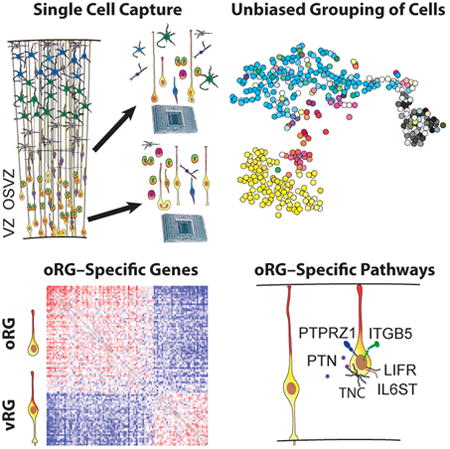
Summary
Radial glia, the neural stem cells of the neocortex, are located in two niches: the ventricular zone and outer subventricular zone. Although outer subventricular zone radial glia may generate the majority of human cortical neurons, their molecular features remain elusive. By analyzing gene expression across single cells, we find that outer radial glia preferentially express genes related to extracellular matrix formation, migration, and stemness, including TNC, PTPRZ1, FAM107A, HOPX, and LIFR. Using dynamic imaging, immunostaining, and clonal analysis, we relate these molecular features to distinctive behaviors of outer radial glia, demonstrate the necessity of STAT3 signaling for their cell cycle progression, and establish their extensive proliferative potential. These results suggest that outer radial glia directly support the subventricular niche through local production of growth factors, potentiation of growth factor signals by extracellular matrix proteins, and activation of self-renewal pathways, thereby enabling the developmental and evolutionary expansion of the human neocortex.
Graphical abstract
Introduction
The human neocortex contains 16 billion neurons of diverse types that develop from an initially uniform neuroepithelium. In the ventricular zone (VZ), radial glia undergo interkinetic nuclear migration and possess apical processes that contact the ventricle and form adherens junctions. Apical complex proteins transduce signals from the cerebrospinal fluid that are critical for the survival, proliferation, and neurogenic capacity of ventricular radial glia (vRG) (Lehtinen et al., 2011). However, the majority of human radial glia are located in the outer subventricular zone (OSVZ) (Lewitus et al., 2013). These outer radial glia (oRG) retain basal processes but lack apical junctions and undergo a distinct migratory behavior, mitotic somal translocation, directly preceding cell division (Hansen et al., 2010). Thus vRG and oRG cells reside in distinct niches defined by differences in anatomical location, provision of growth factors, cell morphology, and behavior (Fietz et al., 2010). Although oRG cells may generate the majority of cortical neurons (Lewitus et al., 2013; Smart et al., 2002), the molecular features sustaining neural stem cell properties of oRG cells in the OSVZ niche are largely unknown and the long-term proliferative capacity of these cells has not been examined.
Understanding the molecular programs specifically employed by oRG cells would provide insights into mechanisms of cortical development and support strategies to generate this cell type in vitro. Previous studies have attempted to identify genes uniquely expressed in oRG cells using a variety of transcriptional profiling strategies, including comparisons between microdissected samples (Fietz et al., 2012; Miller et al., 2014) and between cell populations expressing particular surface proteins (Florio et al., 2015; Johnson et al., 2015), but the difficulty of isolating bona fide oRG cells has made clear definition of their gene expression profiles challenging.
To specifically compare molecular features of radial glia in the VZ and OSVZ, we performed RNA sequencing of single cells captured from these two zones without additional enrichment steps. We then classified cells by analyzing thousands of genes that vary across cells and isolated radial glia from other cell types in silico (Pollen et al., 2014). We find that the proneural gene networks recently attributed to oRG cells are largely restricted to intermediate progenitor cells. Within classically defined radial glia, we discover molecular distinctions between vRG and oRG cells. The transcriptional state enriched in oRG cells includes genes involved in extracellular matrix production, epithelial-to-mesenchymal transition, and stem cell maintenance. Surprisingly, we find components of the LIFR/STAT3 self-renewal pathway are selectively expressed by oRG but not vRG cells, and we confirm that STAT3 signaling is necessary for oRG cell cycle progression. We further find that single oRG cells have the capacity to produce hundreds of deep and upper cortical layer neurons. Based on these results, we propose that oRG cells directly support the development of an enlarged OSVZ neural stem cell niche through the local production of growth factors, the expression of extracellular matrix proteins that potentiate growth factor signaling, and the activation of the LIFR/STAT3 signaling pathway.
Results
Molecular Diversity of Cells in the Cortical Germinal Zones
To analyze molecular features of cells in the germinal zones during human cortical neurogenesis, we captured single cells from microdissected VZ and SVZ specimens of human cortex at gestational week 16-18 (GW16-18) and generated sequencing libraries (schematic Figure 1A). We subsequently analyzed 393 single cells from three individuals in which we detected at least 1000 genes (Table S1). To classify cells, we performed principal component analysis (PCA) and used expectation-maximization clustering to group cells based on their position in PC space (Figure S1, Experimental Procedures). Based on the expression of known marker genes, we interpreted groups to represent cells along the cortical excitatory lineage and inhibitory interneurons generated in the ventral telencephalon (Figures 1B, 1C, 1D, and S1, Table S2).
Figure 1. Molecular Diversity of Single Cells from Human Cortical Germinal Zone.
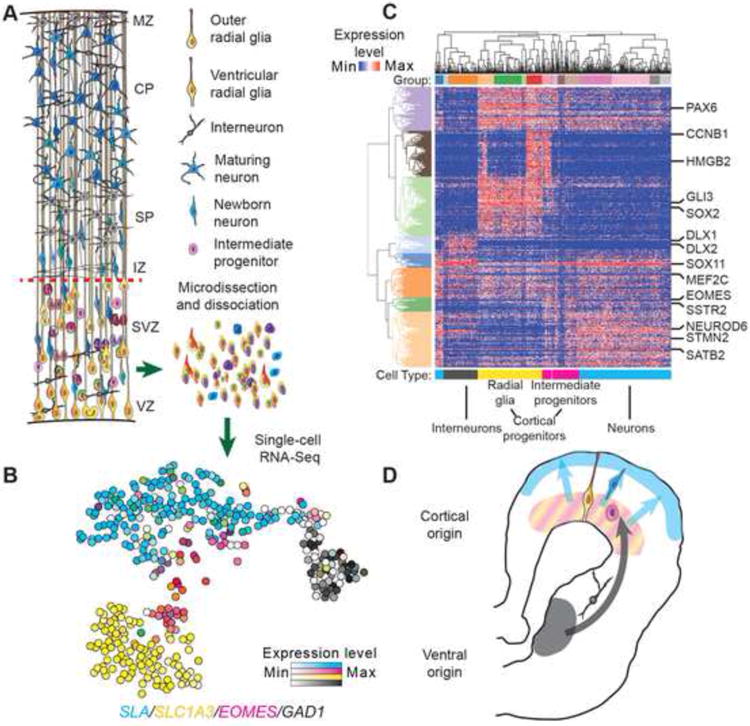
(A) Schematic representation of major cell populations of developing cortex. VZ- ventricular zone, SVZ- subventricular zone, IZ- intermediate zone, SP- subplate, CP- cortical plate, MZ- marginal zone. (B) Representation of transcriptional heterogeneity of germinal zone cells profiled by single cell mRNA-Seq. Cells are arranged according to their position determined using t-distributed stochastic neighbor embedding. (C) Heatmap showing gene expression levels for 1% of genes most strongly contributing to PC1-4. Select marker genes are highlighted. Groups represent clusters with highest approximately unbiased p-values following multiscale bootstrapping of hierarchical clustering based on expectation-maximization cluster assignments (see also Figure S1). (D) Interpretation of distinct cortical and ventral telencephalic lineages detected among germinal zone cells.
We further examined groups of cells expressing known markers of the cortical excitatory neuron lineage (schematic, Figure 2A). Four groups robustly expressed markers of human radial glia SLC1A3, PAX6, SOX2, PDGFD, GLI3 (yellow bar, Figure 2A). Another four groups retained a reduced level of PAX6, SOX2 and SLC1A3 expression, but also expressed early neuronal markers such as STMN2 and NEUROD6. These groups were also characterized by the absence of canonical radial glia marker expression, including VIM and HES1, and by the specific expression of canonical and novel intermediate progenitor markers EOMES (TBR2), ELAVL4, NEUROG1, NEUROD1, NEUROD4, PPP1R17, and PENK (magenta bar, Figures 2A, 2B, and S1) (Hevner et al., 2006; Kawaguchi et al., 2008). We found that the vast majority of cells expressing EOMES and PPP1R17 mRNA also expressed EOMES protein with variable SOX2 expression (Figure S2). Immunostaining for PPP1R17 revealed diverse morphologies of these cells, including multipolar cells with short processes as well as unipolar and bipolar cells with one or two radially or tangentially oriented processes. Regardless of morphology, these progenitor cells did not express the classical molecular signature of radial glia (Figure S2). Thus, our analysis provides a clear distinction between radial glia and intermediate progenitor cells. Future studies may relate the molecular heterogeneity of intermediate progenitors and the relative expression of radial glial and proneural genes to the diverse and dynamic morphological features reported in OSVZ progenitors (Betizeau et al., 2013).
Figure 2. Major Sources of Transcriptional Heterogeneity Among Radial Glia.
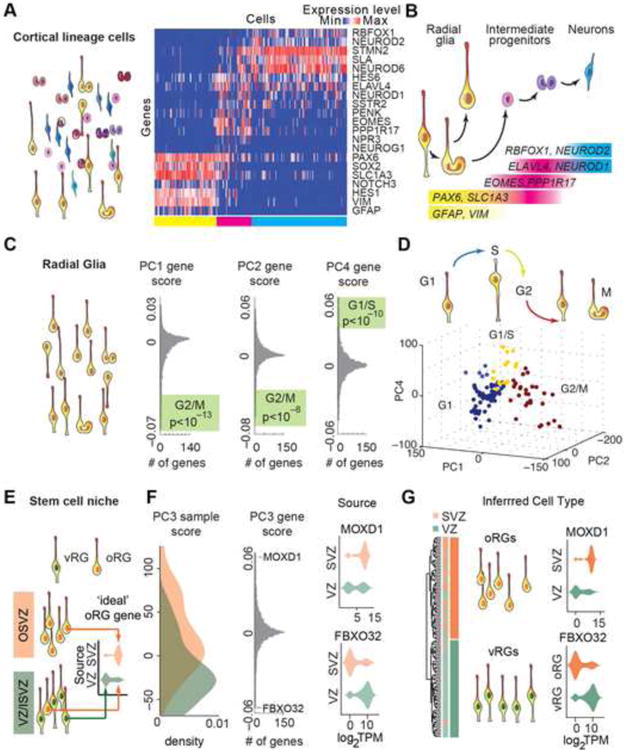
(A) Analysis of cell type identity of in silico sorted cortical lineage cells (schematic) – heatmap illustrates expression of identity genes across cells. Colored bars below heatmap highlight membership of cells into major groups (Figure S1). (B) Interpretation of sequential gene expression changes during cortical excitatory neuron differentiation (see also Figure S2). (C) Schematic illustrating in silico sorting of classically defined radial glia from the set of cortical lineage cells. Radial glia themselves display heterogeneity with respect to cell cycle progression, morphology, anatomical position and behavior, and PCA reveals major sources of transcriptional variation among radial glia. Histogram of PCA gene loading scores with gene ontology enrichments highlighted (adjusted p-value, Fisher exact test). (D) Interpretation of cell cycle phases for radial glia based on clustering according to sample scores along PC1, 2 and 4 (see also Figure S3). (E) Schematic of the anatomical sources of radial glia and violin plots illustrate prediction that VZ and adjacent inner SVZ contain a mixed population of vRG and oRG cells. (F) Histogram of VZ and OSVZ radial glia sample scores and gene loading scores along PC3. Radial glia from OSVZ (n=39) have significantly higher PC3 scores than radial glia from VZ (n=68) across 3 samples (p < 10-4, Welch t-test, see also Figure S3). Violin plots show distribution of expression values for strongly loading genes across cells from VZ and OSVZ sources. (G) Interpretation of oRG and vRG identity based on clustering cells according to top 1% of genes loading PC3. Schematic highlights the grouping of radial glia by inferred cell type rather than by anatomical source. Violin plots show distribution of expression values within inferred cell types.
Major Sources of Transcriptional Variation among Radial Glia Relate to Cell Cycle and Stem Cell Niche
We next analyzed variation in gene expression across 107 cells from the four groups that robustly expressed canonical markers of radial glia. We anticipated that cell cycle would be the major source of transcriptional variation across proliferative populations (Pollen et al., 2014). Indeed, genes involved in cell cycle regulation, mitosis, and DNA replication explained most variation along PC1, PC2 and PC4 (Figure 2C, Table S3). Clustering radial glia based on variation along these axes revealed cell groups representing G1, G1/S checkpoint, and G2/M checkpoint (Figure 2D, Figure S3). During interkinetic nuclear migration, the cell bodies of radial glia migrate away from the ventricle during G1, towards the ventricle during S/G2 phase, and divide at the ventricular surface. We examined the expression of a novel predicted G1 marker, CRYAB, and found that cells expressing CRYAB transcript were displaced from the ventricle, rarely expressed the G2/M marker phospho-histone H3, and rarely incorporated BrdU, a label for DNA replication, consistent with the G1 specificity of this transcript (Figure S3). Thus, differentiation and cell cycle are major sources of transcriptional heterogeneity among cells in the germinal zone, and single cell analysis reveals novel molecular features of these states.
We hypothesized that differences in stem cell niche occupancy would also contribute to variation among radial glia. Descriptions of cell behavior and morphology suggest that the VZ and adjacent inner SVZ contain mixed populations of vRG cells and oRG cells destined to migrate to the OSVZ (Reillo et al., 2011) (Figure 2E). In contrast, the OSVZ contains a more pure population of oRG cells that lack apical processes (Fietz et al., 2010; Hansen et al., 2010), although the morphology of these cells may also be dynamic and diverse (Betizeau et al., 2013; Gertz et al., 2014). We found that the anatomical source of radial glia was significantly associated with the position of cells along PC3 (Figure 2F, Figure S3). Indeed, many of the genes with strong positive and negative loading scores along PC3 showed differential expression patterns between radial glia collected from VZ and SVZ (Figure 2F). By clustering radial glia based on the 1% of genes most strongly loading PC3, we identified two transcriptionally distinct groups, one almost purely composed of cells from VZ that we interpreted as vRG cells and another composed of cells from both VZ and SVZ, that we interpreted as oRG cells (Figure 2G).
Predicted Markers Relate to Position, Morphology, and Behavior of oRG Cells
To relate these distinct transcriptional states to the stem cell niches of the developing neocortex, we first searched for genes likely to distinguish predicted radial glia subtypes. We measured the specificity of genes by their correlation with an ideal marker gene uniformly expressed in only one putative radial glia subpopulation across all 393 cells (Figures 3A, S3). We identified 67 candidate marker genes strongly correlated with the oRG population, 33 candidate genes strongly correlated with the vRG population, and 31 genes strongly correlated with both radial glia populations (Figure 3A, green, orange, and yellow boxes respectively, Table S3). In support of these predictions, we observed that candidate vRG markers showed higher expression in the VZ while candidate oRG markers showed higher expression in the SVZ across tissue samples collected from developing human cortex, and their expression levels were inversely correlated across radial glia cells (Figures 3B and 3C) (Miller et al., 2014).
Figure 3. Candidate oRG Markers Relate to Position, Morphology, and Behavior of Cell Type.
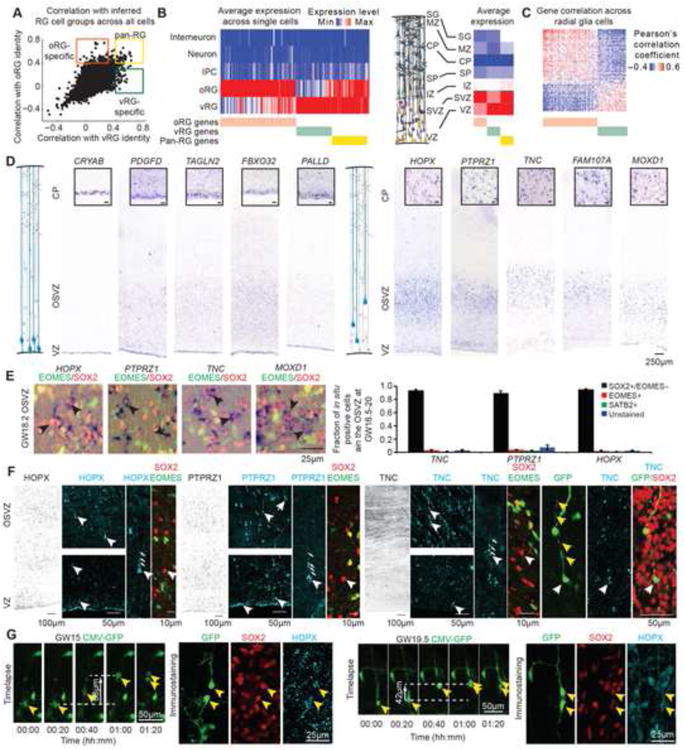
(A) Scatterplot highlighting specificity of genes to inferred radial glia subpopulations. Specificity is calculated by Pearson correlation with an idealized marker gene expressed only in candidate oRG cells (Y-axis) or candidate vRG cells (X-axis). Orange, green, and yellow boxes highlight genes with predicted specificity for oRG, vRG, and all radial glia cells (pan-RG) respectively (see also Figure S3). (B) Left heatmap showing expression of oRG, vRG, and pan-RG genes across inferred cell types and their average expression in microdissected cortical tissue samples (right heatmap) (Miller et al., 2014). (C) Similarity matrix of oRG and vRG specific gene expression levels across radial glia cells. (D) Representative examples of in situ hybridization staining for candidate vRG and oRG markers in human cortical tissue sections at GW18.2. Inset shows higher magnification of positively stained region (scale bar 25μm). (E) In situ hybridization combined with immunostaining for identity markers. Black arrows highlight cells expressing marker transcript and SOX2 but not EOMES (see also Figure S4 containing further examples). Bar chart shows quantification of molecular identity of OSVZ cells positive for oRG marker mRNA. Values represent mean ± s.e.m., n=3 biological replicates, GW16.7, GW18.2 and GW20. (F) Immunostaining of GW18 cortex for candidate oRG proteins and identity markers. White arrows highlight examples of immunopositive radial glia with staining in basal fiber. Yellow arrows indicate varicosities of the basal fiber of an oRG cell infected with adenovirus-GFP, with TNC immunostaining in the cell body. (G) Timelapse imaging of oRG cells undergoing MST and post-staining for HOPX, SOX2, and GFP. Yellow arrows highlight GFP-expressing cells. See also Figure S4.
To further investigate candidate marker genes, we performed in situ hybridization at GW17-19, stages of peak neurogenesis. We found that expression of vRG candidates, CRYAB, PDGFD, TAGLN2, FBXO32, and PALLD, was strongest in the VZ, while expression of oRG candidates, HOPX, PTPRZ1, TNC, FAM107A, and MOXD1, was strongest in the OSVZ (Figure 3D). Quantification revealed that the vast majority of cells expressing TNC, PTPRZ1, or HOPX also expressed the radial glia marker SOX2, but not the intermediate progenitor marker EOMES or neuronal marker SATB2 (Figures 3E and S4). Immunostaining revealed expression of HOPX, PTPRZ1, and TNC proteins in cells with basal fibers that lacked EOMES expression, linking this molecular identity to the typical morphology of oRG cells (Figure 3F). To next relate this molecular identity to distinctive oRG behaviors, we performed timelapse imaging of organotypic cortical slices from GW15, GW17, GW18, GW18.7, and GW19.5 specimens infected with GFP-expressing adenovirus, and then examined the expression of the most specific oRG marker, HOPX. We observed that cells undergoing mitotic somal translocation behavior can generate SOX2/HOPX double–positive daughter cells with long basal processes characteristic of oRG cells throughout neurogenesis (Figures 3G and S4). Together, these results connect the molecular identity determined from single cell RNA sequencing to the anatomical location, morphology, and behavior of oRG cells.
Specific Expression of Stemness Pathways Suggest Mechanism for Maintaining the OSVZ Stem Cell Niche
Because oRG cells lack direct access to trophic factors distributed by the cerebrospinal fluid (Lehtinen et al., 2011), we explored whether genes enriched in oRG cells relate to known functional categories related to growth factor signaling (Figure 4A). We found that protein products of many genes enriched in oRG cells mediate interactions with the extracellular matrix (Figures 4B, 4C, and 4D, Table S3). These proteins included TNC, PTPRZ1, SDC3, HS6ST1, and ITGB5, which cooperate to promote neural stem cell maturation by controlling local concentrations of fibroblast and epidermal growth factors (Barros et al., 2011; Garwood et al., 2004; Milev et al., 1998; Szklarczyk et al., 2015). Furthermore, the PTPRZ1 ligand PTN was expressed in both radial glia populations, but was the most significantly up-regulated gene in oRG cells (Table S4). We confirmed by immunostaining that TNC and PTPRZ1 are expressed in a subset of ITGB5 positive oRG cells (Figure 4E). Many of the cell surface proteins enriched in oRG cells are also highly overexpressed in glioblastomas compared with normal human astrocytes including TNC, PTPRZ1, and LGALS3BP (Autelitano et al., 2014; Nie et al., 2015), and PTN stimulation of PTPRZ1 is sufficient to stimulate the coordinated processes of epithelial-to-mesenchymal transition in a glioblastoma cell line (Perez-Pinera et al., 2006). Therefore, we investigated whether human glioblastoma samples contain cell populations that co-express oRG markers. In a recent study of five primary glioblastoma samples, PTN, PTPRZ1, and FABP7 were the genes most correlated with a stemness signature across single tumor cells (Patel et al., 2014). We extended this analysis to all predicted oRG and vRG markers (Figure 4F) and found that oRG markers were more highly correlated with both the stemness and general radial glia signatures than vRG markers (Figure 4G), suggesting that oRG-enriched genes are associated with stemness pathways and growth factor signaling in both the developing OSVZ stem cell niche and in primary glioblastoma.
Figure 4. Pathways Enriched in oRG Cells Relate to Growth Factor Signaling and Maintenance of Proliferation.
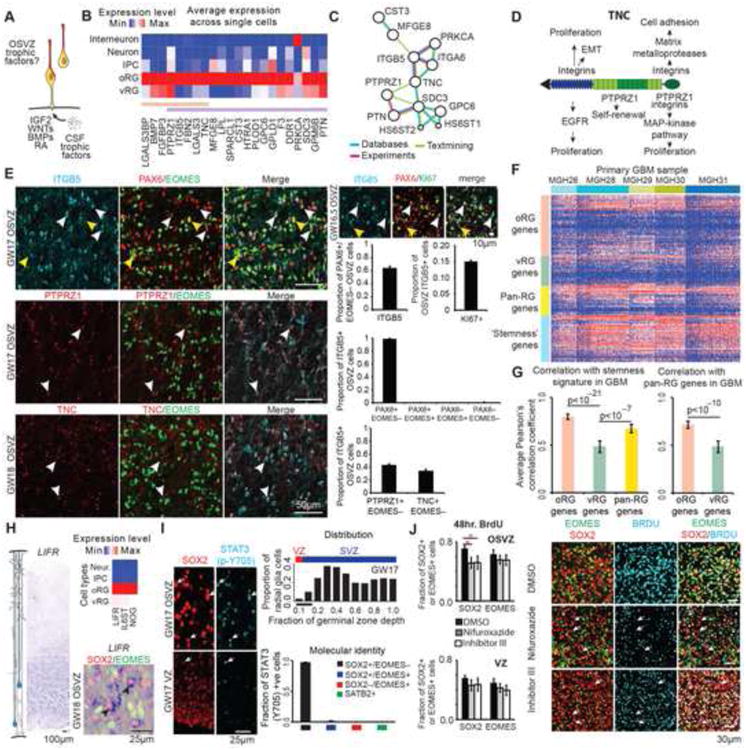
(A) Schema – oRG cells lack apical processes that transduce signals mediated by trophic factors in ventricular cerebrospinal fluid (CSF). (B) Heatmap of extracellular matrix gene expression across inferred cell types. Orange bar highlights genes with high oRG specificity across all cells and purple bar highlights genes differentially expressed by DESeq. (C) Functional protein association network generated using String-db (Szklarczyk et al., 2015). (D) Model of TNC protein, interaction partners and downstream pathways. (E) Immunolabeling of human OSVZ tissue sections reveals expression of ITGB5 by radial glia, including proliferating KI67+ and KI67-(top row, examples indicated with arrows) but not EOMES+ intermediate progenitors (yellow arrowheads denote PAX6+,EOMES+ intermediate progenitor cells, which do not express ITGB5, see also Figure S2). Top right panels show immunolabelling for ITGB5 and proliferation marker KI67 (white arrows indicate proliferating radial glia expressing ITGB5 and yellow arrow indicates non-actively proliferating radial glia expressing ITGB5). Bottom left rows show expression of TNC and PTPRZ1 in a subset of ITGB5+ radial glia (examples indicated with arrowheads). Bar charts represent quantification (mean ± s.e.m) across three biological replicates between GW16.5 and GW18. (F) Heatmap of oRG, vRG, pan-RG, and glioblasoma multiforme (GBM) stemness gene expression signatures across 598 cells from five primary GBM tumors (Patel et al., 2014). (G) Bar graphs show the oRG signature has the strongest correlation with the GBM stemness signature and with the pan-RG signature across GBM tumor samples. p-values report significance of difference between standardized correlation coefficients, error bars represent 95% confidence interval. (H) Heatmap showing average expression of selected genes across inferred cell types, and validation of LIFR expression in oRG cells by in situ hybridization (examples indicated by arrows). (I) Immunostaining of human tissue section for phosphorylated STAT3 (p-Y705) and SOX2 (see also Figure S5). Top bar graph represents quantification of nuclear immunostaining for phosphorylated STAT3 across germinal zone depth starting from ventricular edge (0.0) to the basal edge of the OSVZ (1.0). Bottom graph represents the molecular identity of STAT3+ Y705+ cells quantified in the OSVZ of three biological replicates between GW16.5 and GW18. Data represent mean ± s.e.m.. (J) Inhibition of STAT3 phosphorylation in organotypic OSVZ slice cultures reduces BrdU incorporation by radial glia. Images show representative examples of immunostained experimental sections. Bar graphs display quantification of BrdU incorporation by radial glia and intermediate progenitors in presence of STAT3 phosphorylation inhibitors or control DMSO (n=4, * - p<0.05, paired Student's t-test, error bars represent standard error of mean). Arrows indicate examples of cells expressing SOX2, but not EOMES or SATB2 that did not incorporate BrdU over 48 hour period in inhibitor treated slices.
We next explored whether additional signaling pathways were up-regulated in oRG cells. We found that LIFR and its co-receptor IL6ST (GP130) were specifically expressed in oRG cells (Figures 4H and 4I). LIF signaling through LIFR and IL6ST phosphorylates STAT3 at tyrosine 705 (p-Y705) to promote stem cell maintenance (Huang et al., 2014). Immunostaining revealed that p-Y705-STAT3 was specifically localized at the nuclei of oRG cells and not detected in other cell types (Figures 4I and S5). To investigate the function of STAT3 in oRG cells, we pharmacologically blocked STAT3 phosphorylation in human slice cultures. After two days, we observed a reduction in the proportion of oRG cells that incorporated BrdU (Figure 4J), consistent with the proposed role of phosphorylated STAT3 in neural stem cell maintenance (Hong and Song, 2015; Huang et al., 2014). In addition, we found that expression of constitutively–active Stat3 in developing mouse cortex increased the proportion of electroporated cells expressing Sox2, but not Eomes, compared with expression of only the fluorescent reporter (Figure S5). Together, these results support a role for STAT3 (p-Y705) in maintaining stemness of human oRG cells.
Extensive Neurogenic Capacity of oRG Cells
We next examined the differentiation potential and proliferative capacity of human oRG cells (Figure 5A). Many oRG-enriched genes are also associated with astrocytes later in development, including TNC, ITGB5, DIO2, and ACSBG1 (Cahoy et al., 2008), and LIFR signaling through STAT3 can promote gliogenesis in combination with BMP signaling (Bonaguidi et al., 2005). However, oRG cells did not express other astrocyte markers such as AQP4, CA2, IL33, and ALDOC, which we observed in putative astrocytes later in development (Figure S5). In addition, oRG cells strongly expressed NOG (Figure 4H), which inhibits BMP signaling and promotes neurogenesis in GFAP-expressing progenitors (Bonaguidi et al., 2005; Bonaguidi et al., 2008; Lim et al., 2000). We further noted that many genes up-regulated in oRG cells relative to vRG cells were also strongly expressed by cortical neurons such as NPY, RTN1, CTNND2, SEZ6L, and NRCAM (Figures 5B, S6), suggesting a relationship between neurons and oRG cells.
Figure 5. Extensive Proliferative and Neurogenic Capacity of oRG Cells.
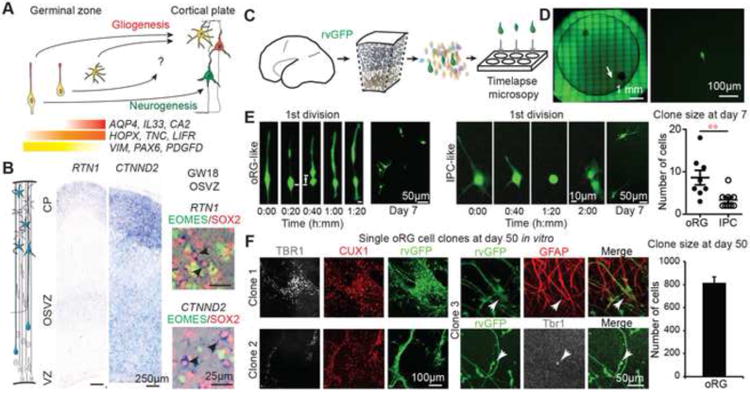
(A) Schema representing progenitor cell competence of radial glia, oRG cells and glial progenitors (See also Figure S5). (B) In situ hybridization for genes expressed by oRG and other cell types but depleted in vRG cells and immunolabelling of GW18 human cortical sections (see also Figure S6). Black arrows indicate examples of mRNA expressing oRG cells. (C) Experimental design for single cell clonal lineage analysis of oRG cells labeled with retrovirus GFP (rvGFP). (D) Image and magnification of a well containing a single purified GFP-positive cell. Timelapse imaging of single cell with arrow highlighting MST preceding first division. (E) Images show movie frames capturing the initial division and a resulting clone after 7 days in culture of a cell exhibiting oRG specific mitotic somal translocation (left) and a cell undergoing an initial stationary division with fiber retraction characteristic of intermediate progenitors and a resulting clone after 7 days in culture (right). Chart shows clone sizes at 7 days for cells classified as oRGs and IPCs based on first division and morphology. (F) Immunostaining for neuronal or glial markers of 3 oRG cell clones after 50 days in vitro. Bar chart represents quantification of the average clone size of oRG cells. See also Figure S6.
To further examine the neurogenic potential of oRG cells, we isolated single cells from the germinal zone by fluorescence activated cell sorting and cultured them on a feeder cell layer (Figure 5C and 5D). Using timelapse imaging, we followed individual cells for one week in vitro. Cells that displayed the distinctive somal translocation directly preceding the first division were classified as oRGs, while cells that retracted processes and underwent stationary first division were classified as intermediate progenitors. Consistent with their stem-like character, single oRG cells gave rise to larger clones than intermediate progenitor cells (Figure 5E). We followed three oRG clones in vitro for an additional six weeks and they generated hundreds of daughter cells, including deep and upper layer neurons, as well as glial cells (Figures 5F and S6). Thus, human oRG cells express self-renewal pathways not detected in vRG cells during peak upper cortical layer neurogenesis, generate diverse neural daughter cells, and have the capacity for extensive proliferation.
oRG Signature Emerges in VZ and is Conserved in Primates
To further investigate the developmental and evolutionary origin of the transcriptional signature of oRG cells, we examined the expression of oRG marker genes across different stages of human corticogenesis and in different species. Surprisingly, we observed strong expression of transcripts encoding oRG marker genes in the VZ at early stages of human cortical development. This expression became progressively restricted to the OSVZ at later stages, with FAM107A persisting the longest in the VZ (Figures 6A, 6B, S7). TNC, FAM107A, and PTPRZ1 protein expression followed a similar developmental progression, but TNC and PTPRZ1 antibodies only labeled subsets of radial glia cells in the VZ (Figure 6C). The coordinated expression of these genes in the VZ around GW13.5 coincides with the elaboration of the OSVZ (Bayatti et al., 2008; Martinez-Cerdeno et al., 2012; Shitamukai et al., 2011; Zecevic et al., 2005) and may represent a molecular program involved in oRG specification.
Figure 6. Molecular Signature of oRG Cells Emerges in VZ.
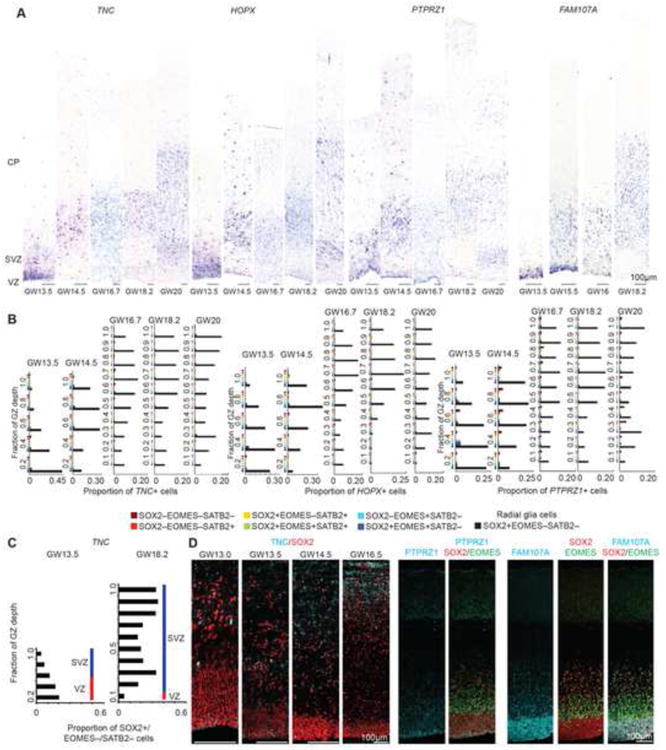
(A) In situ hybridization for genes enriched in oRG cells across multiple developmental time points corresponding to human cortical neurogenesis. Sections were stained with antibodies against SOX2, EOMES and SATB2 to identify major cell populations (see also Figure S4 and S7). (B) The proportion of oRG marker cells that express all combinations of protein markers was quantified across 10 bins that span the germinal zone throughout neurogenesis. In all cases, oRG marker transcripts are almost exclusively expressed by SOX2+/EOMES–/SATB2– cells (black bars). Bar graphs highlight that at GW13.5 oRG markers predominantly label radial glia in the VZ, but by GW14.5 oRG markers predominantly label radial glia cells in the SVZ. (C) The proportion of SOX2+/EOMES–/SATB2– that expressed TNC was quantified across the span of the germinal zone at GW13.5 and GW18. (D) Immunolabelling of early developmental time points for oRG markers and SOX2. At GW13.5, TNC and PTPRZ1 protein show limited expression in radial glia close to the ventricular edge, while FAM107A protein is widely expressed in GW13.5 VZ. See also Figure S7.
During mouse development oRG cells are rare and there is not a distinct OSVZ. To investigate whether human oRG markers are expressed in mouse radial glia, we used recently published gene coexpression networks (Lui et al., 2014) and found that on average, human oRG markers are less likely to show specific expression in mouse radial glia than general radial glia markers (Figure 7A). Nonetheless, TNC expression has previously been detected in radial glia of the mouse ventral pallium (Garcion et al., 2004; Gotz et al., 1998; Wiese et al., 2012). Immunostaining for TNC and PTPRZ1 revealed that both proteins are most strongly detected in the VZ and SVZ of the lateral and ventral pallium (Figure 7B), where mouse oRG cells are most common (Wang et al., 2011b). Closer examination of SOX2–positive cells in the SVZ revealed examples of putative mouse oRG cells expressing TNC and PTPRZ1 (Figure 7B, insets). More widespread transcription of TNC, PTPRZ1, and HOPX throughout the mouse cortical VZ coincides with the conclusion of cortical neurogenesis (Figure S7). Thus conserved elements of the oRG signature may reflect regional and temporal heterogeneity of mouse radial glia, but many genes enriched in human oRG cells are not expressed in mouse radial glia, including MOXD1 (Wang et al., 2011a), FAM107A, FBN2, BMP7, HS6ST2, LGALS3, and TKTL1 (Figure S7). In contrast to mouse, the developing macaque cortex contains a large OSVZ region and prominent oRG population (Betizeau et al., 2013; Smart et al., 2002). Using microarray data from developing macaque cortex, we found that the expression of oRG marker genes in macaque development mirrors that of human development (Figure 7C). We confirmed this pattern for select oRG marker genes by analyzing primary tissue samples. We detected expression of TNC, PTPRZ1, and HOPX in macaque VZ early in development, but found that OSVZ expression of these markers, along with FAM107A, predominates at later stages of corticogenesis (Figure 7D). Together, our data indicate that major elements of the transcriptional signature of human oRG cells are conserved in primates.
Figure 7. Conservation of oRG Marker Gene Expression in Primates.
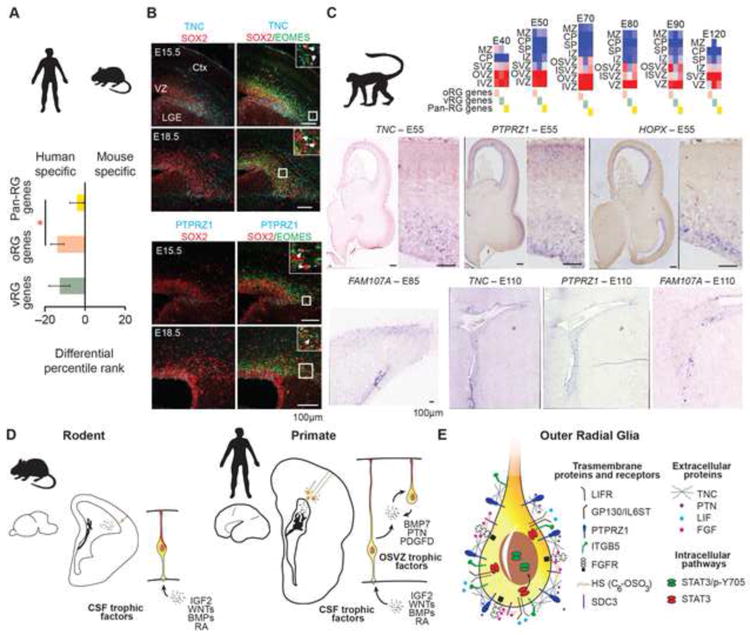
(A) Bar graph indicates average differential percentile rank of pan-RG, oRG, and vRG genes for radial glia gene coexpression network specificity in human and mouse. Compared with pan-RG genes (n=29), human oRG genes (n=66) show reduced conservation with mouse radial glia signature (p < 0.05, Wilcoxon rank sum test). (B) Mouse E15.5 and E18.5 cortical sections immunoreacted for Tnc and Ptprz1 along with Sox2 and Eomes. Inset images show a magnified view of SVZ region and arrows highlight examples of Sox2–positive, Eomes–negative cells that co-label for Tnc or Ptprz1. LGE- lateral ganglionic eminence, Ctx-cortex. (C) Heatmaps show average expression level of oRG, vRG and pan-RG genes in distinct regions of developing macaque cortex. IVZ – inner VZ, OVZ – outer VZ; The NIH Blueprint Non-Human Primate (NHP) Atlas. In situ hybridization of macaque cortex showing expression of oRG marker genes mirrors human trajectory. (D) Radial glia in small rodent brains are concentrated along the ventricle and access cerebrospinal fluid trophic factors directly via apical processes. In contrast, large primate brains contain numerous radial glia in the OSVZ. Local production of growth factors by radial glia may provide additional trophic support to oRG cells in the OSVZ niche. (E) Increased expression of extracellular matrix proteins that potentiate growth factor signaling and activation of LIFR/STAT3 pathway may further maintain stemness in oRG cells. See also Figure S7.
Discussion
Our study identifies neuronal differentiation, cell cycle progression, and anatomical position as major sources of transcriptional variation across single cells sampled from germinal niches of the developing human cortex. The transcriptional state associated with neuronal differentiation involves reduced expression of classical radial glia markers such as VIM and HES1, and up-regulation of proneural transcription factors such as NEUROG1, NEUROD4, and EOMES, and neuropeptide signaling genes PENK, SSTR2, and OXTR. This transcriptional state was recently attributed to heterogeneity among oRG cells (Johnson et al., 2015). However, based on expression of mRNA, EOMES protein, and the novel marker PPP1R17, which reveals diverse multipolar morphologies, we interpret this transcriptional state to represent intermediate progenitor cells.
In contrast, we identify a novel transcriptional state, independent of neuronal differentiation, that distinguishes oRG from vRG cells by analyzing the major sources of variation among classically-defined radial glia. We relate this transcriptional state to the position, morphology, and dynamic behavior of oRG cells. Together, this multimodal characterization establishes an integrative identity for oRG cells. These neural stem cells are characterized by the expression of novel markers including HOPX, TNC, ITGB5, as well as pan-radial glia markers such as VIM, HES1, and ATP1A2; the presence of a basal, but not apical fiber; mitotic-somal translocation behavior; and extensive proliferative and neurogenic capacity. This cell type is most abundant in the OSVZ stem cell niche, for which it was named, but also resides in the inner SVZ and VZ, and the transcriptional state first emerges in the VZ during early cortical neurogenesis. The oRG marker genes may enable the construction of molecular tools for selectively visualizing, manipulating, or purifying oRG cells in tissue and for evaluating the identity of human cortical progenitor cells generated from pluripotent stem cells. In addition, these genes may provide insights into the cell types affected in neurodevelopmental disorders.
Beyond simply marking oRG cells, the genes we identify belong to common pathways that suggest mechanisms by which human oRG cells actively maintain the OSVZ as a neural stem cell niche. Many of these genes promote growth factor signaling including TNC, PTPRZ1, ITGB5, SDC3, HS6ST1, IL6ST, and LIFR (Sim et al., 2006; Wiese et al., 2012). For example, TNC potentiates FGF signaling to support the maturation of neural stem cells (Garcion et al., 2004), while integrin signaling along the basal fiber promotes radial glia identity (Fietz et al., 2010). Interestingly, TNC contains EGF-like repeats, and multiple binding domains for PTPRZ1, syndecans, integrins, and other cell surface receptors (Besser et al., 2012; von Holst, 2008). Thus, TNC expression in oRG cells may couple key protein networks regulating growth factor signaling, migration, and self-renewal. In addition, LIFR/STAT3 signaling is known to maintain radial glia neural stem cell identity (Bonaguidi et al., 2005), and we show that p-Y705-STAT3 signaling is necessary for normal cell cycle progression in oRG cells, but is surprisingly absent in vRG cells. Although STAT3 signaling can also contribute to gliogenesis, we speculate that expression of NOG may inhibit gliogenesis as observed in the rodent adult neurogenic germinal zones (Bonaguidi et al., 2008; Lim et al., 2000). We directly examined the neural stem cell properties of oRG cells using single cell clonal analysis. We find that single oRG cells at mid-neurogenesis can generate clones of nearly 1000 neuronal and glial daughter cells. This highlights the remarkable proliferative capacity of human oRG cells compared to mouse radial glia that typically generate 10-100 daughter cells throughout the neurogenic period (Gao et al., 2014; Qian et al., 2000; Vasistha et al., 2014).
The cell behaviors that distinguish oRG from vRG cells – loss of adhesion, delamination, and rapid migratory bursts preceeding cell division – have been compared to epithelial-to-mesenchymal transition (Itoh et al., 2013). Recent work suggests that oRG and glioma cells both undergo myosin II-dependent migratory movements (Beadle et al., 2008; Ostrem et al., 2014). Interestingly, many of the genes and proteins we detected in oRG cells have been implicated in invasive migratory behavior, including genes expressed in the VZ when oRG cells first emerge. For example, TNC, ITGB5, and PTN/PTPRZ1 signaling promote multiple aspects of epithelial-to-mesenchymal transition (Bianchi et al., 2010; Katoh et al., 2013; Perez-Pinera et al., 2006), PRKCA is necessary for the up-regulation of SNAI1 and down-regulation of CLDN1 during these transitions (Kyuno et al., 2013), and FAM107A establishes focal adhesions and increases glioma invasiveness (Le et al., 2010). The expression of these genes suggests possible mechanisms by which oRG cells emerge from the VZ and undergo mitotic somal translocation. More generally, we found the oRG transcriptional signature to be enriched in cells from primary glioblastoma and conserved in macaque suggesting that the development of invasive glioblastoma and the evolutionary expansion of the OSVZ may recruit common sets of genes controlling migration and self-renewal.
Sequencing of single cell mRNA while retaining cell position information provides a general method for identifying distinct subpopulations whose molecular identity may relate to microenvironment. Here, we explored variation in radial glia gene expression while considering stem cell niche as a covariate. Our results revealed novel molecular features of neural stem cell populations previously distinguished only by cell behavior, morphology, and position. Together with recent findings (Fietz et al., 2012; Lui et al., 2014), these results highlight three mechanisms that may maintain stemness of the expanded oRG population in the OSVZ stem cell niche: local production of trophic factors such as PTN and BMP7 by radial glia, expression of extracellular matrix proteins that potentiate growth factor signaling, and activation of the LIFR/p-STAT3 signaling pathway (Figures 7D and 7E). Because the oRG population is thought to be responsible for the majority of human cortical neurogenesis, and OSVZ size correlates with the evolutionary expansion of the brain, future studies can investigate the role of these genes in neurodevelopmental disorders and cortical evolution.
Experimental Procedures
Single Cell Analysis
Micro-dissected VZ and SVZ tissue samples were dissociated using Papain (Worthington). Single cell sequencing libraries were generated using the C1™ Single-Cell Auto Prep Integrated Fluidic Circuit (Fluidigm), the SMARTer® Ultra Low RNA Kit (Clontech), and the Nextera® XT DNA Sample Preparation Kit (Illumina) and deposited (dbGaP: phs000989.v1.p1). Reads were aligned using Tophat2 and the expression of RefSeq genes was quantified by the featureCounts routine. Gene expression values were normalized based on library size as counts per million reads (CPM). Libraries with fewer than 1000 genes detected above 1 CPM were eliminated as outliers. Cells were assigned to groups using PCA and Expectation-Maximization clustering and groups were interpreted based on the expression of known marker genes and tissue validation. The specificity of genes to each group was determined using the Pearson's correlation, and confirmed with DESeq2. The expression of cell type markers was evaluated in silico using the Allen Institute Prenatal LMD Microarray Atlas (Miller et al., 2014) and NIH Blueprint NHP Atlas, as well as human and mouse gene coexpression networks (Lui et al., 2014) and single cell glioblastoma data (Patel et al., 2014), and in tissue using immunohistochemistry and in situ hybridization as described in Supplemental Experimental Procedures.
STAT3 Signaling
To examine the function of decreased STAT3 signaling in oRG cells, we cultured fetal cortical slices for 48hr. in the presence of inhibitors and then performed immunostaining. In utero electroporation was performed at E13.5 of a mutated form of STAT3, which mimics the Y705 phosphorylation state driven by the EF1a promoter (Addgene, 24983). Timed-pregnant Swiss-Webster mice were obtained from Simonsen Laboratories and maintained according to protocols approved by the UCSF Institutional Animal Care and Use Committee.
Single Cell Clonal Analysis
Cells dissociated from cortical germinal zone were infected cells with a pNIT-GFP retrovirus for 2-4 hours, cultured on matrigel (BD Biosciences) for 3 days in media containing DMEM (Invitrogen, 11965), 1% B-27 supplement (Invitrogen, 12587-010), 1% N-2 supplement (17502-048), and recombinant human FGF-basic (10ng/ml, Peprotech, AF-100-18B). We then used FACS (ARIA, BD Biosciences) to sort GFP-expressing cells at 1 cell/well into 96 well plates pre-seeded with feeder cells. We used time lapse microscopy to identify the mitotic behavior of the initial cell divisions for each clone. After one week, the cells were cultured for six weeks in media without FGF.
Supplementary Material
Acknowledgments
We thank Stephen Noctor, Marc Unger, Bob Jones, Beatriz Alvarado, Tricia Sun, Mercedes Paredes, Becky Anderson, Raphael Kohler, Jerry Wang, Shaohui Wang, Yingying Wang, and Simone Mayer for providing resources, suggestions and technical assistance, and the staff at the San Francisco General Hospital for providing access to donated tissue samples. A.A.P. is supported by a Damon Runyon Cancer Research Foundation postdoctoral fellowship (DRG-2166-13). A.D. is partially supported by UCSF-CTSI UL1 TR000004. This research was supported by NIH awards U01 MH105989 and R01NS075998 to ARK.
Footnotes
A.A.L., J.A.W., and J.S. have a financial interest in Fluidigm Corporation as employees and/or stockholders.
Author contributions: A.A.P., T.J.N. and A.R.K. conceived and supervised the project. A.A.P., T.J.N., J.S., A.A.L, and J.A.W. performed single cell RNA-Seq experiments. J.C. and C.R.N. designed and performed single cell clonal lineage experiments. T.J.N., A.A.P., and H.R. designed and performed tissue validation and functional experiments with input from A.R.K., and D.A.L.. Bioinformatic analysis was performed by A.A.P., T.J.N. H.R., C.S., M.C.O., S.J.L., and A.D.. Manuscript was prepared by A.A.P. and T.J.N. with input from all authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
The NIH Blueprint Non-Human Primate (NHP) Atlas, NIH Contract HHSN-271-2008-00047-C to the Allen Institute for Brain Science (Seattle, WA).
- Autelitano F, Loyaux D, Roudieres S, Deon C, Guette F, Fabre P, Ping Q, Wang S, Auvergne R, Badarinarayana V, et al. Identification of novel tumor-associated cell surface sialoglycoproteins in human glioblastoma tumors using quantitative proteomics. PloS one. 2014;9:e110316. doi: 10.1371/journal.pone.0110316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros CS, Franco SJ, Muller U. Extracellular matrix: functions in the nervous system. Cold Spring Harbor perspectives in biology. 2011;3:a005108. doi: 10.1101/cshperspect.a005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayatti N, Moss JA, Sun L, Ambrose P, Ward JF, Lindsay S, Clowry GJ. A molecular neuroanatomical study of the developing human neocortex from 8 to 17 postconceptional weeks revealing the early differentiation of the subplate and subventricular zone. Cerebral cortex. 2008;18:1536–1548. doi: 10.1093/cercor/bhm184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle C, Assanah MC, Monzo P, Vallee R, Rosenfeld SS, Canoll P. The role of myosin II in glioma invasion of the brain. Molecular biology of the cell. 2008;19:3357–3368. doi: 10.1091/mbc.E08-03-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser M, Jagatheaswaran M, Reinhard J, Schaffelke P, Faissner A. Tenascin C regulates proliferation and differentiation processes during embryonic retinogenesis and modulates the de-differentiation capacity of Muller glia by influencing growth factor responsiveness and the extracellular matrix compartment. Developmental biology. 2012;369:163–176. doi: 10.1016/j.ydbio.2012.05.020. [DOI] [PubMed] [Google Scholar]
- Betizeau M, Cortay V, Patti D, Pfister S, Gautier E, Bellemin-Menard A, Afanassieff M, Huissoud C, Douglas RJ, Kennedy H, et al. Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron. 2013;80:442–457. doi: 10.1016/j.neuron.2013.09.032. [DOI] [PubMed] [Google Scholar]
- Bianchi A, Gervasi ME, Bakin A. Role of beta5-integrin in epithelial-mesenchymal transition in response to TGF-beta. Cell cycle. 2010;9:1647–1659. doi: 10.4161/cc.9.8.11517. [DOI] [PubMed] [Google Scholar]
- Bonaguidi MA, McGuire T, Hu M, Kan L, Samanta J, Kessler JA. LIF and BMP signaling generate separate and discrete types of GFAP-expressing cells. Development. 2005;132:5503–5514. doi: 10.1242/dev.02166. [DOI] [PubMed] [Google Scholar]
- Bonaguidi MA, Peng CY, McGuire T, Falciglia G, Gobeske KT, Czeisler C, Kessler JA. Noggin expands neural stem cells in the adult hippocampus. The Journal of neuroscience. 2008;28:9194–9204. doi: 10.1523/JNEUROSCI.3314-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. The Journal of neuroscience. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fietz SA, Kelava I, Vogt J, Wilsch-Brauninger M, Stenzel D, Fish JL, Corbeil D, Riehn A, Distler W, Nitsch R, et al. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nature neuroscience. 2010;13:690–699. doi: 10.1038/nn.2553. [DOI] [PubMed] [Google Scholar]
- Fietz SA, Lachmann R, Brandl H, Kircher M, Samusik N, Schroder R, Lakshmanaperumal N, Henry I, Vogt J, Riehn A, et al. Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proceedings of the National Academy of Sciences. 2012;109:11836–11841. doi: 10.1073/pnas.1209647109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florio M, Albert M, Taverna E, Namba T, Brandl H, Lewitus E, Haffner C, Sykes A, Wong FK, Peters J, et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science. 2015;347:1465–1470. doi: 10.1126/science.aaa1975. [DOI] [PubMed] [Google Scholar]
- Gao P, Postiglione MP, Krieger TG, Hernandez L, Wang C, Han Z, Streicher C, Papusheva E, Insolera R, Chugh K, et al. Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell. 2014;159:775–788. doi: 10.1016/j.cell.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcion E, Halilagic A, Faissner A, ffrench-Constant C. Generation of an environmental niche for neural stem cell development by the extracellular matrix molecule tenascin C. Development. 2004;131:3423–3432. doi: 10.1242/dev.01202. [DOI] [PubMed] [Google Scholar]
- Garwood J, Garcion E, Dobbertin A, Heck N, Calco V, ffrench-Constant C, Faissner A. The extracellular matrix glycoprotein Tenascin-C is expressed by oligodendrocyte precursor cells and required for the regulation of maturation rate, survival and responsiveness to platelet-derived growth factor. The European journal of neuroscience. 2004;20:2524–2540. doi: 10.1111/j.1460-9568.2004.03727.x. [DOI] [PubMed] [Google Scholar]
- Gertz CC, Lui JH, LaMonica BE, Wang X, Kriegstein AR. Diverse behaviors of outer radial glia in developing ferret and human cortex. The Journal of neuroscience. 2014;34:2559–2570. doi: 10.1523/JNEUROSCI.2645-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz M, Stoykova A, Gruss P. Pax6 controls radial glia differentiation in the cerebral cortex. Neuron. 1998;21:1031–1044. doi: 10.1016/s0896-6273(00)80621-2. [DOI] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Hodge RD, Daza RA, Englund C. Transcription factors in glutamatergic neurogenesis: conserved programs in neocortex, cerebellum, and adult hippocampus. Neuroscience research. 2006;55:223–233. doi: 10.1016/j.neures.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Hong S, Song MR. Signal transducer and activator of transcription-3 maintains the stemness of radial glia at mid-neurogenesis. The Journal of neuroscience. 2015;35:1011–1023. doi: 10.1523/JNEUROSCI.2119-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Yan H, Ye S, Tong C, Ying QL. STAT3 phosphorylation at tyrosine 705 and serine 727 differentially regulates mouse ESC fates. Stem cells. 2014;32:1149–1160. doi: 10.1002/stem.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Moriyama Y, Hasegawa T, Endo TA, Toyoda T, Gotoh Y. Scratch regulates neuronal migration onset via an epithelial-mesenchymal transition-like mechanism. Nature neuroscience. 2013;16:416–425. doi: 10.1038/nn.3336. [DOI] [PubMed] [Google Scholar]
- Johnson MB, Wang PP, Atabay KD, Murphy EA, Doan RN, Hecht JL, Walsh CA. Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nature neuroscience. 2015;18:637–646. doi: 10.1038/nn.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh D, Nagaharu K, Shimojo N, Hanamura N, Yamashita M, Kozuka Y, Imanaka-Yoshida K, Yoshida T. Binding of alphavbeta1 and alphavbeta6 integrins to tenascin-C induces epithelial-mesenchymal transition-like change of breast cancer cells. Oncogenesis. 2013;2:e65. doi: 10.1038/oncsis.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi A, Ikawa T, Kasukawa T, Ueda HR, Kurimoto K, Saitou M, Matsuzaki F. Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development. 2008;135:3113–3124. doi: 10.1242/dev.022616. [DOI] [PubMed] [Google Scholar]
- Kyuno D, Kojima T, Yamaguchi H, Ito T, Kimura Y, Imamura M, Takasawa A, Murata M, Tanaka S, Hirata K, et al. Protein kinase Calpha inhibitor protects against downregulation of claudin-1 during epithelial-mesenchymal transition of pancreatic cancer. Carcinogenesis. 2013;34:1232–1243. doi: 10.1093/carcin/bgt057. [DOI] [PubMed] [Google Scholar]
- Le PU, Angers-Loustau A, de Oliveira RM, Ajlan A, Brassard CL, Dudley A, Brent H, Siu V, Trinh G, Molenkamp G, et al. DRR drives brain cancer invasion by regulating cytoskeletal-focal adhesion dynamics. Oncogene. 2010;29:4636–4647. doi: 10.1038/onc.2010.216. [DOI] [PubMed] [Google Scholar]
- Lehtinen MK, Zappaterra MW, Chen X, Yang YJ, Hill AD, Lun M, Maynard T, Gonzalez D, Kim S, Ye P, et al. The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron. 2011;69:893–905. doi: 10.1016/j.neuron.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewitus E, Kelava I, Huttner WB. Conical expansion of the outer subventricular zone and the role of neocortical folding in evolution and development. Frontiers in human neuroscience. 2013;7:424. doi: 10.3389/fnhum.2013.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA, Tramontin AD, Trevejo JM, Herrera DG, Garcia-Verdugo JM, Alvarez-Buylla A. Noggin antagonizes BMP signaling to create a niche for adult neurogenesis. Neuron. 2000;28:713–726. doi: 10.1016/s0896-6273(00)00148-3. [DOI] [PubMed] [Google Scholar]
- Lui JH, Nowakowski TJ, Pollen AA, Javaherian A, Kriegstein AR, Oldham MC. Radial glia require PDGFD-PDGFRbeta signalling in human but not mouse neocortex. Nature. 2014;515:264–268. doi: 10.1038/nature13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Cerdeno V, Cunningham CL, Camacho J, Antczak JL, Prakash AN, Cziep ME, Walker AI, Noctor SC. Comparative analysis of the subventricular zone in rat, ferret and macaque: evidence for an outer subventricular zone in rodents. PloS one. 2012;7:e30178. doi: 10.1371/journal.pone.0030178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milev P, Monnerie H, Popp S, Margolis RK, Margolis RU. The core protein of the chondroitin sulfate proteoglycan phosphacan is a high-affinity ligand of fibroblast growth factor-2 and potentiates its mitogenic activity. The Journal of biological chemistry. 1998;273:21439–21442. doi: 10.1074/jbc.273.34.21439. [DOI] [PubMed] [Google Scholar]
- Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie S, Gurrea M, Zhu J, Thakolwiboon S, Heth JA, Muraszko KM, Fan X, Lubman DM. Tenascin-C: a novel candidate marker for cancer stem cells in glioblastoma identified by tissue microarrays. Journal of proteome research. 2015;14:814–822. doi: 10.1021/pr5008653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrem BE, Lui JH, Gertz CC, Kriegstein AR. Control of outer radial glial stem cell mitosis in the human brain. Cell reports. 2014;8:656–664. doi: 10.1016/j.celrep.2014.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P, Alcantara S, Dimitrov T, Vega JA, Deuel TF. Pleiotrophin disrupts calcium-dependent homophilic cell-cell adhesion and initiates an epithelial-mesenchymal transition. Proceedings of the National Academy of Sciences. 2006;103:17795–17800. doi: 10.1073/pnas.0607299103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Shuga J, Wang X, Leyrat AA, Lui JH, Li N, Szpankowski L, Fowler B, Chen P, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nature biotechnology. 2014;32:1053–1058. doi: 10.1038/nbt.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Shen Q, Goderie SK, He W, Capela A, Davis AA, Temple S. Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron. 2000;28:69–80. doi: 10.1016/s0896-6273(00)00086-6. [DOI] [PubMed] [Google Scholar]
- Reillo I, de Juan Romero C, Garcia-Cabezas MA, Borrell V. A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cerebral cortex. 2011;21:1674–1694. doi: 10.1093/cercor/bhq238. [DOI] [PubMed] [Google Scholar]
- Shitamukai A, Konno D, Matsuzaki F. Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. The Journal of neuroscience. 2011:3683–3695. doi: 10.1523/JNEUROSCI.4773-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim FJ, Lang JK, Waldau B, Roy NS, Schwartz TE, Pilcher WH, Chandross KJ, Natesan S, Merrill JE, Goldman SA. Complementary patterns of gene expression by human oligodendrocyte progenitors and their environment predict determinants of progenitor maintenance and differentiation. Annals of neurology. 2006;59:763–779. doi: 10.1002/ana.20812. [DOI] [PubMed] [Google Scholar]
- Smart IH, Dehay C, Giroud P, Berland M, Kennedy H. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cerebral cortex. 2002;12:37–53. doi: 10.1093/cercor/12.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic acids research. 2015;43:D447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasistha NA, Garcia-Moreno F, Arora S, Cheung AF, Arnold SJ, Robertson EJ, Molnar Z. Cortical and Clonal Contribution of Tbr2 Expressing Progenitors in the Developing Mouse Brain. Cerebral cortex. 2014 doi: 10.1093/cercor/bhu125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Holst A. Tenascin C in stem cell niches: redundant, permissive or instructive? Cells, tissues, organs. 2008;188:170–177. doi: 10.1159/000112848. [DOI] [PubMed] [Google Scholar]
- Wang WZ, Oeschger FM, Montiel JF, Garcia-Moreno F, Hoerder-Suabedissen A, Krubitzer L, Ek CJ, Saunders NR, Reim K, Villalon A, et al. Comparative aspects of subplate zone studied with gene expression in sauropsids and mammals. Cerebral cortex. 2011a;21:2187–2203. doi: 10.1093/cercor/bhq278. [DOI] [PubMed] [Google Scholar]
- Wang X, Tsai JW, LaMonica B, Kriegstein AR. A new subtype of progenitor cell in the mouse embryonic neocortex. Nature neuroscience. 2011b;14:555–561. doi: 10.1038/nn.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese S, Karus M, Faissner A. Astrocytes as a source for extracellular matrix molecules and cytokines. Frontiers in pharmacology. 2012;3:120. doi: 10.3389/fphar.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecevic N, Chen Y, Filipovic R. Contributions of cortical subventricular zone to the development of the human cerebral cortex. The Journal of comparative neurology. 2005;491:109–122. doi: 10.1002/cne.20714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


