Abstract
Significant advances have been made in developing novel therapeutics for cancer treatment, and targeted therapies have revolutionized the treatment of some cancers. Despite the promise, only about five percent of new cancer drugs are approved, and most fail due to lack of efficacy. The indication is that current preclinical methods are limited in predicting successful outcomes. Such failure exacts enormous cost, both financial and in the quality of human life. This primer explores the current status, promise and challenges of preclinical evaluation in advanced mouse cancer models and briefly addresses emerging models for early-stage preclinical development.
EXPLOSION OF CANCER THERAPIES AND CHALLENGES TO CLINICAL SUCCESS
Ever-increasing knowledge of cancer biology has yielded countless possibilities for diagnostic and therapeutic strategies (Fig. 1), while at the same time revealing enormous disease complexities that challenge clinical success. Such challenges include tumor microenvironment complexities, intra- and inter-tumor molecular and biological heterogeneity, systemic and tumoral immune and metabolic response heterogeneity, and the ability of drug-resistant stem-like cancer initiating cells to repopulate treated cancers (Pattabiraman and Weinberg, 2014). Too often, experimental targeted therapies designed to assimilate known disease complexity have proven ineffective, only to highlight the limitations in our understanding. In contrast to most experimental targeted therapies, encouraging advancements have been made using a number of cell-based and targeted immunotherapies, which have produced sustained responses in patients (Page et al., 2014). However, only a fraction of patients respond to these therapies.
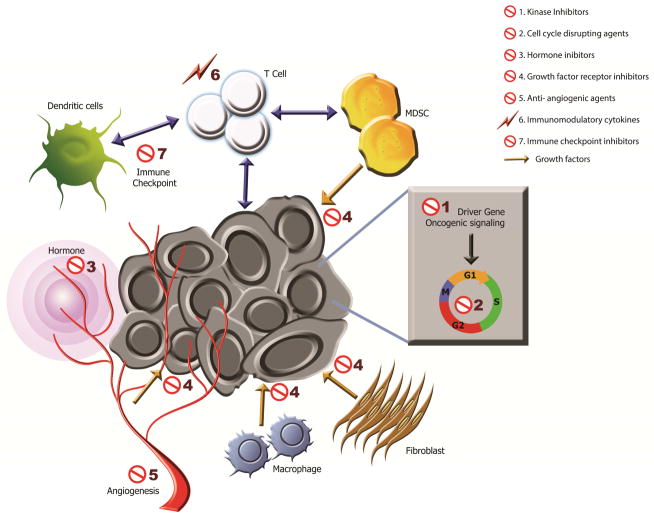
Fig. 1. Targeting the tumor and its microenvironment.
Genetic alterations produce oncogenes that drive signaling pathways in cancer cells facilitating survival and growth. However, tumor cells also cooperate with stromal cells, including vessels, fibroblasts and various immune cells, to acquire growth factors, an energy supply and protection from host defenses. These key autonomous and stromal mechanisms constitute potential therapeutic targets both locally, and for immune cells also in the circulating blood and distant immune organs. 1) Cancer cell growth driven by a mutated kinase (“Driver Gene”) can be targeted by small-molecule inhibitors. 2) Oncogenic signaling promoting uncontrolled cell cycling can be disrupted (e.g., anti-metabolites, anti-microtubule agents, DNA-damaging agents). 3 & 4) Growth of cancer cells stimulated by release of either host-derived hormones (3, concentric purple circles from blood) or growth factors (4, yellow arrows from myeloid-derived suppressor cells [MDSC], fibroblasts, macrophages, blood vessels) can be targeted by hormone inhibitors (e.g., anti-hormones or biosynthesis inhibitors) or growth factor receptor inhibitors, respectively. 5) Tumor growth requires development of new vasculature for enhanced nutrient demands, which can be blocked by anti-angiogenic agents. 6 & 7) Tumor cells can shift the inflammatory response to an immunosuppressive mode (e.g., activation of CTLA-4 and PD-1 in T cells or PD-L1 in cancer cells). The immunosuppressive environment can be reversed via treatment of immunomodulatory cytokines (6, lightning bolt; e.g., IL-2, IL-15) or immune checkpoint inhibitors (7, e.g., anti-CTLA-4, anti-PD-1, or anti-PD-L1), resuming anti-cancer activity of T cells. The cellular interactions in immune responses are marked by double-headed arrows. (Artwork by Jonathan Marie)
Over the last decade, cancer classification has shifted from relying solely on histiopathologic properties to including key molecular attributes that can predict therapeutic outcomes. That certain molecular aberrations are targets for effective therapy first led to clinical practice when a leukemia (APL) bearing the PML-RARα translocation was shown to be sensitive to retinoic acid (tretinoin) (Quignon et al., 1997), which targets the RARα component to effect leukemic cell differentiation. Since that time, targeted therapies have become the standard of care for CML [imatinib (Gleevec), which inhibits BCR-ABL] and for Her2+ breast cancer [tratuzumab (Herceptin), which inhibits Her2). Although these successes establish the promise of targeted therapies, most attempts to attain similar results targeting known molecular drivers have failed, and the reasons are often elusive because of human research limitations. Some general principles have been recognized that emphasize the need for preclinical platforms approximating human cancers. For, example, in each of the noted successes, single potent cancer drivers present in a significant fraction of patient malignancies were targeted; however, when a minor fraction of patients are responsive, all-comer clinical trial data may mask the responders. This was first demonstrated in non-small cell lung cancer (NSCLC) patient trials that initially failed to show significant responsiveness to EGFR-targeted tyrosine kinase inhibitors; however, the ~10% of patients whose tumors actually harbored activating EGFR mutations were uniquely sensitive (Lynch et al., 2004) (Paez et al., 2004). Now, screening of lung cancers for such mutations prior to therapy is routine practice. Lung cancer is the most prevalent US cancer; if limited to clinical trials, accurate identification of therapies effective in a fraction of less-common cancer types may not be possible. Nonetheless, when a specific target was known, stratification of patients has identified additional effective therapies, such as inhibitors for BRAF mutant melanomas and ALK translocation-positive NSCLCs (Pagliarini et al., 2015). Unfortunately, patients treated with single targeted therapies inevitably relapse with cancers that are resistant to the original drug.
Another challenge in targeting single drivers is the feedback response upon molecular network disruption that prevents efficacy or causes increased severity. Understanding such molecular responses can aid in the discovery of more effective combination therapies. In addition, unbiased molecular queries are showing promise in identifying signatures that correspond to prognosis and/or therapeutic outcomes. For example, in some cases, unique transcriptome signatures stratify cancers into distinct therapeutic and/or prognostic categories and thus improve patient management (e.g., (Rosenwald et al., 2002)). Thus far, this approach has been used primarily for determining which patients require aggressive chemotherapy treatment, hence reducing the frequency of over-treatment. Oncotype DX and FDA-approved MammaPrint tests, both based on distinguishing transcriptome signatures, are now utilized in the clinic to identify the low risk breast cancer patients to be excluded from aggressive treatment. Yet, accuracy is not optimal, and numerous challenges currently prevent broad implementation of molecular signature diagnostics (van't Veer and Bernards, 2008). Additionally, the hope is that molecular signatures can be identified via unbiased compound or molecular screens that will dictate specific effective treatments even when the targets are unknown.
Thus, although clearly impactful, the use of cancer molecular constitution to guide clinical practice is in its infancy, and research to identify parameters that hone specificity and improve accuracy is ongoing. If confined to human research, achieving maximum effectiveness is likely impossible due to low frequencies of each molecular subtype within most cancers and limitations associated with clinical trials. More challenging is understanding the impact of complex and varied inherited genetic constitution on clinical outcomes with subsequent conversion to clinical practice of (Hood and Friend, 2011). In this regard, the sophistication of complex trait evaluation in mice using the collaborative and diversity crosses may offer a path to discovery (Churchill et al., 2004; Svenson et al., 2012).
The above summary provides only a cross-section of the therapeutic and diagnostic possibilities currently under investigation, and the reader is referred to current review articles for more comprehensive information (Chin et al., 2011; Hood and Friend, 2011; Yap et al., 2013). Ultimately, the current limitation to improving cancer patient care within reasonable timeframes may not be the availability of potentially efficacious therapies; rather, a major blockade is the lack of a fully developed and integrated set of reliable preclinical technologies that can navigate complex variables in therapeutic responses and diagnostic accuracy. To optimally develop efficacious therapies, preclinical research must utilize a diversity of models that collectively incorporate the biology and genetics dictating therapeutic outcomes for specific cancers, and yet achieve sufficient throughput. Here we summarize the value and constraints of mouse cancer models, highlight recent progress indicating promise, summarize non-mammalian and ex vivo preclinical models, and explore the needs for, and challenges to, developing robust multi-faceted preclinical platforms for routine use.
MOUSE CANCER MODELS IN PRECLINICAL RESEARCH
Murine cancer models designed to capture the complexities of human cancers currently offer the most advanced preclinical opportunity for navigating diverse mechanisms that provide rationale for therapeutic development (Van Dyke and Jacks, 2002). One approach is to probe pathobiology mechanisms to design effective treatments by perturbation with molecularly targeted therapies (Olive and Tuveson, 2006). Additionally, the models are being used/developed as preclinical efficacy determination platforms to guide clinical trial designs (Singh et al., 2012). However, the application of complex cancer models to clinical research directives is an emerging science, currently executed in individual settings and with limited resources. Significant research, ideally in a team-directed, multi-institutional effort, is required to hone existing technologies into integrated preclinical workflows to optimally accelerate positive clinical outcomes.
A variety of approaches to mouse cancer modeling are now available (Fig. 2), and each has strengths and weaknesses (Table 1). Here, we address the limitations of standard Cell line- Derived Xenograft (CDX) models, describe genetically and biologically engineered mouse cancer models [Genetically Engineered Mouse (GEM), GEM-Derived Allograft (GDA), Patient-Derived Xenograft (PDX) models], review values and constraints, and highlight recent progress. Thus far, results indicate promise in understanding cancer pathobiology and in the enhancement of clinical efficacy prediction, but also underscore the need for further development to achieve consistent reliability.
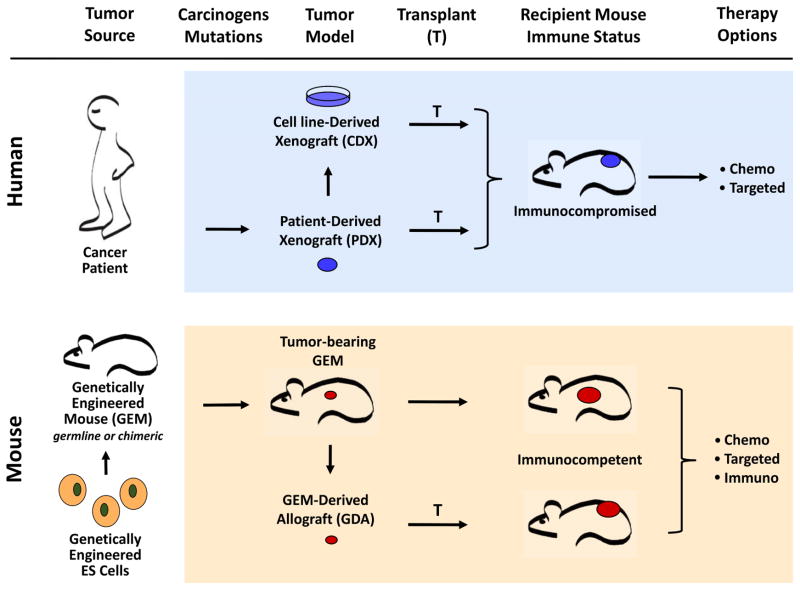
Fig. 2. Current state of preclinical cancer modeling.
Preclinical mouse models can be defined according to the species source of tumor, how it is created, and how it is manipulated. Upper Panel) Tumors derived from human patients, and other non-murine species, can be directly transplanted into immunocompromised mice to form Patient-Derived Xenograft (PDX) models. Alternatively, these same tumors can produce established cell lines maintained in vitro as cell cultures, and transplanted into immunocompromised mice to form Cell line-Derived Xenograft (CDX) models. Since the hosts of these tumors need be immunocompromised, they are useful only for testing the efficacy of chemotherapeutics (Chemo) and targeted small-molecule inhibitors (Targeted). Xenograft models derived from canine patients also belong to this category, but are not shown here. Lower Panel) Mice can be engineered to generate tumors of human relevance with respect to histopathology, etiology and molecular wiring. Offspring of such Genetically Engineered Mice (GEM) can serve directly as preclinical models themselves, in which case the tumor is treated at its precise point of origin. Notably, model building can be streamlined by using non-germline approaches, one of which is to genetically modify ES cells and study the arising chimeric mice without time-consuming breeding schemes. Alternatively, tumors harvested from GEMs can be transplanted and expanded into fully immunocompetent syngeneic hosts, forming GEM-Derived Allograft (GDA) models. Syngeneic models allow preclinical studies of not only chemotherapeutic and small-molecule drugs, but also of all varieties of immunotherapeutic agents (Immuno).
Table 1.
Comparison of clinical and preclinical model properties
| Immune Status of the Host |
Micro- environment Context |
Human Relevance |
Tissue Availability |
Disease Complexity |
Experimental Robustness |
Initiation/ Progression |
Feasibility in Pathway Engineering |
Cost | |
|---|---|---|---|---|---|---|---|---|---|
| Clinical Trials | Functional | Natural | Standard | Highly Limited | High | Low | Intact | Irrelevant | Very High |
| Cancer Cell Line-Derived Xenografts (CDX) | Deficient | Xenogeneic | Situational* (passage number dependent) | Expandable | Low | High | Bypassed | Yes | Low |
| Patient cancer-Derived Xenografts (PDX) | Deficient | Xenogeneic | High | Expandable/Limited | Moderate | High | Bypassed | No | High |
| Genetically Engineered Mice (GEM) | Functional | Natural | High/Variable (model dependent) | Limited | High | Moderate/Variable | Intact | Yes | High |
| GEM-Derived Allografts (GDA) | Functional | Allogeneic | Low/Variable | Expandable | Moderate | High | Bypassed | Yes | Moderate |
CDX models have been shown to be limited as predictors of clinical outcome. However, they continue to be valuable for evaluating resistance mechanisms, for identifying non-targeted cytotoxic agents, for assessing drug toxicity, and as a platform to triage potentially effective targeted therapies. In general, the longer cells are in culture, the further they drift from normal tumor evolution, and the less relevant they become.
Traditional Mouse Models in Therapeutic Development
Historically, preclinical mouse models have co-evolved with cancer therapy development (Fig. 3). The earliest models were built through transplantation of murine tumors into immunocompetent host mice (DeVita and Chu, 2008; Talmadge et al., 2007). These early mouse-in- mouse isograft models served as workhorses for drug screening during the 1960s-70s, and were successful in identifying a number of effective cytotoxic drugs such as vincristine and procarbazine (DeVita and Chu, 2008). During the 1980s, researchers explored mechanisms of metastasis using selected murine and human tumor cell lines. A series of investigations by Fidler and colleagues demonstrated that metastasis is not random but site-selective (Fidler and Hart, 1982), and that metastatic patterns are injection site-dependent, supporting the establishment of “orthotopic” models (Talmadge et al., 2007). Since then, cancer therapeutic development has relied upon the more tractable CDX transplantation models, in which tumors develop after subcutaneous injection of in vitro-established human cancer cells into immunocompromised mice (Fig. 2). The cell lines have been selected over many passages for rapid 2-D growth on plastic in serum-containing media. The NCI60 call line panel (DeVita and Chu, 2008; Talmadge et al., 2007) provided a valuable resource from which most CDXs were generated, and recent efforts have greatly expanded the repertoire (Reinhold et al., 2015). These models are easily established in a wide variety of laboratory settings and have been successfully used to identify an abundance of cytotoxic drugs leading to chemotherapy treatments that still dominate clinical cancer management (Fig. 3).
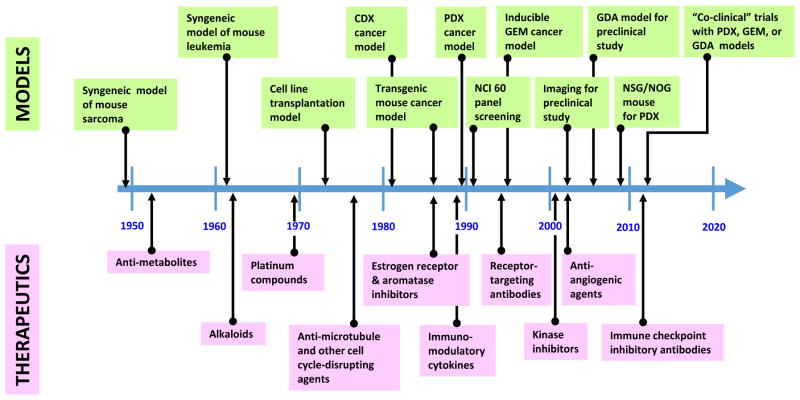
Fig. 3. Timeline of key preclinical cancer model developments since 1950.
As the conceptual targets of cancer treatment progressed from actively dividing cells to oncogenic signaling and immune checkpoints, preclinical models (green boxes, upper part) and cancer therapies (pink boxes, lower part) co-evolved accordingly. This evolution was highly dependent on technical advances, resulting in waves of activity. For example, recent development of fully immunocompromised mice and diverse syngeneic GEM models has significantly promoted PDX and GDA models, respectively, for preclinical cancer studies.
Unfortunately, CDXs have failed to predict human efficacy for most therapies targeted to cancer-driving proteins (Johnson et al., 2001), as evidenced by the low FDA approval rate of 5– 7% for targeted therapeutics (Sharpless and Depinho, 2006). With an average time from discovery to clinical practice of 12 years, at an average estimated cost of $0.5–2.0 billion (Adams and Brantner, 2006) and an immeasurable human price, this low yield forestalls even a goal to chronically manage, rather than cure, cancers. The observation that most cancer therapeutics fail in clinical phase II and III efficacy assessment indicates that current standard preclinical practice inadequately addresses complex challenges to successful treatment, such as host immune responses, cancer heterogeneity and drug resistance. Consequently, the system cannot be used to optimize a multitude of variables known to influence therapeutic outcomes, such as combinatorial therapies, dosing schedules and drug delivery methods (Al-Lazikani et al., 2012). CDXs continue to be valuable in identifying non-targeted cytotoxic agents and in primary assessment of drug toxicity (Teicher, 2006), for analyzing resistance mechanisms (Garraway and Janne, 2012) and in triaging potentially effective targeted therapies for evaluation in more representative models.
Mouse Models Designed after Patient Cancers
Mice and humans are believed to have diverged from each other ~87 million years ago (Bailey et al., 2013), so naturally there are numerous significant similarities between the two species, and also many marked disparities, including differences in immune systems and drug metabolism. Based on the premise that many cancers have been cured in mice and not in people, many argue that mice are inappropriate for use in therapeutic development (Leaf, 2004). However, it is critical to understand that “cures” have been attained only in CDX models, thus dismissal of all mouse cancer models as irrelevant is unwarranted. Human cancers are enormously complex, and their evolutionary etiology generates vast diversity among and within them, thus challenging the attainment of successful treatments. However, as knowledge of cancer complexities has increased, so has the ability to design mouse models that better represent cancer patients. PDX and GEM models develop tumors with the greatest similarity to human diseases yet achieved, and the past 5 years have seen an increase in their employment in preclinical research. As with all models, each approach has its strengths and limitations (Table 1). Early studies suggest promise for improved guidance in the development of successful clinical treatments (Table 2), and yet also emphasize the need for further scrutiny and refinement. The following provides a balanced consideration of model advantages and limitations, their ramifications in obtaining optimally accurate preclinical data, and the logistical requirements for achieving efficiency, accuracy and reproducibility.
Table 2.
Representative clinically relevant mouse trials
| Trial Design | Cancer Type | Model Type | Engineered Drivers | Drugs/Treatment | Significance | Relevant Publications |
|---|---|---|---|---|---|---|
| Preclinical | Hematopoietic (APL) | GEM | PML-RARα fusion PLZF-RARα fusion |
Retinoic acid | Demonstrated the efficacy of retinoic acid plus As2O3 in specific APL subtypes, validated in clinic | (Ablain and de The, 2014; Pandolfi, 2001) |
| Preclinical | Pancreas (Neuro-endocrine) | GEM | RIP1-Tag2 | Sunitinib | Demonstrated the efficacy of Sunitinib plus Imatinib, validated in clinic | (Pietras and Hanahan, 2005; Raymond et al., 2011); FDA approved for pancreatic cancer treatment in 2011 |
| Preclinical | Medulla-blastoma | GEM | Ptc1+/− P53−/− |
GDC-0449 (SMO inhibitor) | Demonstrated the efficacy of an Shh pathway small molecule inhibitor, validated in clinic | (Romer et al., 2004; Rudin et al., 2009) |
| Preclinical | Pancreas (Neuro-endocrine) | GEM | RIP1-Tag2 | Erlotinib Rapamycin |
Demonstrated efficacy of combining drugs targeting EGFR and mTOR | (Chiu et al., 2010) |
| Co-clinical | Pancreas (PDA) | GEM | LSL-KrasG12D LSL-Trp53R172H Pdx-1-Cre |
Gemcitabine Nab-Paclitaxel |
Provided mechanistic insight into clinical cooperation between Gemcitabine and Nab-Paclitaxel | (Frese et al., 2012; Goldstein et al., 2015) |
| Co-clinical | Pancreas (PDA) | GEM | LSL-KrasG12D LSL-Trp53R172H Pdx-1-Cre |
CD40 monoclonal antibody Gemcitabine |
Demonstrated that targeting stroma was effective in treatment of metastatic PDA | (Beatty et al., 2013) |
| Co-clinical | Lung (NSCLC) | GEM | KRASG12D p53fl/fl Lkb1fl/fl |
Selumetinib Docetaxel |
Validation of improved response of adding Selumetinib to Docetaxel treatment | (Chen et al., 2012; Janne et al., 2013) |
| Co-clinical | Lung (NSCLC) | GEM | EML4-ALK fusion | Crizotinib Docetaxel Pemetrexed |
GEM model predicted clinical outcome of drug combinations | (Chen et al., 2014; Lunardi and Pandolfi, 2015) |
| Co-clinical | Various Sarcomas | PDX | N/A | Various chemotherapies | PDX testing predicted clinical outcome of drug combinations | (Stebbing et al., 2014) |
| Postclinical | Ovarian (SEOC) | GDA; PDX | RB/p53-deficient BRCA1/2-deficient |
Olaparib Cisplatin |
Validation of treatment efficacy in BRCA mutant tumors in both GDA and PDX models | (Kortmann et al., 2011; Szabova et al., 2014) |
| Postclinical | Pancreas (Neuro-endocrine) | GDA | RIP1-Tag2 | Anti-VEGFR1 & anti-VEGFR2 antibodies | Identification of mechanisms of resistance to anti-angiogenic therapies | (Casanovas et al., 2005) |
| Biomarker | Lung (NSCLC) | GEM; Carcinoge n-induced | Various Models | N/A | Used in-depth quantitative MS-based proteomics to profile plasma proteins | (Hanash and Taguchi, 2011) |
| Biomarker | Pancreas (PDA) | GEM | KrasG12D Ink4a/Arffl/fl Pdx-1-Cre |
N/A | Used in-depth proteomic analyses to identify candidate markers applicable to human cancer | (Faca et al., 2008) |
Patient-Derived Xenograft (PDX) Models
Relative to CDX models, immunocompromised mice bearing subcutaneous surgically-derived clinical tumor samples (PDX models) are better aligned with human disease, since intact tissue that preserves tumor architecture is transferred directly to recipient mice and not compromised by in vitro adaptation (Fig. 2). PDXs are the only models harboring bona fide tumor targets directly from the patient, and hence their use in drug discovery is expanding rapidly. Promise for such models, first developed by (Fiebig et al., 1984), was demonstrated when chemotherapeutic agents, such as alkaloids and anti-metabolites, were shown to elicit similar responses in mice and patients (Mattern et al., 1988). In contrast, a study of responses to numerous cytotoxic agents in NCI60-based CDX models showed that the predictive value for efficacy was much less impressive (Johnson et al., 2001). Unfortunately, early studies utilizing PDXs were limited by difficulties in collecting clinical samples and in achieving sufficient take rates.
The recent resurgence of PDX model use for therapeutic evaluation has been fueled by significant improvements in clinical sample access and transplantation technology. Cancers established as PDXs can, in early passages, retain the stromal composition and histologic and molecular heterogeneity characteristic of those in patients (Hidalgo et al., 2014; Tentler et al., 2012). Since these properties critically impact therapeutic responses and biomarker specificity, PDX models provide a preclinical venue for addressing some of the most challenging barriers to successful patient therapy. Furthermore, human target specificity allows for direct evaluation of lead human-specific therapeutics, such as antibodies, in clinical development.
Methodologies for PDX establishment and characterization are detailed elsewhere (Hidalgo et al., 2014; Tentler et al., 2012; Zhang et al., 2013). For some cancers, such as certain melanomas, lung and colorectal cancers, transplant take rates can reach ≥75%, and the time required for tumor growth can be as little as 2–4 months. However, these attributes vary widely depending on sample type and amount (e.g., fresh biopsy tissue, fine needle aspirate, circulating tumor cells), tumor origin, molecular properties and recipient strain (Mattern et al., 1988). Consequently, some cancers, such as neuroendocrine, luminal ER+ breast, and prostate cancers (Rosfjord et al., 2014) are underrepresented. Notably, PDX engraftibility appears to significantly correlate with clinical aggressiveness (Ilie et al., 2015 ).
Relative to subcutaneous transplants, cancers orthotopically transferred into organs of origin are more likely to maintain tumor microenvironment characteristics that impact therapeutic outcomes (Talmadge et al., 2007). However, orthotopic PDX production is technically challenging, and, for most cancer types, tumor growth and responses must be monitored via expensive and often laborious longitudinal imaging. Thus, subcutaneous models are currently exclusively used for preclinical studies.
Production of PDX cohorts is by serial tumor transplantation, and, given the likelihood of change with each passage, therapeutic studies are most representative in low-passage models. Additionally, human stromal components are maintained for only 2–3 passages, with mouse stromal elements becoming dominant thereafter (Rosfjord et al., 2014). Unfortunately, if limited to early passage use, each model represents a limited resource. Hence, most preclinical studies utilize models that have been expanded, banked, and developed into significantly-sized cohorts. The extent of sacrifice in accurately predicting efficacy is presently undefined and likely depends on the mechanism of therapeutic activity. As such, in propagating PDXs, parental tumor traits should be routinely monitored, and deviations must be considered in interpreting therapeutic and biomarker data.
To circumvent immune rejection, human cancers must be transplanted into immunocompromised mice. Commonly used recipients, such as nude, SCID, and NOD/SCID strains, vary in the extent of immune impairment (detailed in Supplementary Information). IL- 2Rγ-deficient NOD/SCID mice (NSG and NOG strains) are the most severely impaired, and often yield improved take rates. Critically, the requirement for immunocompromised hosts precludes assessment of arising therapies designed to modulate immune function (e.g., immune checkpoint inhibitors α-CTLA-4, α-PD-1, α-PD-L1). Moreover, therapeutic responses in general are likely influenced by preexisting cancer-dependent immune phenotypes and immune responses elicited upon therapy-induced tumor perturbation (Zitvogel et al., 2008). The extent to which compromised immune systems limit predictive value for a given therapeutic approach will be determined as comparisons between PDX and clinical outcomes are expanded. Technologies to “humanize” the mouse immune system by transplanting purified human CD34+ hematopoietic stem cells into myeloablated NSG/NOG recipients (e.g., “BLT” mice: http://jaxservices.jax.org/invivo/humanized-BLT-mice.html) and other chimeric strategies have been developed (Legrand et al., 2009; Shultz et al., 2014). However, the high cost of recipient mice, limitations on human bone marrow acquisition, engraftment variability, and technical demands currently preclude use of these models in preclinical therapeutic discovery.
Despite the challenges to routine preclinical application, several PDX studies have proven effective in paralleling human outcomes (Malaney et al., 2014), in exploring drug resistance mechanisms (Das Thakur et al., 2013) and in identifying targets for second-line treatment (Girotti et al., 2015). Programs are also underway to employ PDX models in individualized precision cancer care. To date, this approach has been most successfully applied to pediatric patients with advanced sarcomas who have demonstrated the predicted response, sometimes to drugs not previously associated with this indication (Tentler et al., 2012). Patient-specific studies are currently limited by expense and relatively long and unpredictable times for establishing test animals. Since current clinical trials generally involve patients who have undergone prior failed treatments, results may not always be obtainable in a beneficial timeframe.
Genetically Engineered Mouse (GEM) Cancer Models
Of all murine cancer models, GEMs provide the most complete representation of cancer development; cancers develop from initiation through progression, co-evolve with intrinsic stroma, and possess an intact immune system (Figs. 1, 2). However, GEM models are the most challenging to work with effectively, and species differences must be carefully considered in experimental designs and interpretations. Extensive experience and infrastructure are required to ensure the use of optimally accurate models and to achieve sufficiently populated well-controlled preclinical studies. Yet, GEM cancer models provide the only opportunity to evaluate drug delivery, therapeutic response and biomarker expression for cancers evolving within their natural microenvironment (autochthonous cancers). These complex dynamic processes contribute to overall disease properties, and in particular, constitute a source of the inter- and intra-tumoral heterogeneity that challenges successful therapeutic development. Additionally, the accuracy of some therapeutic interventions, such as those targeting the immune system, may depend on the constitution of evolutionary, rather than transplanted, disease. Indeed, overall, GEMs and GEM-derived models are currently the only preclinical platform for evaluation and optimization of immunomodulatory therapies. Although some immune properties differ in mouse and human, there is significant conservation (Bailey et al., 2013); moreover, many differences can be managed via data interpretation or minimized by using genetically engineered “humanized” models (Scheer et al., 2013). Finally, autochthonous GEMs are the only viable models for evaluating prevention therapies.
Several reports show that well-designed GEM studies can contribute to improved clinical trials (Table 2), not only in identifying potentially efficacious therapies but also in predicting both positive and detrimental effects in molecular subclasses. A major power of GEM approaches is in the flexibility to create models with precise molecular specificity. With increasing sophistication, several strategies [summarized below and detailed elsewhere (Abate-Shen et al., 2014)] have been employed over the past three decades to significantly enrich our understanding of cancer mechanisms. A plethora of genes frequently altered in human cancers have been validated as disease drivers in GEMs, thereby facilitating the evaluation of cancer evolutionary mechanisms and kinetics, susceptible cell and molecular targets, relative cancer cell and microenvironment roles, and mechanisms of invasion and metastasis. Indeed, entire natural disease histories can be mapped (Stiedl et al., 2015; Van Dyke and Jacks, 2002).
In the process of basic discovery, countless GEM cancer models representing a variety of histiocytic cancer types driven by multiple independent drivers have been produced, and many are currently used in preclinical evaluations. Although no model can perfectly capture the human condition, several GEM models tractable for preclinical studies develop cancers with remarkable molecular and pathologic similarity to their human counterparts. However, since most established GEMs were created to address basic mechanisms, many do not accurately model human disease and/or are intractable for effective preclinical evaluation. Furthermore, each engineering approach can elicit untoward anomalies. Such circumstances can be accommodated in the interpretation of mechanistic studies, but are the basis for exclusion of many models for effective preclinical research. Thus, choosing appropriate models as subjects for preclinical discovery requires a deep understanding of cancer biology and genetics and also of engineering modalities. The following provides a reasonable guide for optimizing the value of GEM-based preclinical platforms.
Germline GEMs
An extensive array of technologies is employed to engineer the mouse germline with great precision. By editing the genome of embryonic stem (ES) cells or zygotes, mice can be programmed for cell type-specific disruption of tumor suppressor genes via direct mutation or expression of interfering non-coding RNAs (RNAi) (Walrath et al., 2010), and for oncogene expression at physiological or cancer-analogous levels. Furthermore, mice can be “humanized” by engineering the expression of drug targets in relevant cell types (Scheer et al., 2013) so that human-specific targeted therapies, such as antibody-based drugs, can be tested in GEM models. While traditional methods for constructing locus-specific genetic changes require significant lead times for engineering, the recent development of rapid sequence-targeted approaches (Mou et al., 2015) has significantly reduced this time to weeks instead of many months. In particular, clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 technology, which is efficient and versatile, is accelerating germline engineering and also facilitating rapid somatic engineering (see below).
Depending on the strategy, expression of an engineered “event” can be constitutive or inducible, although gene induction with cell-type and temporal specificity provides the best possibility for accurately modeling disease development. Inducibility is achieved by combining cell-specific expression of transcription factors (e.g., doxycycline-modulated tet-transactivators) or recombinases (Cre-lox or Flp-FRT) with cognate cis elements linked to a target gene, or by expressing proteins fused with a hormone-responsive domain (e.g., the tamoxifen-inducible estrogen receptor domain) (see Supplementary Information). When multiple distinct inducible systems are combined within the same cancer model, cancer-specific mutations can be induced sequentially in order to map and emulate cancer evolution [e.g., (Young et al., 2011)] and thus to generate increasingly relevant preclinical models. Reversible inducibility can be achieved with each of these technologies, although small molecule-mediated modulation of transcription factors and hormone-responsive domains are the most tractable for toggling expression on and off (Abate-Shen et al., 2014; Texido, 2013). This approach facilitates the identification of events required to sustain tumor growth (“oncogene addiction”) [e.g., (Soucek et al., 2008)] and thus of 15 potential therapeutic targets [e.g., (Kwong et al., 2012)]. Tumor responses to the shutdown of oncogenes or restoration of functional tumor suppressors within tumors, or appropriate effector cells, indicate the potential efficacy of targeted therapies, while genetic ablation in the entire animal predicts the overall toxic effects of specific inhibitors. However, since off-target effects will not register in this approach, results only indicate whether a given therapy is potentially efficacious.
A critical, often overlooked, consideration when building GEM cancer models is the incorporation of known environmental etiologies. However, there are notable examples wherein certain environmental factors were validated as etiologic agents and thus produced representative cancer models, including HPV E6/E7-induced cervical cancer ((Riley et al., 2003), UV accelerated melanomas [BRAFV600E, (Cao et al., 2013), mutant HRAS (Kannan et al., 2003) and HGF/MET (Noonan et al., 2001) models], and Helicobacter-fueled gastrointestinal cancers (Rogers and Fox, 2004). Exposure of GEM cancer models to environmental mutagens can be used to approximate the mutation load of many human cancers (e.g., (Westcott et al., 2015)), which influences therapeutic outcomes such as in drug-resistant relapse and neoantigen load-dependent immunomodulation.
The extent to which findings in GEMs extend to patients depends on engineering mice based on our understanding of human cancer etiologic drivers, cellular origins, heterogeneity, pathogenesis, and clinical properties. To recapitulate human cancer development, clinically relevant driver gene(s) or pathways must be perturbed in relevant target cells. For adult cancers, gene expression should be targeted to adult, rather than developing, organs. Furthermore, for optimal modeling, cancers should progress in a relevant sequence, since the order of events impacts properties of evolving tumors. Ideally, both initiation and progression to aggressive cancer should be evaluated using individual and relevant combinations of molecular aberrations thought to be causal in humans. High phenotypic penetrance and consistency among animals within a lineage are essential for tractability.
The accuracy of disease modeling depends on actually achieving the specificity envisioned in experimental designs, which is not always realized because of technical limitations and/or gaps in current knowledge. Of course, engineered sequences must be validated, but it is also critical that expected transcriptional specificities be confirmed. Unless targeted to specific genomic locations, transgenes insert randomly, and expression can be dramatically altered depending on insertion sites. Furthermore, transgenes may not carry all necessary regulatory signals. Hence, several founder lines should be established and fully characterized before selecting accurate representative lines for modeling cancers. Even targeted genetic changes have the potential to alter gene regulation. Thus, specificity, levels, and range of expression must be evaluated for each model; aberrant expression usually alters disease and can also yield ectopic phenotypes that hinder tractability and invalidate data. Yet, a surprising number of existing engineered strains, including those driving inducible expression, are not fully characterized. Hence, when choosing cancer models for preclinical studies, it is essential that expression and disease patterns are well established and accurately represented (see Supplementary Information).
Non-germline GEM models
While autochthonous GEM models have great utility, most are not tractable for large-scale screening of multiple anti-cancer drug candidates due to high cost, long timelines, extensive complex breeding, and/or difficulties in obtaining synchronous tumorigenesis. Preclinical analysis of metastatic lesions is particularly challenging; primary tumors arise stochastically with no reliable timetable, as in humans, and multiple tumors often develop. Thus, extensive longitudinal tomographic imaging is required to enroll mice bearing similarly sized tumors for therapeutic evaluation. Such procedures require specialized expertise and can be too expensive and time-consuming for first-line drug screening. Several strategies to produce “non-germline” GEMs have been developed that bypass breeding, reduce expense, and, in some cases, improve flexibility, uniformity and timelines (Heyer et al., 2010).
GEM-Derived Allograft (GDA) Models
GDAs marry the genetic and biologic human cancer similarities of GEM models with the relative ease of transplantation technology of PDXs (Heyer et al., 2010). Without in vitro manipulation, tissue fragments derived from tailor-made GEM tumors are expanded by transplantation, orthotopically or subcutaneously, into immunocompetent syngeneic hosts (Fig. 2). Thus, tumors can be banked to facilitate large cohort production, and efficacy studies can be performed in industry-friendly timeframes (~3–8 weeks), allowing for increased throughput. Indeed, a battery of treatments can be evaluated in GDAs prior to (preclinical) or parallel with (“co-clinical”) clinical trials (Table 2). As with GEMs, immune systems are fully functional in GDAs, and interactions among tumor cells and their intrinsic microenvironments are maintained.
GDAs are particularly amenable to the evaluation of metastatic disease, which is responsible for most cancer-related deaths and is rarely assessed preclinically. In GDAs, metastases occur from single primary tumors, which can be resected to allow time for metastatic progression (Day et al., 2012). This approach also emulates clinical care standards for many cancers and facilitates comparing therapeutic responses of both primary and metastatic disease derived from the same GEM cancer.
As with PDXs, serial passaging increases the likelihood that tumor properties will deviate from parental samples due to further evolution and/or selection of sub-compartment growth; thus, transplanted tumors should be monitored for molecular and biological similarity to founding tumors. Additionally, since transplanted tumors do not evolve in situ, GDAs cannot legitimately be used for prevention studies, and some therapeutic outcomes may differ between autochthonous GEMs and GDAs. Given the potential tradeoff of accuracy for tractability, candidate therapies efficacious in GDAs should be subsequently validated in the original GEM models prior to clinical studies.
Stem Cell-derived Chimeras and Somatic Models
Mice chimeric for genetically engineered cells are created through implantation of GEM-derived or genetically manipulated ES cells into pre-implantation embryos. Since oncogenic alleles are engineered ex vivo in ES cells, many mice with the desired genetic composition can be generated in the absence of complex, laborious and long-term breeding schemes. The potential value of this approach was first highlighted in the production, analysis and preclinical evaluation of lung adenocarcinoma (Zhou et al., 2010). Once constructed, ES cells harboring the desired alleles can be derived from blastocysts produced by a penultimate cross. In turn, this bankable resource can be used to generate mice chimeric for mutant and wild type cells (Premsrirut et al., 2011), facilitating conditional RNAi-mediated knockdown of target expression via manipulation of ES cells, which can then be used to generate chimeric mouse cohorts (Dow et al., 2012).
Notably, the advent of CRISPR/Cas9 technology, and with it the ability to perform complex gene editing with relative ease and speed, has dramatically enhanced the value of non-germline GEM approaches. Several groups have precisely modified oncogenes and tumor suppressor genes directly in somatic cells of adult mice, significantly improving the feasibility and flexibility of this genetic engineering approach (Chen et al., 2015; Dow et al., 2015; Maddalo et al., 2014; Platt et al., 2014). These models also better mimic human cancer relative to standard germline GEMs in that tumors typically arise from fewer cells in the context of normal stroma
In a variation of the non-germline GEM approach, genetically engineered stem or progenitor cells can be transplanted into syngeneic mice, where they can home to appropriate tissue targets and become the cells of origin for developing tumors (Heyer et al., 2010). These models are especially amenable for studying hematopoietic cancers, where the stem cells are well characterized and the host can be prepared for receiving transplanted cells by using irradiation to create a favorable niche for the engineered hematopoietic stem/progenitor cells to colonize. Successes have also been reported for other cancers (Heyer et al., 2010).
LOGISTICS FOR OPTIMIZING PRECLINICAL STUDIES
Extensive complexities that impede successful drug development in cancer patients dictate that faithful murine cancer models must themselves be complex. Both PDX- and GEM-based models offer this opportunity. However, their very complexity warrants that informative models are generated and characterized with substantial knowledge of cancer mechanisms and modeling limitations, rigorous animal maintenance and production, routine phenotypic and genetic monitoring, appropriate strategies for therapeutic response evaluation, and consideration of multiple variables that impact data interpretation. To achieve routine therapeutic and biomarker development that positively influence patient care, preclinical studies must be (1) well-powered with significant cohort sizes and several evaluation parameters, (2) goal-oriented and efficiently executed, and (3) highly reproducible.
Experimental Considerations
Once models that optimally represent human disease have been selected, clinical relevance relies on experimental parameters that are comparable and/or translatable to human practice. These include, but are not limited to, dosing levels and schedules, drug pharmacology, response evaluation methods, and endpoint choices. Therapeutic agents’ pharmacokinetics (PK) and ability to modify targets when known (pharmacodynamics; PD) should be measured in tumor-bearing mice. The fate of administered drugs is largely determined by drug metabolizing enzymes essential for their absorption, distribution, metabolism, and excretion (ADME). Therefore, the differences that exist between the central metabolizing enzymes in mice and humans, the cytochrome P450 (CYP) family, constitute a confounding factor in extrapolating drug PKs and the responses they elicit. Since the maximum tolerated dose (MTD) of many drugs in mice is significantly higher than in humans, it is essential to evaluate efficacy by using doses achievable in patients. However, this is possible only when human PKs are known; for example, for repurposing FDA-approved drugs, for preclinical evaluation of combination therapies that comprise single phase II agents, and for co-clinical experimentation wherein mouse and human evaluations are performed in parallel, such that clinical toxicity results are available. Even when appropriate human dosing is known, there is no simple formula for approximating comparable doses to achieve the same PK in mice, and instead experimental determination is required (Sparreboom et al., 1996). Yet, when evaluating numerous agents, this approach is not possible; rather, subsequent coordination of clinical results and further preclinical dose escalation experiments are needed for optimal response assessment.
In an effort to apply a genetic solution to the PK problem, a number of humanized CYP GEMs have been developed (Gonzalez, 2004; Scheer and Wolf, 2014). Despite these advances, the humanized alleles have not yet been incorporated into GEM/PDX cancer models. Such an undertaking will require significant resources, substantial time, and community effort to generate and evaluate revised models. Nonetheless, the investment will be worthwhile if the gap between laboratory mice and patients is narrowed.
The choice of preclinical experimental endpoints to determine therapeutic responses is also critical for achieving outcomes most representative of those in patients (Talmadge et al., 2007). In prevention studies, efficacy is based on disease-free or minimized status. For intervention therapy, efficacy is justified by overall survival and should not be judged solely on tumor growth inhibition. The importance of survival endpoints is highlighted by a pancreatic cancer clinical trial designed based on short-term GEM studies demonstrating reduced tumor volumes in response to sonic hedgehog pathway inhibition combined with gemcitabine (standard of care) compared to gemcitabine alone (Olive et al., 2009). Unfortunately, the trial terminated early due to increased disease dissemination and poor patient survival. However, subsequent survival studies in the GEM model replicated the clinical result, demonstrating that initial drug effects did not predict survival outcomes. Hence, the model appropriately predicted patient responses, but only with a meaningful endpoint (Rhim et al., 2014).
Tumor growth and therapeutic responses in subcutaneous transplant models, such as CDXs, PDXs and GDAs can be monitored by standard caliper measurement. Tumor growth in autochthonous and orthotopic transplant models (other than skin and breast models) and in all metastatic models must be monitored by longitudinal imaging strategies (Wang et al., 2015). High-resolution 3-D images are compiled from sectional images generated by tomographic scanning of signals from X-ray (CAT), magnetic field-excited atoms (MRI), and injected radioactive tracers (SPECT; PET) (see Supplementary Information). Tomographic imaging requires specific expertise for accurate execution and is relatively expensive and time-consuming. Optical imaging, which detects visualized wavelengths generated from excited fluorescent chromophores [e.g., jellyfish green fluorescent protein (GFP)] or firefly luminescent reactions (e.g., luciferase), can be employed for detection in real-time and is cost- and time-effective; however, these methods do not produce accurate tomographic data and are limited by tissue absorption. Notably, traceable marker proteins required for optical imaging are xenogeneic with respect to mammals and can induce immune responses in immunocompetent mice, which can result in inconsistent activity, graft rejection and/or inhibition of metastasis, confounding data interpretation. Hence, effective employment of xenogeneic reporters is restricted to short-term studies or studies in immunocompromised models, limiting their usefulness in preclinical science (Steinbauer et al., 2003). However, this problem can be circumvented, at least in part, by employing host mice genetically engineered to express respective markers at an early age, which elicits tolerance and thus recognition as “self” (Day et al., 2014).
Several additional points associated with preclinical trial design are worth emphasizing. Tumor mass is a critical factor in preclinical studies; vastly different outcomes can result from initiating drug dosing when tumors are different sizes. Moreover, human tumors are typically much larger than their mouse counterparts, which could affect how preclinical data translates to the clinic. It is also vital to run preclinical trials with a sufficient number of animals in each experimental arm to achieve statistically significant results; ensuring statistical power must be considered a priority for any preclinical study. Therefore, it is prudent to consult biostatisticians prior to finalizing study designs. Finally, the influence of genetic background on tumor behavior can be significant, and must be considered when designing model systems. Generation and analysis of mouse cancer models within the collaborative cross, a large panel of inbred mouse strains (Churchill et al., 2004), could also provide important insights into the impact of complex germline genetics on tumor predisposition and drug response.
Infrastructure
Critical work establishing the utility of murine cancer models in preclinical research has taken place in independent laboratories over the last 20 years. However, because of severe resource limitations, the absolute need to perpetuate basic investigator-driven mechanistic discovery, and an increasingly competitive environment wherein success is measured by individual merit, the opportunity for laboratories to execute preclinical studies beyond the pilot level is limited. Recent reports indicate that most preclinical outcomes at this level are not reproduced when studies are not conducted with robust experimental standards, such as inclusion of appropriate positive and negative controls, execution with sufficient statistical power, attention to pharmacological considerations, and implementation of blind evaluations (Begley and Ellis, 2012; Begley and Ioannidis, 2015). Adherence to all these standards is simply not possible in individual laboratories under current conditions. To increase accessibility to preclinical evaluation in murine cancer models, several institutions have established core facilities that perform studies using dedicated staff and common methodologies. These cores represent a necessary step to improve reproducibility in preclinical outcomes. Yet, most core facilities do not have the resources to instate the full range of skills and technologies indicated in “Experimental Considerations” above to ensure optimal quality and replication of clinical approaches. Additionally, conducting well-powered blinded studies requires a sizable dedicated staff, which is generally not achievable in academic cores. Finally, global improvement of murine preclinical research must include the generation of an increased range of well-characterized, technically tractable and optimally accurate models vetted for preclinical evaluation, along with the development of exportable standard operating procedures (SOPs).
To address these needs, over the past decade several organized efforts have been established that are dedicated to: (1) improving the accuracy and reproducibility of preclinical drug development platforms; (2) developing and exporting SOPs and models; (3) understanding cancer pathobiology through targeted therapeutics; and/or (4) applying the outcomes of optimized preclinical therapeutic and biomarker studies to clinical research for improved patient care. Common attributes in each case include: (1) a sufficient number of dedicated staff covering a broad range of expertise; (2) access to sophisticated instrumentation and technology for a full range of small animal imaging modalities, histological and molecular pathology, genomic technologies, pharmacological methods, model generation, and appropriate maintenance and quality control for a large “bank” of models; and (3) data management strategies. Examples of such organizations include the Center for Advanced Preclinical Research (CAPR; Center for Cancer Research, National Cancer Center and the Frederick National Laboratory for Cancer Research, https://ccr.cancer.gov/capr-home); Mouse Clinic for Cancer and Aging research (MCCA; Netherlands Cancer Institute and the European Research Institute for the Biology of Aging, http://www.mccanet.nl/); Center for Co-Clinical Trials (MD Anderson, http://www.cancermoonshots.org/platforms/center-for-co-clinical-trials/); and the Co-Clinical Project: Informing Clinical Trials Using Preclinical Mouse Models (Harvard Medical School). Similar efforts focused specifically on pancreatic ductal carcinoma include the Mouse Hospitals (Columbia Medical School, http://www.olivelab.org/mouse-hospital.html, and Cold Spring Harbor Laboratories).
EMERGING AND FUTURE PROSPECTS
This PRIMER focuses on the attributes and limitations of murine cancer models that currently best emulate our existing understanding of human cancers, an ever-expanding awareness of which is required to drive development of effective preclinical platforms. The high cost and low yield of efficacious therapies, despite clinical evaluation of countless potential therapeutics, motivate the use and development of preclinical PDX and GEM in the guidance of clinical research. Ultimately, collective employment of a variety of model systems will likely be required to successfully impact clinical outcomes.
Optimal mouse studies are sufficiently cumbersome so as to preclude the simultaneous evaluation of numerous drugs and unbiased libraries; high-throughput in vitro screening systems are essential precursors to in vivo evaluations. Despite their limitations, cancer cell lines have proven valuable in uncovering mechanisms of acquired drug resistance for in vitro drug screens (Torrance et al., 2001), and several technologies such as RNAi and CRISPR/Cas9 methods have enhanced their versatility (Corcoran et al., 2013; Shalem et al., 2014). However, cancer cell-line screens identify only drugs that target intrinsic cancer cell functions. Targeting tumor stroma or microenvironment/tumor cell interactions requires the use of in vitro systems that approximate the composition of cancers that preserve important cancer constituents, cell-cell interactions, and architectural features. To this end, several ex vivo platforms have been developed, including spheroids, organoids, microtumors (tumor tissue in synthetic matrix), and tissue slices (Burdett et al., 2010; Mendoza et al., 2010; Yamada and Cukierman, 2007). While optimization and validation of emerging ex vivo models in drug screening is ongoing, many may be incorporated into early phases of drug development, resulting in efficient triage and increased success in vivo.
In addition to ex vivo systems, non-murine whole organism drug screens have shown promise for early triage (Gao et al., 2014). Due to their relatively small size, low cost and high fecundity, invertebrates such as flies (Drosophila) and nematodes (C. elegans) have shown promise. Furthermore, zebrafish (Danio rerio) are particularly well suited for high throughput screens because of rapid extra-uterine development, embryonic transparency and recently developed pigment deficiency to facilitate imaging (Barriuso et al., 2015). Using automated high content and high throughput platforms, zebrafish can be used for chemical, genetic, and pathway-based screens (Lieschke and Currie, 2007). Notably, data generated from zebrafish models have been used in clinical trials. For example, the pyrimidine biosynthesis enzyme DHODH was identified in zebrafish screens as a novel melanoma drug target, and a clinical trial is underway in which patients are being treated with the DHODH inhibitor leflunomide (Hagedorn et al., 2014; White et al., 2011). Zebrafish have also been used as hosts for human and mouse xenografts to monitor invasiveness, angiogenesis and drug responses in real time (Zhang et al., 2015). However, as with xenotransplantation of human cells into mice, inappropriate tumor-host interactions could limit the relevance and translational value of fish models.
Optimization of preclinical models that can impact clinical practice will require overcoming challenges in several arenas (Table 3). However, achieving this goal will undoubtedly require expansion and integration of organized efforts by many factions. The sophistication of such preclinical studies requires expertise in many disparate fields and necessitates involvement of scientists in the public sector, who often possess critical expertise and mechanisms not available in the private sector. However, communication and data-sharing among investigators and organizations, though essential for efficient optimization of effective preclinical standard operating procedures, are limited. A future priority will be to develop interactive web-based systems to house and mine experimental databases and SOPs for community sharing. Such organized initiatives will begin to meet the significant and immediate need to revolutionize the accuracy of preclinical assessment and to develop and utilize PDX- and GEM-based disease models in research to increase the number of effective treatments reaching clinical trials and thus, cancer patients.
Table 3.
Future challenges and possible solutions for mouse preclinical cancer trials
| Issue | Challenges | Possible Solutions |
|---|---|---|
| Model improvement | More precise spatial and temporal control of genetic alterations in mouse tissues | Improve technologies for genomic editing (e.g., CRISPR) and regulating gene activity |
| Human relevance of stroma, immune system and therapeutic targets in mouse cancer models | “Humanize” genes via genetic engineering and immune system by reconstitution with human hematopoietic stem cells | |
| Recapitulation of the tumor heterogeneity found in human cancers | Introduce environmental etiological factors (e.g., UV in skin cancer models); allow tumor evolution by avoiding inappropriately dominant oncogenic drivers | |
| Study setting | Difficulties in diagnosis and treatment of large cohorts of mice as individual patients | Synchronize tumorigenesis by adopting inducible GEM or transplantable GDA systems |
| Disease progression and clinically relevant endpoints in preclinical study | Improve biomarkers and imaging techniques for tumor tracking; adopt clinically-relevant endpoints (e.g., progression-free survival) | |
| Integration of pathologic, genomic, bioinformatic, molecular and immunological analyses | Develop/share improved and standardized protocols; organize workflows with core facilities | |
| Extrapolation to human disease | Evaluating effects of life style on therapeutic outcomes | Consider gender, diet, and exposure to environmental factors in protocol development; consider effects of microbiota |
| Physiological difference between mouse and human | “Humanize” aspects of mice; consider scaling law in PD/PK, life span, hemodynamics, etc. |
In summary, we now have a wealth of model systems that show early promise in establishing robust preclinical assessment platforms for improving clinical success. Each system has specific and sometimes unique value, and all will undoubtedly play a significant role in varied aspects of future preclinical studies. At this junction, systematic comparisons in the prediction of human outcomes by distinct model systems has not been carried out and is needed in order to construct sound preclinical operating principles. The selection of models for a given study will undoubtedly depend on the required purpose. While 2-D cell cultures are useful for identifying cancer cell-intrinsic vulnerabilities, 3-D ex vivo methods incorporate assessment of multicellular interactions. Non-mammalian animals further offer reasonable throughput in complex biological systems, while PDX and GEM models provide the best representation of tumor microenvironments, physiological responses, and disease pathology. GEMs further allow for evaluation of immune system interventions and of responses unique to in situ developed disease. Ultimately, the complementary use of many of these models and continual efforts to improve their effectiveness will propel preclinical studies to a new era of cancer therapeutics development. This is a uniquely exciting era wherein preclinical models, rather than serving simply to confirm clinical outcomes, have the potential to routinely fuel optimized clinical success.
Supplementary Material
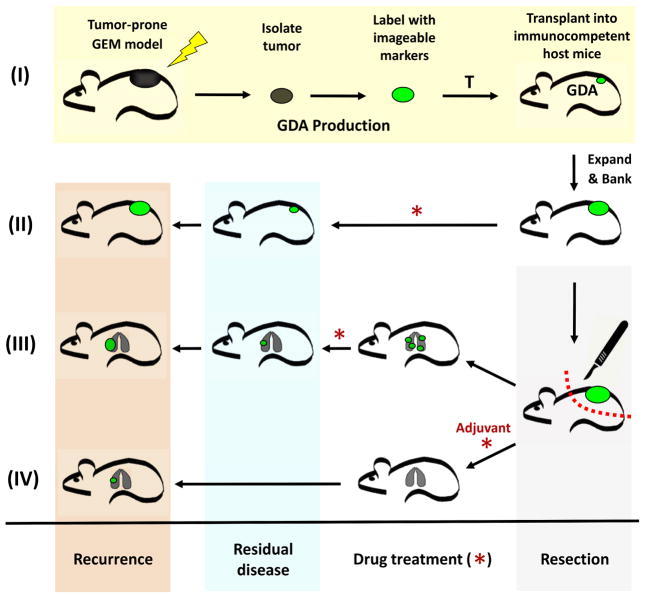
Fig. 4. Generation and application of metastatic GDA models.
(I) GDAs are derived from tumors arising in mice genetically tailored to produce human-relevant models. Relevance can be further enhanced by including appropriate etiological agents (lighting bolt). Arising tumors are resected, labeled with imageable markers, and directly transplanted into fully immunocompetent syngeneic mice at either subcutaneously or at orthotopic. Harvested tumors can be labeled with a variety of imageable markers to monitor growth and drug response, and to FACS purify for analysis. Once successfully transplanted, GDAs can be expanded for banking and/or preclinical studies. Mice bearing GDAs can be treated directly with individual or combination drugs (*) to study therapeutic efficacy at the “primary” tumor site (II). (III and IV) Alternatively, GDAs can be resected using survival surgery, and treatments focused on metastatic disease, simulating first-line treatment in human patients following primary tumor resection. GDA models allow for interventive treatment of metastatic disease once detected (III), or preventive adjuvant treatment initiated immediately following surgical resection (IV). GDA models are thus well suiting for studying primary or metastatic disease, with interventive or preventive approaches using pathway-targeted small molecule and/or immunotherapeutic agents.
Acknowledgments
We thank Mr. Jonathan Marie (www.thefrenzybear.com) for Figure 1 artwork, Drs. James Doroshow [Developmental Therapeutic Program (DTP), National Cancer Institute], Melinda Hollingshead (DTP), and Neal Goodwin (Champions Oncology, Inc.) for guidance on PDX studies, and Yurong Song for contributions to Table 2.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abate-Shen C, Politi K, Chodosh L, Olive K. Mouse Models of Cancer: A Laboratory Manual. CSHL Press; 2014. [Google Scholar]
- Ablain J, de The H. Retinoic acid signaling in cancer: The parable of acute promyelocytic leukemia. International journal of cancer Journal international du cancer. 2014;135:2262–2272. doi: 10.1002/ijc.29081. [DOI] [PubMed] [Google Scholar]
- Adams CP, Brantner VV. Estimating the cost of new drug development: is it really 802 million dollars? Health affairs (Project Hope) 2006;25:420–428. doi: 10.1377/hlthaff.25.2.420. [DOI] [PubMed] [Google Scholar]
- Al-Lazikani B, Banerji U, Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nat Biotechnol. 2012;30:679–692. doi: 10.1038/nbt.2284. [DOI] [PubMed] [Google Scholar]
- Bailey M, Christoforidou Z, Lewis MC. The evolutionary basis for differences between the immune systems of man, mouse, pig and ruminants. Vet Immunol Immunopathol. 2013;152:13–19. doi: 10.1016/j.vetimm.2012.09.022. [DOI] [PubMed] [Google Scholar]
- Barriuso J, Nagaraju R, Hurlstone A. Zebrafish: A New Companion for Translational Research in Oncology. Clin Cancer Res. 2015;21:969–975. doi: 10.1158/1078-0432.CCR-14-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty GL, Torigian DA, Chiorean EG, Saboury B, Brothers A, Alavi A, Troxel AB, Sun W, Teitelbaum UR, Vonderheide RH, et al. A phase I study of an agonist CD40 monoclonal antibody (CP-870,893) in combination with gemcitabine in patients with advanced pancreatic ductal adenocarcinoma. Clin Cancer Res. 2013;19:6286–6295. doi: 10.1158/1078-0432.CCR-13-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature. 2012;483:531–533. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- Begley CG, Ioannidis JPA. Reproducibility in science: improving the standard for basic and preclinical research. Circ Res. 2015;116:116–126. doi: 10.1161/CIRCRESAHA.114.303819. [DOI] [PubMed] [Google Scholar]
- Burdett E, Kasper FK, Mikos AG, Ludwig JA. Engineering tumors: a tissue engineering perspective in cancer biology. Tissue Eng Part B Rev. 2010;16:351–359. doi: 10.1089/ten.TEB.2009.0676. [DOI] [PubMed] [Google Scholar]
- Cao J, Wan L, Hacker E, Dai X, Lenna S, Jimenez-Cervantes C, Wang Y, Leslie NR, Xu GX, Widlund HR, et al. MC1R is a potent regulator of PTEN after UV exposure in melanocytes. Molecular cell. 2013;51:409–422. doi: 10.1016/j.molcel.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanovas O, Hicklin DJ, Bergers G, Hanahan D. Drug resistance by evasion of antiangiogenic targeting of VEGF signaling in late-stage pancreatic islet tumors. Cancer cell. 2005;8:299–309. doi: 10.1016/j.ccr.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, Scott DA, Song J, Pan JQ, Weissleder R, et al. Genome-wide CRISPR Screen in a Mouse Model of Tumor Growth and Metastasis. Cell. 2015;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Akbay E, Mikse O, Tupper T, Cheng K, Wang Y, Tan X, Altabef A, Woo S-A, Chen L, et al. Co-clinical trials demonstrate superiority of crizotinib to chemotherapy in ALK-rearranged non-small cell lung cancer and predict strategies to overcome resistance. Clin Cancer Res. 2014;20:1204–1211. doi: 10.1158/1078-0432.CCR-13-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Cheng K, Walton Z, Wang Y, Ebi H, Shimamura T, Liu Y, Tupper T, Ouyang J, Li J, et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature. 2012;483:613–617. doi: 10.1038/nature10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat Med. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- Chiu CW, Nozawa H, Hanahan D. Survival benefit with proapoptotic molecular and pathologic responses from dual targeting of mammalian target of rapamycin and epidermal growth factor receptor in a preclinical model of pancreatic neuroendocrine carcinogenesis. J Clin Oncol. 2010;28:4425–4433. doi: 10.1200/JCO.2010.28.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, Beavis WD, Belknap JK, Bennett B, Berrettini W, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Corcoran RB, Cheng KA, Hata AN, Faber AC, Ebi H, Coffee EM, Greninger P, Brown RD, Godfrey JT, Cohoon TJ, et al. Synthetic lethal interaction of combined BCL-XL and MEK inhibition promotes tumor regressions in KRAS mutant cancer models. Cancer cell. 2013;23:121–128. doi: 10.1016/j.ccr.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Thakur M, Salangsang F, Landman AS, Sellers WR, Pryer NK, Levesque MP, Dummer R, McMahon M, Stuart DD. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013;494:251–255. doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day CP, Carter J, Bonomi C, Hollingshead M, Merlino G. Preclinical therapeutic response of residual metastatic disease is distinct from its primary tumor of origin. International journal of cancer Journal international du cancer. 2012;130:190–199. doi: 10.1002/ijc.25978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day CP, Carter J, Weaver Ohler Z, Bonomi C, El Meskini R, Martin P, Graff-Cherry C, Feigenbaum L, Tuting T, Van Dyke T, et al. "Glowing head" mice: a genetic tool enabling reliable preclinical image-based evaluation of cancers in immunocompetent allografts. PLoS One. 2014;9:e109956. doi: 10.1371/journal.pone.0109956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVita VT, Jr, Chu E. A history of cancer chemotherapy. Cancer research. 2008;68:8643–8653. doi: 10.1158/0008-5472.CAN-07-6611. [DOI] [PubMed] [Google Scholar]
- Dow LE, Fisher J, O'Rourke KP, Muley A, Kastenhuber ER, Livshits G, Tschaharganeh DF, Socci ND, Lowe SW. Inducible in vivo genome editing with CRISPR-Cas9. Nat Biotechnol. 2015;33:390–394. doi: 10.1038/nbt.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow LE, Premsrirut PK, Zuber J, Fellmann C, McJunkin K, Miething C, Park Y, Dickins RA, Hannon GJ, Lowe SW. A pipeline for the generation of shRNA transgenicmice. Nat Protoc. 2012;7:374–393. doi: 10.1038/nprot.2011.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faca VM, Song KS, Wang H, Zhang Q, Krasnoselsky AL, Newcomb LF, Plentz RR, Gurumurthy S, Redston MS, Pitteri SJ, et al. A mouse to human search for plasma proteome changes associated with pancreatic tumor development. PLoS Med. 2008;5:e123. doi: 10.1371/journal.pmed.0050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler IJ, Hart IR. Biological diversity in metastatic neoplasms: origins and implications. Science. 1982;217:998–1003. doi: 10.1126/science.7112116. [DOI] [PubMed] [Google Scholar]
- Fiebig HH, Schuchhardt C, Henss H, Fiedler L, Lohr GW. Comparison of tumor response in nude mice and in the patients. Behring Inst Mitt. 1984:343–352. [PubMed] [Google Scholar]
- Frese KK, Neesse A, Cook N, Bapiro TE, Lolkema MP, Jodrell DI, Tuveson DA. nab-Paclitaxel potentiates gemcitabine activity by reducing cytidine deaminase levels in a mouse model of pancreatic cancer. Cancer discovery. 2012;2:260–269. doi: 10.1158/2159-8290.CD-11-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Chen L, Huang C. Anti-cancer drug discovery: update and comparisons in yeast, Drosophila, and zebrafish. Curr Mol Pharmacol. 2014;7:44–51. doi: 10.2174/1874467207666140702113629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Janne PA. Circumventing cancer drug resistance in the era of personalized medicine. Cancer discovery. 2012;2:214–226. doi: 10.1158/2159-8290.CD-12-0012. [DOI] [PubMed] [Google Scholar]
- Girotti MR, Lopes F, Preece N, Niculescu-Duvaz D, Zambon A, Davies L, Whittaker S, Saturno G, Viros A, Pedersen M, et al. Paradox-Breaking RAF Inhibitors that Also Target SRC Are Effective in Drug-Resistant BRAF Mutant Melanoma. Cancer cell. 2015;27:85–96. doi: 10.1016/j.ccell.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein D, El-Maraghi RH, Hammel P, Heinemann V, Kunzmann V, Sastre J, Scheithauer W, Siena S, Tabernero J, Teixeira L, et al. nab-Paclitaxel plus gemcitabine for metastatic pancreatic cancer: long-term survival from a phase III trial. Journal of the National Cancer Institute. 2015;107 doi: 10.1093/jnci/dju413. [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ. Cytochrome P450 humanised mice. Human genomics. 2004;1:300–306. doi: 10.1186/1479-7364-1-4-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn EJ, Durand EM, Fast EM, Zon LI. Getting more for your marrow: boosting hematopoietic stem cell numbers with PGE2. Exp Cell Res. 2014;329:220–226. doi: 10.1016/j.yexcr.2014.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanash S, Taguchi A. Application of proteomics to cancer early detection. Cancer journal. 2011;17:423–428. doi: 10.1097/PPO.0b013e3182383cab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer J, Kwong LN, Lowe SW, Chin L. Non-germline genetically engineered mouse models for translational cancer research. Nature reviews Cancer. 2010;10:470–480. doi: 10.1038/nrc2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo M, Amant F, Biankin AV, Budinska E, Byrne AT, Caldas C, Clarke RB, de Jong S, Jonkers J, Maelandsmo GM, et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer discovery. 2014;4:998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood L, Friend SH. Predictive, personalized, preventive, participatory (P4) cancer medicine. Nat Rev Clin Oncol. 2011;8:184–187. doi: 10.1038/nrclinonc.2010.227. [DOI] [PubMed] [Google Scholar]
- Ilie M, Nunes M, Blot L, Hofman V, Long-Mira E, Butori C, Selva E, Merino-Trigo A, Venissac N, Mouroux J, et al. Setting up a wide panel of patient-derived tumor xenografts of non-small cell lung cancer by improving the pre analytical steps. Cancer medicine. 2015;4:201–211. doi: 10.1002/cam4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janne PA, Shaw AT, Pereira JR, Jeannin G, Vansteenkiste J, Barrios C, Franke FA, Grinsted L, Zazulina V, Smith P, et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. The Lancet Oncology. 2013;14:38–47. doi: 10.1016/S1470-2045(12)70489-8. [DOI] [PubMed] [Google Scholar]
- Johnson JI, Decker S, Zaharevitz D, Rubinstein LV, Venditti JM, Schepartz S, Kalyandrug S, Christian M, Arbuck S, Hollingshead M, et al. Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. Br J Cancer. 2001;84:1424–1431. doi: 10.1054/bjoc.2001.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan K, Sharpless NE, Xu J, O'Hagan RC, Bosenberg M, Chin L. Components of the Rb pathway are critical targets of UV mutagenesis in a murine melanoma model. Proc Natl Acad Sci U S A. 2003;100:1221–1225. doi: 10.1073/pnas.0336397100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortmann U, McAlpine JN, Xue H, Guan J, Ha G, Tully S, Shafait S, Lau A, Cranston AN, O'Connor MJ, et al. Tumor growth inhibition by olaparib in BRCA2 germline-mutated patient-derived ovarian cancer tissue xenografts. Clin Cancer Res. 2011;17:783–791. doi: 10.1158/1078-0432.CCR-10-1382. [DOI] [PubMed] [Google Scholar]
- Kwong LN, Costello JC, Liu H, Jiang S, Helms TL, Langsdorf AE, Jakubosky D, Genovese G, Muller FL, Jeong JH, et al. Oncogenic NRAS signaling differentially regulates survival and proliferation in melanoma. Nat Med. 2012;18:1503–1510. doi: 10.1038/nm.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaf C. Why we're losing the war on cancer (and how to win it) Fortune. 2004;149:76–82. 84–76, 88 passim. [PubMed] [Google Scholar]
- Legrand N, Ploss A, Balling R, Becker PD, Borsotti C, Brezillon N, Debarry J, de Jong Y, Deng H, Di Santo JP, et al. Humanized mice for modeling human infectious disease: challenges, progress, and outlook. Cell Host Microbe. 2009;6:5–9. doi: 10.1016/j.chom.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nature reviews Genetics. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- Lunardi A, Pandolfi PP. A co-clinical platform to accelerate cancer treatment optimization. Trends in molecular medicine. 2015;21:1–5. doi: 10.1016/j.molmed.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. The New England journal of medicine. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han Y-C, Ogrodowski P, Crippa A, Rekhtman N, de Stanchina E, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaney P, Nicosia SV, Dave V. One mouse, one patient paradigm: New avatars of personalized cancer therapy. Cancer letters. 2014;344:1–12. doi: 10.1016/j.canlet.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattern J, Bak M, Hahn EW, Volm M. Human tumor xenografts as model for drug testing. Cancer Metastasis Rev. 1988;7:263–284. doi: 10.1007/BF00047755. [DOI] [PubMed] [Google Scholar]
- Mendoza A, Hong S-H, Osborne T, Khan MA, Campbell K, Briggs J, Eleswarapu A, Buquo L, Ren L, Hewitt SM, et al. Modeling metastasis biology and therapy in real-time in the mouse lung. J Clin Invest. 2010;120:2979–2988. doi: 10.1172/JCI40252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou H, Kennedy Z, Anderson DG, Yin H, Xue W. Precision cancer mouse models through genome editing with CRISPR-Cas9. Genome Med. 2015;7:53. doi: 10.1186/s13073-015-0178-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan FP, Recio JA, Takayama H, Duray P, Anver MR, Rush WL, De Fabo EC, Merlino G. Neonatal sunburn and melanoma in mice. Nature. 2001;413:271–272. doi: 10.1038/35095108. [DOI] [PubMed] [Google Scholar]
- Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, Madhu B, Goldgraben MA, Caldwell ME, Allard D, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA. The use of targeted mouse models for preclinical testing of novel cancer therapeutics. Clin Cancer Res. 2006;12:5277–5287. doi: 10.1158/1078-0432.CCR-06-0436. [DOI] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Page DB, Postow MA, Callahan MK, Allison JP, Wolchok JD. Immunemodulation in cancer with antibodies. Annual review of medicine. 2014;65:185–202. doi: 10.1146/annurev-med-092012-112807. [DOI] [PubMed] [Google Scholar]
- Pagliarini R, Shao W, Sellers WR. Oncogene addiction: pathways of therapeutic response, resistance, and road maps toward a cure. EMBO reports. 2015;16:280–296. doi: 10.15252/embr.201439949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfi PP. Oncogenes and tumor suppressors in the molecular pathogenesis of acute promyelocytic leukemia. Human molecular genetics. 2001;10:769–775. doi: 10.1093/hmg/10.7.769. [DOI] [PubMed] [Google Scholar]
- Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells - what challenges do they pose? Nat Rev Drug Discov. 2014;13:497–512. doi: 10.1038/nrd4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras K, Hanahan D. A multitargeted, metronomic, and maximum-tolerated dose" chemo-switch" regimen is antiangiogenic, producing objective responses and survival benefit in a mouse model of cancer. J Clin Oncol. 2005;23:939–952. doi: 10.1200/JCO.2005.07.093. [DOI] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premsrirut PK, Dow LE, Kim SY, Camiolo M, Malone CD, Miething C, Scuoppo C, Zuber J, Dickins RA, Kogan SC, et al. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell. 2011;145:145–158. doi: 10.1016/j.cell.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quignon F, Chen Z, de The H. Retinoic acid and arsenic: towards oncogene-targeted treatments of acute promyelocytic leukaemia. Biochim Biophys Acta. 1997;1333:M53–61. doi: 10.1016/s0304-419x(97)00025-5. [DOI] [PubMed] [Google Scholar]
- Raymond E, Dahan L, Raoul J-L, Bang Y-J, Borbath I, Lombard-Bohas C, Valle J, Metrakos P, Smith D, Vinik A, et al. Sunitinib malate for the treatment of pancreatic neuroendocrine tumors. The New England journal of medicine. 2011;364:501–513. doi: 10.1056/NEJMoa1003825. [DOI] [PubMed] [Google Scholar]
- Reinhold WC, Varma S, Rajapakse VN, Luna A, Sousa FG, Kohn KW, Pommier YG. Using drug response data to identify molecular effectors, and molecular "omic" data to identify candidate drugs in cancer. Hum Genet. 2015;134:3–11. doi: 10.1007/s00439-014-1482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, Dekleva EN, Saunders T, Becerra CP, Tattersall IW, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer cell. 2014;25:735–747. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley RR, Duensing S, Brake T, Munger K, Lambert PF, Arbeit JM. Dissection of human papillomavirus E6 and E7 function in transgenic mouse models of cervical carcinogenesis. Cancer research. 2003;63:4862–4871. [PubMed] [Google Scholar]
- Rogers AB, Fox JG. Inflammation and Cancer. I. Rodent models of infectious gastrointestinal and liver cancer. Am J Physiol Gastrointest Liver Physiol. 2004;286:G361–366. doi: 10.1152/ajpgi.00499.2003. [DOI] [PubMed] [Google Scholar]
- Romer JT, Kimura H, Magdaleno S, Sasai K, Fuller C, Baines H, Connelly M, Stewart CF, Gould S, Rubin LL, et al. Suppression of the Shh pathway using a small molecule inhibitor eliminates medulloblastoma in Ptc1(+/−)p53(−/−) mice. Cancer cell. 2004;6:229–240. doi: 10.1016/j.ccr.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. The New England journal of medicine. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- Rosfjord E, Lucas J, Li G, Gerber H-P. Advances in patient-derived tumor xenografts: from target identification to predicting clinical response rates in oncology. Biochem Pharmacol. 2014;91:135–143. doi: 10.1016/j.bcp.2014.06.008. [DOI] [PubMed] [Google Scholar]
- Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L, Holcomb T, Stinson J, Gould SE, Coleman B, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. The New England journal of medicine. 2009;361:1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer N, Snaith M, Wolf CR, Seibler J. Generation and utility of genetically humanized mouse models. Drug Discov Today. 2013;18:1200–1211. doi: 10.1016/j.drudis.2013.07.007. [DOI] [PubMed] [Google Scholar]
- Scheer N, Wolf CR. Genetically humanized mouse models of drug metabolizing enzymes and transporters and their applications. Xenobiotica. 2014;44:96–108. doi: 10.3109/00498254.2013.815831. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science (New York, N Y ) 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless NE, Depinho RA. The mighty mouse: genetically engineered mouse models in cancer drug development. Nat Rev Drug Discov. 2006;5:741–754. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- Shultz LD, Goodwin N, Ishikawa F, Hosur V, Lyons BL, Greiner DL. Human cancer growth and therapy in immunodeficient mouse models. Cold Spring Harb Protoc. 2014;2014:694–708. doi: 10.1101/pdb.top073585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Murriel CL, Johnson L. Genetically engineered mouse models: closing the gap between preclinical data and trial outcomes. Cancer research. 2012;72:2695–2700. doi: 10.1158/0008-5472.CAN-11-2786. [DOI] [PubMed] [Google Scholar]
- Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparreboom A, van Tellingen O, Nooijen WJ, Beijnen JH. Nonlinear pharmacokinetics of paclitaxel in mice results from the pharmaceutical vehicle Cremophor EL. Cancer research. 1996;56:2112–2115. [PubMed] [Google Scholar]
- Stebbing J, Paz K, Schwartz GK, Wexler LH, Maki R, Pollock RE, Morris R, Cohen R, Shankar A, Blackman G, et al. Patient-derived xenografts for individualized care in advanced sarcoma. Cancer. 2014;120:2006–2015. doi: 10.1002/cncr.28696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbauer M, Guba M, Cernaianu G, Kohl G, Cetto M, Kunz-Schughart LA, Geissler EK, Falk W, Jauch KW. GFP-transfected tumor cells are useful in examining early metastasis in vivo, but immune reaction precludes long-term tumor development studies in immunocompetent mice. Clin Exp Metastasis. 2003;20:135–141. doi: 10.1023/a:1022618909921. [DOI] [PubMed] [Google Scholar]
- Stiedl P, Grabner B, Zboray K, Bogner E, Casanova E. Modeling cancer using genetically engineered mice. Methods Mol Biol. 2015;1267:3–18. doi: 10.1007/978-1-4939-2297-0_1. [DOI] [PubMed] [Google Scholar]
- Svenson KL, Gatti DM, Valdar W, Welsh CE, Cheng R, Chesler EJ, Palmer AA, McMillan L, Churchill GA. High-resolution genetic mapping using the Mouse Diversity out bred population. Genetics. 2012;190:437–447. doi: 10.1534/genetics.111.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabova L, Bupp S, Kamal M, Householder DB, Hernandez L, Schlomer JJ, Baran ML, Yi M, Stephens RM, Annunziata CM, et al. Pathway-specific engineered mouse allograft models functionally recapitulate human serous epithelial ovarian cancer. PLoS One. 2014;9:e95649. doi: 10.1371/journal.pone.0095649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmadge JE, Singh RK, Fidler IJ, Raz A. Murine models to evaluate novel and conventional therapeutic strategies for cancer. Am J Pathol. 2007;170:793–804. doi: 10.2353/ajpath.2007.060929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teicher BA. Tumor models for efficacy determination. Mol Cancer Ther. 2006;5:2435–2443. doi: 10.1158/1535-7163.MCT-06-0391. [DOI] [PubMed] [Google Scholar]
- Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, Arcaroli JJ, Messersmith WA, Eckhardt SG. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol. 2012;9:338–350. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Texido G. Genetically engineered animal models for in vivo target identification and validation in oncology. Methods Mol Biol. 2013;986:281–305. doi: 10.1007/978-1-62703-311-4_18. [DOI] [PubMed] [Google Scholar]
- Torrance CJ, Agrawal V, Vogelstein B, Kinzler KW. Use of isogenic human cancer cells for high-through put screening and drug discovery. Nat Biotechnol. 2001;19:940–945. doi: 10.1038/nbt1001-940. [DOI] [PubMed] [Google Scholar]
- van't Veer LJ, Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature. 2008;452:564–570. doi: 10.1038/nature06915. [DOI] [PubMed] [Google Scholar]
- Van Dyke T, Jacks T. Cancer modeling in the modern era: progress and challenges. Cell. 2002;108:135–144. doi: 10.1016/s0092-8674(02)00621-9. [DOI] [PubMed] [Google Scholar]
- Walrath JC, Hawes JJ, Van Dyke T, Reilly KM. Genetically engineered mouse models in cancer research. Adv Cancer Res. 2010;106:113–164. doi: 10.1016/S0065-230X(10)06004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tseng J-C, Sun Y, Beck AH, Kung AL. Noninvasive imaging of tumor burden and molecular pathways in mouse models of cancer. Cold Spring Harb Protoc. 2015;2015:135–144. doi: 10.1101/pdb.top069930. [DOI] [PubMed] [Google Scholar]
- Westcott PMK, Halliwill KD, To MD, Rashid M, Rust AG, Keane TM, Delrosario R, Jen K-Y, Gurley KE, Kemp CJ, et al. The mutational landscapes of genetic and chemical models of Kras-driven lung cancer. Nature. 2015;517:489–492. doi: 10.1038/nature13898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, Langdon E, Tomlinson ML, Mosher J, Kaufman C, et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011;471:518–522. doi: 10.1038/nature09882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada KM, Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell. 2007;130:601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Yap TA, Omlin A, de Bono JS. Development of therapeutic combinations targeting major cancer signaling pathways. J Clin Oncol. 2013;31:1592–1605. doi: 10.1200/JCO.2011.37.6418. [DOI] [PubMed] [Google Scholar]
- Young NP, Crowley D, Jacks T. Uncoupling cancer mutations reveals critical timing of p53 loss in sarcomagenesis. Cancer research. 2011;71:4040–4047. doi: 10.1158/0008-5472.CAN-10-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Xuan C, Ji Y, Zhang W, Wang D. Zebrafish xenotransplantation as a tool for in vivo cancer study. Familial cancer. 2015 doi: 10.1007/s10689-015-9802-3. [DOI] [PubMed] [Google Scholar]
- Zhang X, Claerhout S, Prat A, Dobrolecki LE, Petrovic I, Lai Q, Landis MD, Wiechmann L, Schiff R, Giuliano M, et al. A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer research. 2013;73:4885–4897. doi: 10.1158/0008-5472.CAN-12-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Rideout WM, 3rd, Zi T, Bressel A, Reddypalli S, Rancourt R, Woo J-K, Horner JW, Chin L, Chiu MI, et al. Chimeric mouse tumor models reveal differences in pathway activation between ERBB family- and KRAS-dependent lung adenocarcinomas. Nat Biotechnol. 2010;28:71–78. doi: 10.1038/nbt.1595. [DOI] [PubMed] [Google Scholar]
- Zitvogel L, Apetoh L, Ghiringhelli F, Kroemer G. Immunological aspects of cancer chemotherapy. Nat Rev Immunol. 2008;8:59–73. doi: 10.1038/nri2216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.