0.01), the last medication administered at least 5 h pre-sampling. The clinical severity of the disease was assessed by a neurologist using the Unified PD Rating Scale (UPDRS) [3]. Controls were recruited among Hadassah Hospital staff and researchers at the Edmond J. Safra Campus (Jerusalem). Blood collection was conducted within a fixed range of hours (10 AM–14 PM). Samples of 9 ml were collected using 4.5 ml EDTA (anti-coagulant) tubes and the leukocytes were filtered from each sample up to 10 min post-extraction.
2.2.2. Leukocyte fractionation
The collected venous blood was filtered using the LeukoLock fractionation and stabilization kit (Ambion, Applied Biosystems, Inc., Foster City, CA) up to 15 post-extraction minutes to enhance inspection accuracy. To ensure high RNA quality, the leukocyte-enriched samples were immediately incubated in RNALater (Ambion) (http://www.affymetrix.com/support/technical/technotes/blood_technote.pdf). Stabilized filters and serum samples were stored at − 80 °C until use.
2.2.3. RNA extraction
RNA extraction followed the manufacturer (Life Technologies) alternative protocol instructions for using the LeukoLock filters. Briefly, cells were flushed (TRI-Reagent Ambion) into 1-bromo-3-chloropropane-containing 15 ml tubes and centrifuged. 0.5 and 1.25 volumes of water and ethanol were added to the aqueous phase. Samples were filtered through spin cartridges, stored in pre-heated 150 μl EDTA; RNA was quantified in Bioanalyzer 2100. Determination of RNA quality and quantity was conducted using the Eukaryote Total RNANano 6000 kit (Agilent). RNA was frozen and stored in − 80 °C immediately after preparation.
2.2.4. cDNA library preparation for microarray interrogation
HJAY profiling of blood leukocytes was conducted using exon array pre-prepared hybridization samples and Gene-Chip Whole Transcript Sense target labeling assay kit (Affymetrix), as per manufacturers' instructions. The high-density HJAYs (Affymetrix) were washed and stained with streptavidin–phycoerythrin and signal amplification was performed using a biotinylated anti-streptavidin antibody. The microarrays were scanned on an Affymetrix GeneChip Scanner 3000 7G scanner, according to the Affymetrix GeneChip Whole Transcript Sense Target Labeling Assay protocol for the GeneChip Exon 1.0 ST arrays. A total of 13 junction microarray samples were obtained (including one re-stained chip).
2.2.5. Microarray probe-set genome annotation and pre-processing
The Affymetrix human junction arrays (HJAYs) were obtained through collaboration with the EURASNET consortium and were used to assess genome-wide changes in expressed exons and junctions. Briefly, these microarrays interrogate 335,663 human transcripts from ~ 25,000 Ensembl genes, 260,488 junctions and 360,569 exons. Using the program AltAnalyze, probe set level RMA [17] expression and DAGB P-values were obtained by calling Affymetrix Power Tools (APT) [17]. AltAnalyze probe set-to-exon associations were obtained by matching the annotated exon sequences (two exons for each junction probe sets) provided by Affymetrix to the reference Ensembl genome (version 62) for Affymetrix annotated gene symbols. In cases where probe sets aligned to an intron, a novel exon annotation indicating the relative intron position was assigned. Alternative exons, junctions, reciprocal probe sets and event-annotations (e.g., alternative cassette exon, alternative promoter) were obtained by comparing the exon-structure of mRNAs from Ensembl, the UCSC genome database and novel junctions assayed by the HJAY array, as previously described [4]. Linear regression analysis was employed by an updated version of AltAnalyze to detect alternative splicing events with a fold change of 2. P < 0.05 was considered significant. Additional methods details are provided at http://www.altanalyze.org/help_main.htm.
2.2.6. Functional prediction analysis of the HJAY detected spliced transcripts
Each detected alternative event (e.g., cassette exon, alternative 3′ end, alternative 5′ end, intron retention, bleeding exon, alternative C-terminal exon) or alternative promoter inclusion event (alternative N-terminal exon or alternative promoter) of a pair of reciprocal junctions detected by the linear regression analysis was examined for putative protein domains or motifs and for miRNA binding sites. Additionally, potential induction of nonsense mediated decay due to the inclusion of a pre-termination codon (PTC) is reported. Alternative event annotations were obtained from both from Ensembl and UCSC Genome Browser databases as previously described for AltAnalyze version 1.0 [5].
2.2.7. Functional enrichment analysis of the HJAY detected spliced transcripts
Functional post-hoc analysis of the alternative isoforms detected by linear regression analysis conducted for each comparison (e.g., patients pre-treatment as compared with controls) using Gene Ontology (GO) Elite [6] was called directly from the adopted AltAnalyze version using Ensembl database. Genes' cut-off parameters included minimal 2-fold change; and t-test raw P-value < 0.05 with minimum number of 3 changed genes defined. GO terms [7] and WikiPathways [8] were ranked by a combination of z-score (cut-off: 1.96) and gene number. Over-representation analysis (ORA) was conducted with 2000 permutations.
2.2.8. Finding alternative splicing events
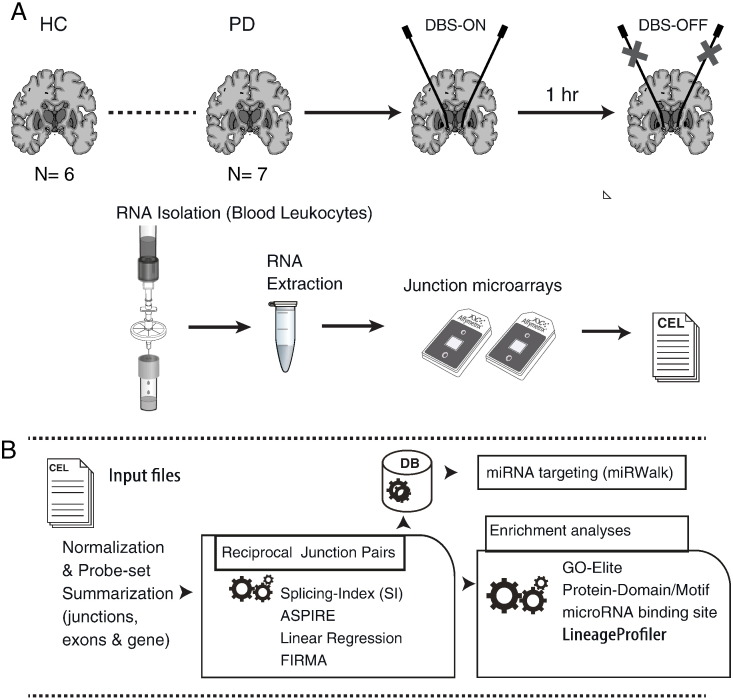
The overall experimental design and computational analysis workflows are shown in Fig. 2. Samples from PD patients pre-DBS were compared to samples from the same patients post-DBS on and following 1 h off electrical stimulation and as compared with healthy control volunteers (overall four experimental groups). To detect high confidence alternatively spliced genomic regions, all of the array-interrogated exons and junctions can be analyzed separately as well as combined (see Fig. 1 for a schematic representation of the array design). All analyses were based on the human genome Ensembl database (http://www.ensembl.org). First, each single microarray data sample is assigned to a group (e.g. control/treatment) and then a comparison between specific groups is defined (the calculated fold change values will correspond to the denominator given in this initial definition). Several analysis methods were applied to quantify alternative splicing on the junction microarray expression data:
- 1
-
2
ASPIRE ([11])
-
3
FIRMA, originally developed for the analysis of exon microarray data [12]
-
4
Linear regression [13]. This robust method was applied on pairs of reciprocal junction pairs (that can either include or exclude the exon found in between them) [13]. The normalized intensity per probe set was calculated as follows: where PI is the intensity of the exon and GE is the gene level expression value in that sample group to obtain a normalized intensity (NI) for each exon. The final linear regression score is the log2 ratio of the slope. This ratio is analogous to a log2 fold change.
Fig. 2.
Experimental design and general analysis flow. (A) RNA was prepared from filtered blood leukocytes which were isolated from whole blood samples of PD patients a day prior to DBS, upon hospitalization from the PD patient participants, and post-DBS, upon clinical stabilization of symptoms following the electrical stimulation, both while being on stimulation (DBS-ON) and following 1 h off electrical stimulation (DBS-OFF). Age- and gender-matched (male) healthy volunteers served as a control group (HC). From each RNA sample, cDNA library was prepared (using the Affymetrix exon microarrays sample preparation protocol) and applied to Affymetrix junction prototype microarrays (HJAYs). (B) The junction array datasets were analyzed using AltAnalyze by applying several measures for alternative splicing, including a linear regression approach. Additionally, enrichment for microRNA (miRNA) binding sites and protein domains in the spliced regions was calculated, and functional analysis was conducted using the GO-Elite module of AltAnalyze. Cellular composition was computed using the software LineageProfiler module. Potential binding by miRNAs was also assessed using the miRWalk repository, for characterization of binding in 3′, 5′ and coding regions of targets that were detected as alternatively spliced.
Fig. 1.
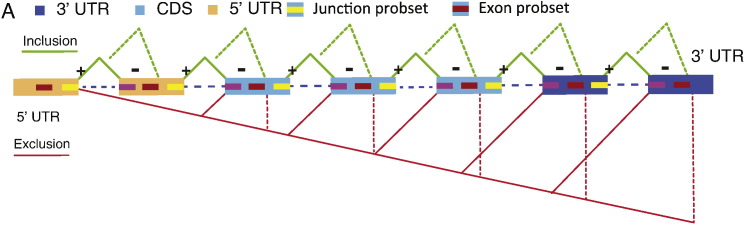
Junction microarrays. A schematic illustration of the genomic regions interrogated by the junction microarray probe-sets. The possible combinations of inclusion and exclusion of junctions and exons interrogating probe-sets are shown. Dashed lines: inter-exonic probe selection region (PSR) combinations with exclusion of junction probe-sets. A splice change is defined where b-directional expression alteration between the two compared conditions occurs in the inclusion and the exclusion probe-sets concurrently, or alternatively — in a PSR and an exclusion probe-set concurrently.
The results were produced for all the probe sets, and may suggest alternative splicing, alternative promoter regulation, or any other variation relative to the constitutive gene expression for a gene (derived from comparisons file). Gene expression values for each sample and group in the input expression file were also reported. Additionally, hierarchical clustering was be applied on the expression signals for classification.
2.2.9. Cellular lineage analysis of the exon microarray data
To identify cellular composition, the LineageProfiler module of AltAnalyze was used. To derive correlation scores to different cell types and tissues (lineages), a database reflecting the most specifically-expressed genes or exons present in each particular lineage, relative to all lineage types examined (ranging from 50 to 150), was created. The resulting database was a small subset of the original, containing the most informative markers. The exon microarray RNA-profile expression data of patients in three states (pre-DBS, post-DBS on and following 1 h off electrical stimulation) and control volunteers was compared to the profile of each lineage only for these markers to derive correlation coefficients and resulting z scores based on the distribution of values for each user RNA-profile. z scores to each lineage were calculated from the distribution of Pearson correlation coefficients, specifically for each sample or condition analyzed. Lineage differences between conditions were specifically evaluated via the AltAnalyze GO-Elite module using the database of lineage specific markers for examined differentially expressed genes. The results were visualized as association scores at the level of hierarchically clustered cell types and curated lineage networks. The results of alternative splicing analyses are described under [14]. The analysis approaches were further adopted and applied to analyze junction level expression data obtained by total RNA sequencing from the same patients pre- and post-treatment [2].
2.2.10. Analysis of protein binding domain composition
Identification of protein domains that were predictably disrupted by alternative splicing changes was conducted through AltAnalyze. To identify alternative protein domains, RNA-seq and microarray probe-set sequences were used to identify which proteins align to, or are missing from, transcripts for each disease, treatment or stimulation cessation spliced gene transcript, and specifically for each spliced isoform.
2.2.11. miRNA: target predictions
Prediction of miRNAs targeting genomic regions that exhibited differential expression in the PD patients compared to control volunteers was conducted using the miRWalk repository. The repository combines prediction data from 8 different prediction programs and adds inspection of all the gene regions including 5′ UTR and coding domains [15]. The analysis was conducted through a construction of a local MySQL database based on these predictions. For each miRNA that was detected as differentially expressed in the deep transcriptome of patients compared to control volunteers and post- compared to pre-DBS, all the predicted targets were searched for and were filtered to those that were detected as alternatively spliced in the human junction microarrays for the corresponding tested condition. Network analysis of miRNAs and alternatively spliced genes predicted as targets was created through the Cytoscape plugin ClueGO [16], [14].
Conflict of interest
We hereby declare that there are no conflicts of interests.
Acknowledgments
The authors thank the Hebrew University of Jerusalem (HUJI) for PhD fellowship support for LS, The Edmond and Lily Safra Center for Brain Sciences, HUJI (ELSC), The Center for Genomic Technologies (HUJI) and the patients and control participant volunteers that have contributed blood samples.
References
- 1.Bergman H., Wichmann T., DeLong M.R. Reversal of experimental parkinsonism by lesions of the subthalamic nucleus. Science. 1990;249(4975):1436–1438. doi: 10.1126/science.2402638. [DOI] [PubMed] [Google Scholar]
- 2.Soreq L. Long non-coding RNA and alternative splicing modulations in Parkinson's leukocytes identified by RNA sequencing. PLoS Comput. Biol. 2014;10(3):e1003517. doi: 10.1371/journal.pcbi.1003517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fahn S.E.R., M.o.t.U.D.C. 1987. Unified Parkinson's disease rating scale. [Google Scholar]
- 4.Salomonis N. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc. Natl. Acad. Sci. U. S. A. 2010;107(23):10514–10519. doi: 10.1073/pnas.0912260107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salomonis N. Alternative splicing in the differentiation of human embryonic stem cells into cardiac precursors. PLoS Comput. Biol. 2009;5(11):e1000553.. doi: 10.1371/journal.pcbi.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zambon A.C. GO-Elite: a flexible solution for pathway and ontology over-representation. Bioinformatics. 2012;28(16):2209–2210. doi: 10.1093/bioinformatics/bts366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashburner M. Gene ontology: tool for the unification of biology. Gene Ontol. Consort. Nat. Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pico A.R. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6(7):e184. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srinivasan K. Detection and measurement of alternative splicing using splicing-sensitive microarrays. Methods. 2005;37(4):345–359. doi: 10.1016/j.ymeth.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Gardina P.J. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ule J. Nova regulates brain-specific splicing to shape the synapse. Nat. Genet. 2005;37(8):844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- 12.Purdom E. FIRMA: a method for detection of alternative splicing from exon array data. Bioinformatics. 2008;24(15):1707–1714. doi: 10.1093/bioinformatics/btn284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugnet C.W. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput. Biol. 2006;2(1):e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soreq L. Small RNA sequencing-microarray analyses in Parkinson leukocytes reveal deep brain stimulation-induced splicing changes that classify brain region transcriptomes. Front. Mol. Neurosci. 2013;6:10. doi: 10.3389/fnmol.2013.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dweep H. miRWalk-database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 2011;44(5):839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Bindea G. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.http://www.affymetrix.com/estore/partners_programs/programs/developer/tools/powertools.affx#1_1.