Abstract
In multiple myeloma, a long non-coding RNA, KIAA0495 (alias PDAM/TP73-AS1), had been found progressively downregulated from normal plasma cell to benign monoclonal gammopathy of undetermined significance to symptomatic myeloma. Herein, by methylation-specific PCR, the putative KIAA0495 promoter was found unmethylated in all healthy controls (N = 14) but methylated in 50 % of myeloma cell lines (N = 10). KIAA0495 methylation was shown inversely correlated with KIAA0495 expression. However, KIAA0495 methylation was detected in none of both primary myeloma samples at diagnosis (N = 61) and at relapse/progression (N = 16). Collectively, despite frequently methylated in cell lines, KIAA0495 methylation appeared unimportant in the pathogenesis or progression of myeloma.
Electronic supplementary material
The online version of this article (doi:10.1186/s12943-015-0444-8) contains supplementary material, which is available to authorized users.
Keywords: Long non-coding RNA, KIAA0495, DNA methylation, Myeloma, Tumour suppressor
Introduction
Long non-coding RNA (lncRNA) is a novel class of functional RNA molecules >200 nucleotides with little or no protein-coding capacity [1, 2]. Based on their genomic locations relative to annotated protein-coding genes, lncRNAs are broadly classified into intergenic, intragenic, antisense, pseudogene, or divergent transcripts [3, 4]. Functionally, lncRNAs play a role in, but not limited to, development, differentiation, and carcinogenesis [2, 5]. Moreover, lncRNAs are found dysregulated and therefore potentially to be oncogenic or tumour suppressive in human cancers [6, 7].
Multiple myeloma is a form of haematological cancer originated from malignant transformation of plasma cells [8]. Clinically, this disease begins with benign monoclonal gammopathy of undetermined significance (MGUS), some of which may undergo asymptomatic smouldering multiple myeloma (SMM), and progresses to symptomatic myeloma [9]. Genetically, despite D-type cyclins are apparently upregulated in all patients, myeloma is a heterogeneous disease characterized by specific gains or losses of chromosomes, or reciprocal translocations, such as t(4;14)(p16.3;q32) and t(14;16)(q32;q23) [10].
DNA methylation refers to the catalytic addition of a methyl (−CH3) group to the cytosine ring of a CpG dinucleotide [11]. Human cancers are characterized by loss of global DNA methylation but gain of methylation at promoter-associated CpG islands (a cluster of CpG dinucleotides) and hence transcriptional silencing of specific tumour suppressor genes or miRNAs [12]. In myeloma, DNA methylation has been shown to mediate silencing of multiple tumour suppressor genes and miRNAs, and has been implicated in the pathogenesis and prognosis of the disease [13, 14].
Interestingly, by gene expression profiling, KIAA0495, a lncRNA transcribed from chromosome 1p36, has been shown to be progressively downregulated from normal plasma cell to MGUS to symptomatic myeloma [15]. Moreover, methylation-mediated silencing of KIAA0495 has been demonstrated in oligodendroglial tumours, leading to enhanced cisplatin resistance via upregulation of anti-apoptotic B-cell CLL/lymphoma 2-like 1 (BCL2L1) [16]. Hence, we hypothesized that DNA methylation may account for the progressive downregulation of KIAA0495 in the pathogenesis of myeloma. Herein, we have studied and reported methylation of KIAA0495 in myeloma cell lines, primary myeloma marrow samples at diagnosis, and at relapse/progression. Materials and methods have been incorporated as Additional file 1.
Findings
Methylation-specific PCR: KIAA0495 methylation in healthy controls and myeloma cell lines
While the putative KIAA0495 promoter region was found embedded in a CpG island (Additional file 2: Figure S1), MSP primers were designed to study methylation of this CpG island in a panel of healthy controls [peripheral (N = 10) and marrow (N = 3) buffy coat, CD138-sorted healthy plasma cell (N = 1)], and myeloma cell lines (N = 10) (Table 1). Direct sequencing analysis of M-MSP products from bisulfite-treated positive control showed expected conversion of unmethylated cytosine into uracil (turned to thymidine after PCR), with methylated cytosine remained unchanged, indicating complete bisulfite conversion and MSP specificity (Fig. 1a). None of 14 healthy controls showed KIAA0495 methylation (Fig. 1b). On the other hand, in myeloma cell lines, KMS-12-PE, LP-1, NCI-H929, OPM-2, and OCI-MY5 were partially methylated, whereas MOLP-8, RPMI-8226, U-266, WL-2, and JJN-3 were completely unmethylated for KIAA0495 (Fig. 1c). Moreover, the MSP methylation statuses of controls and cell lines were confirmed by quantitative bisulfite pyrosequencing (Fig. 1d). These data suggested that methylation of KIAA0495 was tumour-specific, consistent with other tumour suppressor protein-coding genes and non-coding miRNAs in myeloma [17–20].
Table 1.
Primer sequences and PCR reaction conditions
| Primer set | Forward primer (5’ – 3’) | Reverse primer (5’ – 3’) | Product size (bp) | MgCl2/Tm/cycles | Reference |
|---|---|---|---|---|---|
| (I) Methylation-specific polymerase chain reaction (MSP) | |||||
| KIAA0495 | |||||
| M-MSP | TGG AGA TAA CGG GTT TAG AGA AAT C | AAC GAA AAC AAA AAT AAA ACC TTC G | 191 | 1.5 mM/58 °C/40X | -- |
| U-MSP | TGG AGA TAA TGG GTT TAG AGA AAT T | AAA CAA AAA CAA AAA TAA AAC CTT CA | 192 | 2.0 mM/58 °C/40X | -- |
| (II) Reverse transcription-polymerase chain reaction (RT-PCR) | |||||
| KIAA0495 | GCT GCT TGC TGT ACG TGG TG | CGT GGC TGA CAC AAA CTT GC | 178 | 1.5 mM/60 °C/35X | [16] |
| GAPDH | ACC ACA GTC CAT GCC ATC ACT | TCC ACC ACC CTG TTG CTG TA | 452 | 1.5 mM/60 °C/24X | |
Keys: Tm, annealing temperature; M-MSP, MSP for the methylated allele; U-MSP, MSP for the unmethylated allele
Fig. 1.
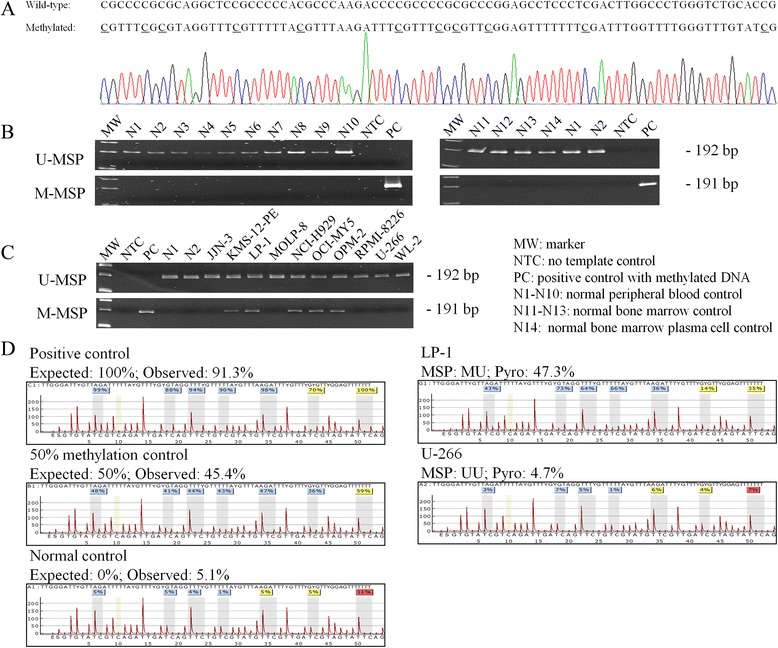
Methylation of KIAA0495 in myeloma. a Sequencing analysis of M-MSP products showed that the cytosine [C] residues of CpG dinucleotides were methylated and remained unchanged after bisulfite conversion, whereas all the other non CpG C residues were unmethylated and converted to thymidine [T], indicating complete bisulfite conversion and specificity of MSP. b M-/U-MSP showed KIAA0495 was completely methylated in positive control with methylated DNA, but completely unmethylated in 14 healthy donor controls, including peripheral buffy coat (N1-N10), marrow buffy coat (N11-N13), and CD138-sorted marrow plasma cells (N14). c M-/U-MSP showed KIAA0495 was partially methylated in KMS-12-PE, LP-1, NCI-H929, OCI-MY5, and OPM-2, whereas it was completely unmethylated in MOLP-8, RPMI-8226, U-266, WL-2, and JJN-3. d Quantitative bisulfite pyrosequencing interrogating the methylation intensity on a stretch of 7 neighboring CpG dinucleotides indicated consistent resutls as shown by the MSP statuses (MM, MU, and UU)
KIAA0495 methylation and expression in myeloma cell lines
To determine whether KIAA0495 methylation was associated with silencing of KIAA0495 expression, the expression level of KIAA0495 was measured by semi-quantitative RT-PCR and quantitative real-time PCR (qPCR), and correlated with the KIAA0495 methylation status as detected by MSP in myeloma cell lines. Of the 10 myeloma cell lines, KIAA0495 methylation was associated with lower expression as detected by both semi-quantitative RT-PCR (P = 0.0079; Fig. 2a) and quantitative real-time PCR (P = 0. 000155; Fig. 2b).
Fig. 2.
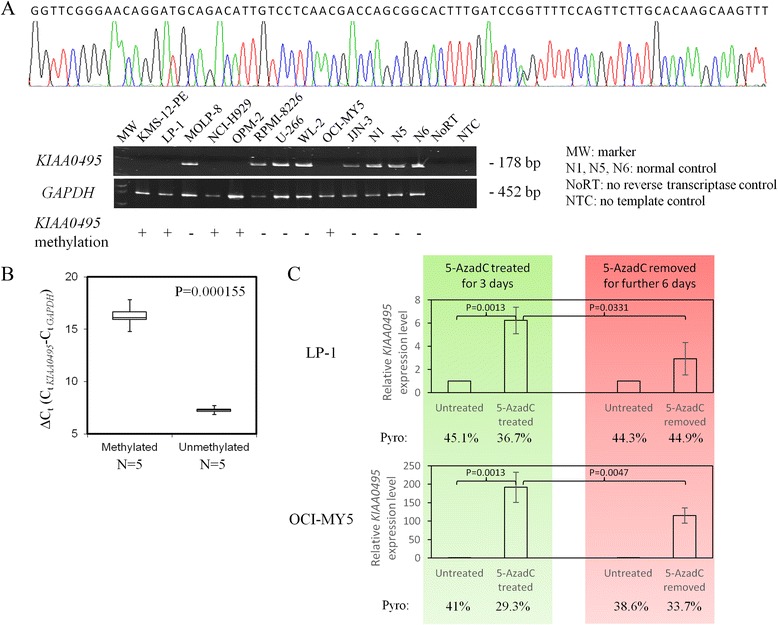
Methylation and expression of KIAA0495 in myeloma cells. a Semi-quantitative RT-PCR analysis showed expression of KIAA0495 in MOLP-8, RPMI-8226, U-266, WL-2, and JJN-3, which were completely unmethylated for KIAA0495, but not in KMS-12-PE, LP-1, NCI-H929, OCI-MY5, and OPM-2, which were partially methylated for KIAA0495 (P = 0.0079). Sequencing analysis confirmed the authenticity of the RT-PCR of KIAA0495. b qRT-PCR analysis showed the expression of KIAA0495 was significantly higher, as indicated by a lower ΔCt, in methylated than completely unmethylated myeloma cell lines (P = 0.000155). c Two partially methylated myeloma cell lines, LP-1 and OCI-MY5, were separately treated with 1 μmol/l of 5-AzadC for 3 days, after which, 5-AzadC was removed and cells were cultured for additional 6 days. On day 3, upon treatment with 5-AzadC, KIAA0495 was demethylated as indicated by the decreased percentage of methylation intensity by quantitative bisulfite pyrosequencing, with concommitent KIAA0495 re-expression as measured by qRT-PCR. On day 9, upon removal of 5-AzadC for 6 days, methylation of KIAA0495 was restored with re-suppression of KIAA0495
Moreover, myeloma cell lines with KIAA0495 methylation were treated with a hypomethylating agent, 5-AzadC, followed by pyrosequencing and qPCR analyses. Upon 5-AzadC treatment, LP-1 and OCI-MY5 cells, which were partially methylated for KIAA0495, showed progressive demethylation of the KIAA0495 promoter, as evidenced by the decreased methylation percentage on a stretch of seven consecutive CpG dinucleotides, together with concomitant re-expression of the KIAA0495 transcript, as illustrated by the increased expression relative to the untreated control (LP-1: P = 0.0013; OCI-MY5: P = 0.0013; Fig. 2c). Nevertheless, when 5-AzadC-treated cells were continually cultured with fresh medium in the absence of 5-AzadC for a further 6 days, methylation of the KIAA0495 promoter was restored, with simultaneous suppression of the KIAA0495 expression (LP-1: P = 0.0331; OCI-MY5: P = 0.0447; Fig. 2c). Taken together, the methylation of KIAA0495 was inversely correlated with KIAA0495 expression level in myeloma cell lines, similar to the methylation-mediated silencing of KIAA0495 demonstrated in glioma cell lines and primary oligodendroglial tumour cells [16], suggesting that methylation of the promoter-associated CpG island emerged to be one of the mechanisms resulting in the regulation of lncRNAs in cancer cells.
Methylation-specific PCR: KIAA0495 methylation in myeloma primary samples at diagnosis and at relapse
To examine if methylation of KIAA0495 was also detected in primary samples, methylation of KIAA0495 was studied in 61 primary samples at diagnosis and 16 primary samples at relapse by MSP. However, none of these samples showed methylation of KIAA0495 (Fig. 3), indicating methylation of KIAA0495 was rarely detected in primary myeloma samples. Hence, similar to the studies of miR-9-1, miR-9-3, and miR-124-1 [21, 22], these data suggested that methylation of certain tumour suppressor non-coding miRNAs or lncRNAs was acquired in vitro during continuous culture of myeloma cells, hence not pathogenic. Therefore, methylation was not the mechanism leading to the progressive downregulation of KIAA0495 from normal plasma cell to MGUS to symptomatic myeloma [15], suggestive of other mechanisms, such as histone modification and miRNA-mediated repression, may come into play. For instance, in gastric cancer cells, histone deacetylation was found to be associated in the downregulation of a tumour suppressor lncRNA, FENDRR [23]. Moreover, in breast cancer cells, miR-21 was shown to target and hence downregulate the expression of a tumour suppressor lncRNA, GAS5 [24]. Furthermore, while KIAA0495 resides on 1p36, which has been shown frequently deleted in newly diagnosed patients with myeloma [25], the loss of expression of KIAA0495 may be associated with chromosome deletion. Last but not least, inactivation of KIAA0495 may also be accounted by haploinsufficiency, which has been demonstrated in the TP53 in myeloma [26].
Fig. 3.
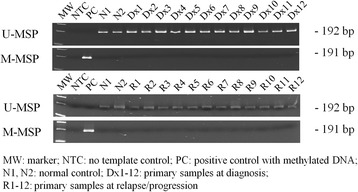
Methylation of KIAA0495 in primary myeloma marrow samples at diagnosis (Dx) and at relapse/progression (R)
Conclusion
Methylation of KIAA0495 is tumour-specific, leading to reversible silencing of KIAA0495 expression in myeloma cell lines. On the other hand, it is rarely detected in primary samples of myeloma at diagnosis or at relapse, and hence not responsible for the progressive downregulation of KIAA0495 from normal plasma cell to MGUS to symptomatic myeloma, as shown by the gene expression profiling, and remains as a possible in vitro event acquired during continuous culture of myeloma cells.
Abbreviations
- lncRNA
Long non-coding RNA
- MGUS
Monoclonal gammopathy of undetermined significance
- miRNA
microRNA
- MSP
Methylation-specific PCR
- M-MSP
Methylated MSP
- U-MSP
Unmethylated MSP
- PCR
Polymerase chain reaction
- RT-PCR
Reverse transcription-PCR
- SMM
Smouldering multiple myeloma
Additional files
Materials and methods. (DOCX 20 kb)
Schematic diagram showing the transcription start sites (TSS) of KIAA0495, distribution of CpG dinucleotides (red lines), and amplicons of methylation-specific PCR (green box) and pyrosequencing (purple box). (TIFF 339 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
KYW and ZZ carried out the experiments. KYW, ZZ and CSC drafted the manuscript. KYW and CSC conceived of the study. All authors participated in the design of the study. All authors read and approved the final manuscript.
Contributor Information
Kwan Yeung Wong, Email: kwanyeung@graduate.hku.hk.
Zhenhai Li, Email: zhhli@connect.hku.hk.
Xiaoqin Zhang, Email: zhxq@connect.hku.hk.
Gilberto Ka Kit Leung, Email: gilberto@hku.hk.
Godfrey Chi-fung Chan, Email: gcfchan@hku.hk.
Chor Sang Chim, Phone: (852) 2255 4769, Email: jcschim@hku.hk.
References
- 1.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 2.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research. Oncogene. 2012;31:4577–87. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kung JTY, Colognori D, Lee JT. Long Noncoding RNAs: Past, Present, and Future. Genetics. 2013;193:651–69. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 6.Gibb E, Brown C, Lam W. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutschner T, Diederichs S. The Hallmarks of Cancer: A long non-coding RNA point of view. RNA Biol. 2012;9:703–19. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12:335–48. doi: 10.1038/nrc3257. [DOI] [PubMed] [Google Scholar]
- 9.Kyle RA, Durie BGM, Rajkumar SV, Landgren O, Blade J, Merlini G, et al. Monoclonal gammopathy of undetermined significance (MGUS) and smoldering (asymptomatic) multiple myeloma: IMWG consensus perspectives risk factors for progression and guidelines for monitoring and management. Leukemia. 2010;24:1121–7. doi: 10.1038/leu.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK, et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009;23:2210–21. doi: 10.1038/leu.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 12.Esteller M. Epigenetics in Cancer. N Engl J Med. 2008;358:1148–59. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 13.Chim CS, Kwong YL, Liang R. Gene hypermethylation in multiple myeloma: lessons from a cancer pathway approach. Clin Lymphoma Myeloma. 2008;8:331–9. doi: 10.3816/CLM.2008.n.048. [DOI] [PubMed] [Google Scholar]
- 14.Wong KY, Huang X, Chim CS. DNA methylation of microRNA genes in multiple myeloma. Carcinogenesis. 2012;33:1629–38. doi: 10.1093/carcin/bgs212. [DOI] [PubMed] [Google Scholar]
- 15.Zhan F, Barlogie B, Arzoumanian V, Huang Y, Williams DR, Hollmig K, et al. Gene-expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis. Blood. 2007;109:1692–700. doi: 10.1182/blood-2006-07-037077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pang JCS, Li KKW, Lau KM, Ng YL, Wong J, Chung NYF, et al. KIAA0495/PDAM Is Frequently Downregulated in Oligodendroglial Tumors and Its Knockdown by siRNA Induces Cisplatin Resistance in Glioma Cells. Brain Pathol. 2010;20:1021–32. doi: 10.1111/j.1750-3639.2010.00405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chim CS, Fung TK, Cheung WC, Liang R, Kwong YL. SOCS1 and SHP1 hypermethylation in multiple myeloma: implications for epigenetic activation of the Jak/STAT pathway. Blood. 2004;103:4630–5. doi: 10.1182/blood-2003-06-2007. [DOI] [PubMed] [Google Scholar]
- 18.Chim CS, Pang R, Fung TK, Choi CL, Liang R. Epigenetic dysregulation of Wnt signaling pathway in multiple myeloma. Leukemia. 2007;21:2527–36. doi: 10.1038/sj.leu.2404939. [DOI] [PubMed] [Google Scholar]
- 19.Wong KY, Liang R, So CC, Jin DY, Costello JF, Chim CS. Epigenetic silencing of MIR203 in multiple myeloma. Br J Haematol. 2011;154:569–78. doi: 10.1111/j.1365-2141.2011.08782.x. [DOI] [PubMed] [Google Scholar]
- 20.Wong KY, Yim RLH, So CC, Jin D-Y, Liang R, Chim CS. Epigenetic inactivation of the MIR34B/C in multiple myeloma. Blood. 2011;118:5901–4. doi: 10.1182/blood-2011-06-361022. [DOI] [PubMed] [Google Scholar]
- 21.Wong KY, So CC, Loong F, Chung LP, Lam WW, Liang R, et al. Epigenetic inactivation of the miR-124-1 in haematological malignancies. PloS one. 2011;6 doi: 10.1371/journal.pone.0019027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Q, Wang LQ, Wong KY, Li ZY, Chim CS. Infrequent DNA methylation of miR-9-1 and miR-9-3 in multiple myeloma. J Clin Pathol. 2015;68:557–61. doi: 10.1136/jclinpath-2014-202817. [DOI] [PubMed] [Google Scholar]
- 23.Xu TP, Huang MD, Xia R, Liu XX, Sun M, Yin L, et al. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J Hematol Oncol. 2014;7:63. doi: 10.1186/s13045-014-0063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C, Xu M, et al. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013;20:1558–68. doi: 10.1038/cdd.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker BA, Leone PE, Chiecchio L, Dickens NJ, Jenner MW, Boyd KD, et al. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood. 2010;116:e56–65. doi: 10.1182/blood-2010-04-279596. [DOI] [PubMed] [Google Scholar]
- 26.Teoh PJ, Chung TH, Sebastian S, Choo SN, Yan J, Ng SB, et al. p53 haploinsufficiency and functional abnormalities in multiple myeloma. Leukemia. 2014;28:2066–74. doi: 10.1038/leu.2014.102. [DOI] [PubMed] [Google Scholar]


