Summary
Mammalian interphase chromosomes interact with the nuclear lamina (NL) through hundreds of large Lamina Associated Domains (LADs). We report a method to map NL contacts genome-wide in single human cells. Analysis of nearly 400 maps reveals a core architecture of gene-poor LADs that contact the NL with high cell-to-cell consistency, interspersed by LADs with more variable NL interactions. The variable contacts tend to be cell-type specific and are more sensitive to changes in genome ploidy than the consistent contacts. Single-cell maps indicate that NL contacts involve multivalent interactions over hundreds of kilobases. Moreover, we observe extensive intra-chromosomal coordination of NL contacts, even over tens of megabases. Such coordinated loci exhibit preferential interactions as detected by Hi-C. Finally, consistency of NL contacts is inversely linked to gene activity in single cells, and correlates positively with the heterochromatic histone modification H3K9me3. These results highlight fundamental principles of single cell chromatin organization.
INTRODUCTION
An important unresolved question in eukaryotic genome biology is how chromosomes are spatially organized inside interphase nuclei. Current evidence suggests that this organization is driven by probabilistic principles (Bickmore, 2013; Cavalli and Misteli, 2013; Gibcus and Dekker, 2013). Systematic fluorescence in situ hybridization (FISH) experiments have revealed that in a homogeneous cell population the nuclear positions of chromosomes are variable with respect to each other and relative to the periphery (Bolzer et al., 2005). However, this positioning is not entirely random; for example, in human lymphoid cells chromosome 18 (chr18) tends to be located near the periphery, while chr19 shows a preference for the nuclear interior (Croft et al., 1999; Cremer et al., 2001).
At a smaller scale, certain individual genomic loci visualized by FISH also exhibit preferences for specific nuclear landmarks such as the nuclear envelope (Marshall et al., 1996; Kosak et al., 2002) and nucleoli (Manuelidis and Borden, 1988; Ochs and Press, 1992), but usually with some degree of random variation. This variability is directly illustrated by in vivo tagging experiments in which loci contacting the nuclear lamina (NL) were tracked over mitosis in a clonal human cell line (Kind et al., 2013). This demonstrated that a sizeable subset of loci that were associated with the NL in mother cells relocated to the nuclear interior in daughter cells, indicating that, at least to some degree, genome contacts with the NL are intrinsically variable.
Complementary to these single-cell microscopy approaches are genome-wide mapping techniques that query the chromosome organization in large pools of cells (van Steensel and Dekker, 2010). For example, the 4C, 5C and Hi-C technologies generate maps of the pair-wise spatial proximity of genomic loci (de Wit and de Laat, 2012; Dekker et al., 2013). Such maps have revealed global patterns that indicate that mammalian interphase chromosomes are partitioned into domains that are roughly 200kb–2Mb in size (Lieberman-Aiden et al., 2009; Dixon et al., 2012; Nora et al., 2012; Rao et al., 2014). Computational models of chromosome polymer folding fitted to 5C and Hi-C data generally suggest that interphase chromosomes adopt multiple configurations that vary from cell to cell (Lieberman-Aiden et al., 2009; Bau et al., 2011; Kalhor et al., 2012; Giorgetti et al., 2014).
Another genome-wide approach to study chromosome architecture is the mapping of interactions with the NL, mostly by means of the DamID technology (Guelen et al., 2008; Peric-Hupkes et al., 2010; Meuleman et al., 2013). The NL provides a very large surface area for potential contacts with the genome. Indeed, DamID studies have estimated that as much as ~35% of the mammalian genome can interact with the NL in any tested cell type, although it has remained unclear how much of the genome contacts the NL in a single cell. Genome–NL interactions occur through about 1,100–1,400 discrete Lamina Associated Domains (LADs), which have a median size of ~0.5Mb and are scattered across all chromosomes. Most genes in LADs are expressed at very low levels, and results of various tethering experiments (Finlan et al., 2008; Reddy et al., 2008; Kind et al., 2013; Therizols et al., 2014) point to a reciprocal relationship between gene positioning at the NL and a repressive chromatin state.
The large size of LADs and their prevalence throughout the genome strongly suggest that LAD–NL interactions play an important role in interphase chromosome architecture. Insights into the single-cell behavior of these interactions will thus enhance our fundamental understanding of chromosome organization. Here, we report a modified version of the DamID technology that is sensitive enough to generate genome-wide maps of NL contacts in single human cells at a resolution of ~100 kb, which is well below the median size of LADs. We generated a total of 395 of such single-cell maps. These maps, complemented by Hi-C analysis and super-resolution microscopy imaging, provide insights into the nature of LAD–NL interactions and uncover principles of cell-to-cell variation in chromosome architecture.
RESULTS
Single-cell DamID methodology
As a model system to develop single-cell DamID we chose the human myeloid leukemia cell line KBM7, which is haploid for all chromosomes except chr8 and a small part of chr15 (Kotecki et al., 1999; Burckstummer et al., 2013). Even though this haploid state is unusual for human somatic cells, it facilitates the interpretation of single-cell genome-wide maps because there is no need to discriminate the homologous chromosomes that are present in diploid cells.
We developed a DamID protocol for single cells as summarized in Figure 1A. We created a KBM7 clone (#14) that expresses an inducible fusion protein consisting of DNA adenine methyltransferase (Dam) and Lamin B1 (LmnB1) (Kind et al., 2013), as well as the Fucci two-color fluorescent reporter system to monitor the cell cycle stage (Sakaue-Sawano et al., 2008). We then induced Dam-LmnB1 protein expression (Figure S1A-B), and 15 hours later we collected single cells at the onset of S-phase by fluorescence-activated cell sorting (FACS) (Figure S1C). This design ensured that the harvested cells had expressed Dam-LmnB1 for most of their recent G1 phase (with lasts on average ~14.9 hours, estimated based on doubling time and FACS data), providing sufficient time for the accumulation of adenine methylation on the genomic loci that contact the NL. Importantly, LmnB1 was strictly confined to the nuclear periphery before and after Dam-LmnB1 induction (Figure S1D).
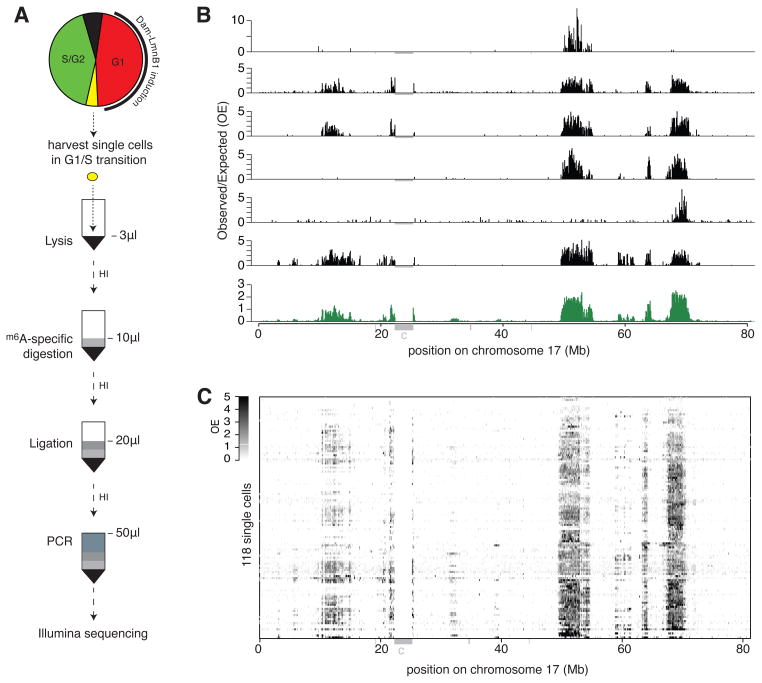
Figure 1. Single-cell DamID mapping of genome–NL interactions.
(A) Schematic representation of the single-cell DamID procedure. HI, heat inactivation step. (B) NL contact maps for chr17 in six individual cells (black) and the average profile of 118 single cells (bottom track, green). OE, observed over expected read count ratio in contiguous 100 kb segments. (C) Grey scale representation of OE values for 118 single cells on chromosome 17. Grey bars underneath the axes in (B–C) mark unmappable regions; “c” is centromeric region. See also Figure S1.
The single FACS-sorted cells were captured directly in a small volume of lysis buffer in a 96-well plate. Subsequent sample processing consisted of only a few steps (Fig. 1A): digestion with DpnI, which is highly specific for Dam-methylated GATC sequences, followed by adaptor ligation (Figure S1E) and a total of 26 cycles of PCR amplification. A key difference with the conventional DamID protocol is that these steps are all done in the same well by sequential addition of reagents, without intermediate purification or concentration steps that could lead to loss of DNA. Gel electrophoresis showed that approximately half of the wells yielded a clear smear of amplified DNA (Figure S1F). These PCR products were prepared for multiplexed Illumina sequencing by ligation of indexed adaptors, and subsequently pooled and sequenced.
Single-cell NL interaction maps
In total, we generated 118 single-cell DamID maps from KBM7 clone #14. After applying quality filters (Figure S1G), we obtained a median of 5*105 reads per cell that could be mapped to a unique genomic location (Figure S1H). On average 92% of the mapped locations started with a GATC motif, which is the recognition sequence of both Dam and DpnI, indicating that the detection is highly specific. For each cell we binned these reads in 100 kb contiguous genomic segments, and then we calculated for each segment an observed over expected (OE) score based on the number of recovered unique reads, the theoretical maximum number of mappable unique reads in the segment, and the total genome-wide read count obtained from each cell. OE scores >1 indicate more Dam-LmnB1 methylation than may be expected by random chance.
Essentially all cells showed a striking domain pattern of OE scores along most chromosomes, as illustrated for chr17 in Figures 1B–C. This pattern is reminiscent of the LAD profiles previously published for populations of cells. However, clear differences can be observed between individual cells (Figure 1B), often with entire Mb-sized domains missing, which is suggestive of cell-to-cell variation in NL contacts.
We note that the chosen segment size of 100 kb is a compromise between resolution and noise. Because the median human LAD size is approximately 0.5 Mb (Guelen et al., 2008), segments of 100 kb are expected to capture most of the LAD organization. Indeed, the OE scores in adjacent segments within each of the 118 single cell samples have a Pearson correlation coefficient of 0.70 ± 0.06 (mean ± s.d.), indicating that neighboring segments report NL interactions in a partially redundant fashion with acceptable noise levels. For reference, at 100 kb resolution our previously published Dam-LmnB1 profiles from pools of human Tig3, hES and HT1080 cells (Guelen et al., 2008; Kind et al., 2013; Meuleman et al., 2013) show a correlation of 0.88–0.90 between neighboring segments. The single-cell correlation of OE scores between adjacent segments is not related to the number of reads per cell (Pearson’s correlation coefficient 0.06, P=0.51), indicating that the latter does not impose a limit to the quality of our data at 100 kb resolution.
Validation of single-cell maps
To further gauge the overall quality of these data, we first reconstructed a population profile by averaging the maps of the 118 single cells and then compared it to a conventional microarray-based DamID profile generated from a large pool of KBM7 cells (Figure 2A). The highly similar domain patterns and an overall Spearman rank correlation coefficient of 0.90 demonstrate that the new protocol captures the same regions of interaction as the previous well-validated protocol.
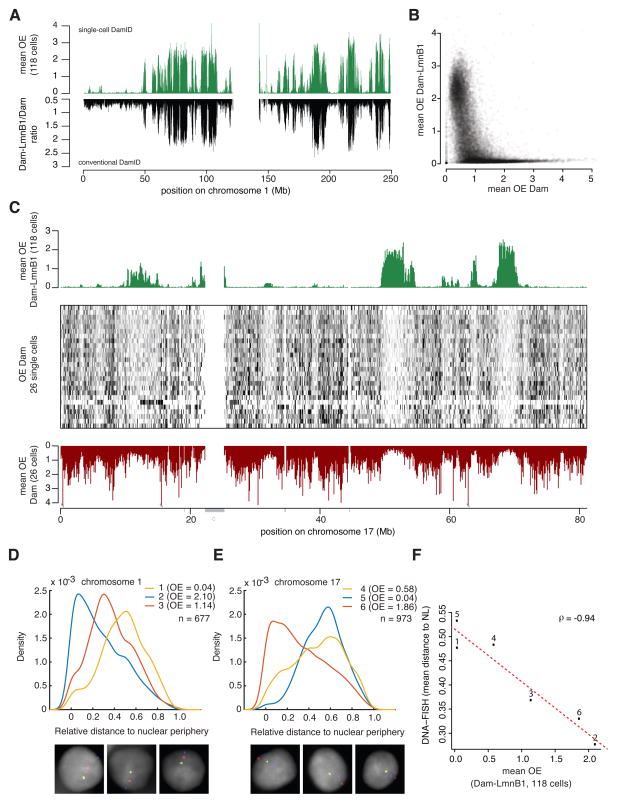
Figure 2. Validation of single-cell DamID maps.
A) Comparison NL contact map for chr1 representing the average of 118 single-cell profiles (top profile) and a conventional DamID map generated with a population of ≈ 1–2x105 cells (bottom profile). Genome-wide Spearman’s ρ = 0.90 between the two methods. (B) Average OE score of 118 single-cell Dam-LmnB1 samples (y-axis) versus the average OE score of 26 single-cell Dam-only samples (x-axis). (C) Comparison of average OE scores obtained with Dam-LmnB1 (top track, average of 118 cells, same as in Figure 1B) and Dam-only (bottom track, average of 26 cells). OE scores for the individual Dam-only cells are shown as grey-scale encoded rows in the center frame. Grey bars underneath the bottom axis mark unmappable regions; “c” is centromeric region. (D–F) Multi-color 3D DNA FISH microscopy with probes for six genomic loci covering a broad range of average OE scores distributed on chr1 (n=677) (D) and chr17 (n=973) (E). Graphs depict the distributions of radial probe positions, with 0 corresponding to the nuclear edge and 1 to the centroid. Three representative nuclei with three-color FISH signals are displayed below the graphs; DNA staining with DAPI is shown in grey. (F) Mean radial positions of the six probes versus the mean DamID OE scores. Numbers 1–6 correspond to probe numbers in (D–E). Dotted line shows linear regression fit.
However, in conventional DamID, a Dam-only control is typically included to normalize for chromatin accessibility and other potential biases (Vogel et al., 2007; Guelen et al., 2008). This normalization is not possible in single cell DamID because the Dam-LmnB1 and Dam-only profiles cannot be obtained from the same cell. Nevertheless, we established a Dam-only and Fucci expressing KBM7 clone and mapped the adenine methylation patterns in 26 single cells. The resulting patterns are very different from the Dam-LmnB1 profiles (Figure 2B–C). In general, regions that have no detectable Dam-LmnB1 signal show clear Dam-only signals – hence these regions are not intrinsically undetectable. Conversely, regions with very high Dam-only signals generally do not show a Dam-LmnB1 signal, indicating that there is no strong bias for accessibility in our Dam-LmnB1 maps. We conclude that leaving out the Dam-only normalization is acceptable for single-cell Dam-LmnB1 profiling.
Next, we performed multi-color fluorescent in situ hybridization (FISH) with probes for six genomic loci covering a broad range of average OE scores (Figure 2D–F). Analysis of hundreds of nuclei revealed a good correspondence between the average distance to the periphery according to FISH and the average OE scores (Spearman’s ρ = 0.94), confirming that our single-cell Dam-LmnB1 profiles provide a view of the spatial organization of the genome relative to the NL. Together these data indicate that single-cell DamID using Dam-LmnB1 generates NL interaction profiles with low noise levels, suitable resolution, acceptable bias, and with good correspondence to localization by FISH.
Cell-to-cell variability and consistency of genome – NL associations
Visual inspection of the collection of single-cell maps suggested that some regions interact more frequently with the NL than others (Figure 1B, C). In order to analyze this systematically, we first converted the OE scores for each cell to a binary NL contact map. For this we used an OE score cutoff of 1, motivated by the bimodal distribution of OE scores (Figure S2A) that suggests that loci are either in a “contact” or “no contact” state. We then calculated for each 100 kb segment the NL contact frequency (CF), defined as the proportion of cells in which this segment contacted the NL. This data processing does not lead to a substantial loss of information content, because the Spearman rank correlation coefficient of the average radial position of the six FISH probes with CF is 0.90 (as compared to 0.94 for OE values, see above), and CF values correlate strongly with the average OE scores (Figure S2B). CF values are also highly robust with respect to sequencing read depth, because subsampling of the single-cell data to the one-but-lowest read count (1.04*105 reads; an average 3.1-fold downsampling) does not affect CF values (Figure S2C).
Strikingly, CFs vary widely across the genome (Figure 3A–B). About 23% of the 100 kb segments show no detectable contact with the NL in any of the 118 cells, and thus are very stably located in the nuclear interior. Conversely, ~15% of the segments have CFs >80%, representing loci that are consistently located at the NL. About 34% of the segments have contact frequencies in the range 20–80% and thus show high cell-to-cell variability in their NL associations. The remaining loci (29%) only occasionally contact the NL (0<CF<20%).
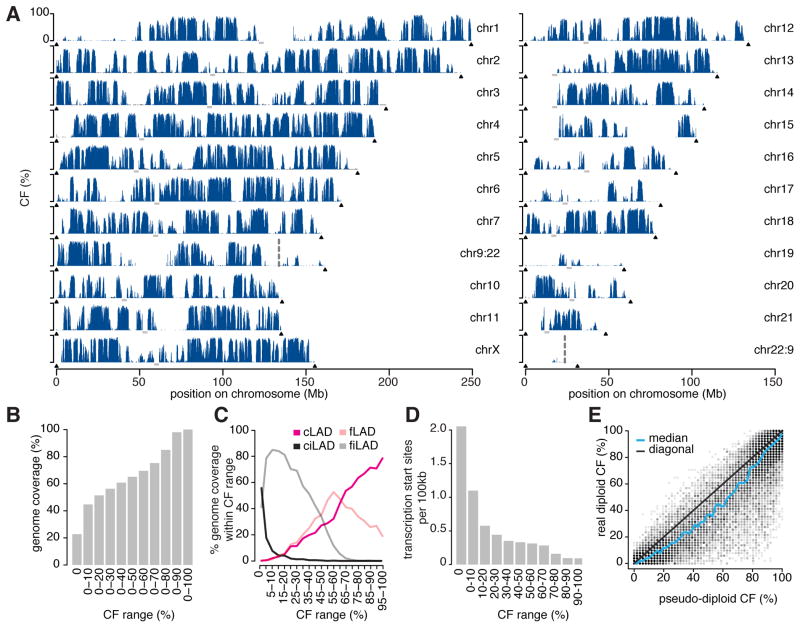
Figure 3. NL contact frequencies are linked to developmental dynamics, gene density, and ploidy.
(A) Estimated contact frequency maps for all chromosomes in clone #14 cells. KBM7 cells carry a balanced translocation between chr9 and chr22 (Burckstummer et al., 2013); vertical dotted lines mark the junctions. Centromeric regions are indicated by grey bars; telomeres by black triangles. chr8 is not shown because it is diploid. (B) Cumulative histogram of genome-wide CF values. (C) Distribution of genomic segments with indicated CFs over constitutive (c) and facultative (f) LADs and inter-LADs (iLADs). (D) Average number of transcription start sites per 100 kb segment, plotted as a function of CF. (E) Comparison of CFs in diploid cells and pseudo-diploid cells. The latter are simulated by combining equal numbers of sequence reads from pairs of haploid cells. See also Figure S2 and Table S1.
These different classes of loci are scattered throughout the genome (Figure 3A), although the smaller chromosomes tend to have a lower density of stable NL contacts than larger chromosomes. An exception to this rule is chr18, which harbors many regions with high CFs. This contrasts in particular with chr19, which only exhibits a few contact sites and very low CFs. This matches previous chromosome painting studies that found chr18 to be preferentially located at the nuclear periphery and chr19 in the nuclear interior (Croft et al., 1999; Cremer et al., 2001). An intriguing pattern is visible on chrX (Figure 3A, bottom left), which has many stable NL contacts along the distal arms, while the centromere-proximal ~40Mb show only variable contacts.
In order to confirm these CF patterns, we used an independently derived KBM7 clone that also expresses Dam-LmnB1 and the Fucci system (clone #5.5), to generate a total of 168 single-cell maps. The genome-wide CF profile of clone #5.5 is highly similar to that of clone #14 (Pearson correlation r = 0.97 (Figure S2D, E). However, the absolute CF values in clone #5.5 are systematically 1.3-fold lower than in clone #14, which we attribute to a somewhat lower activity of the Dam-LmnB1 protein in clone #5.5 (Figure S2F). Nevertheless, the relative CF differences between loci are highly consistent between the two clones. While we cannot rule out that the CF values of clone #14 are an underestimate, the fact that the highest CF values of clone #14 are in the range of ~95%, combined with the tight linearity with clone #5.5, suggests that any underestimate in clone #14 is minor.
Stable and variable NL contacts are linked to degrees of developmental plasticity
Previously we reported that some LADs interact with the NL in a cell-type specific (facultative) fashion, while other LADs do so in a cell-type invariant (constitutive) manner (Peric-Hupkes et al., 2010; Meuleman et al., 2013). We investigated whether this distinct behavior is linked to differences in CF. We used a collection of conventional microarray-based DamID maps of NL interactions in 9 human cell lines of diverse origin (CAdG and BvS, manuscript in preparation; see Extended Experimental Procedures) to classify each 100 kb segment as either constitutive LAD (cLAD), constitutive inter-LAD (ciLAD), facultative LAD (fLAD) or facultative inter-LAD (fiLAD). The latter are inter-LAD regions that are not associated with the NL in KBM7 cells, but do interact with the NL in at least one other cell type.
As expected, ciLADs have low CF values (Figure 3C), consistent with their definition as not associated with the NL. Likewise, cLADs tend to have high CF values. Thus, these constitutive regions are not only invariable between different cell types, but tend to be consistently positioned relative to the NL within one cell type. In contrast, facultative LADs and iLADs have mostly intermediate CF values: fLADs generally have lower CF values than cLADs (P < 2.2*10−16, Wilcoxon test), and fiLADs have higher values than ciLADs (P < 2.2*10−16). The partial overlap in CF distributions of fLADs and fiLADs indicates that the definitions of these LAD classes are not perfect, which may be due to differences in the data types (single-cell vs. population-based DamID maps; sequencing vs. microarray) and resolution. These results uncover a link between CF and the consistency of NL interactions between different cell types.
We previously reported that cLADs have a ~2-fold lower gene density than ciLADs (Meuleman et al., 2013). CF values are more dramatically linked to gene density: regions with CF > 80% have a ~20-fold lower gene density than regions with no NL contacts (Figure 3D). The few genes that are located in such high-CF regions are enriched for Gene Ontology categories very divergent from myeloid cell functions, among them most prominently the olfactory receptor genes (Table S1), which are rarely expressed.
A picture thus emerges in which most cLADs are relatively consistently associated with the NL, providing a structural backbone to chromosomes that is largely invariant between individual cells and also between cell types. In contrast, regions with cell-type specific NL interactions generally interact less consistently with the NL, contributing to cell-to-cell variability in the spatial organization of chromosomes.
Ploidy of KBM7 cells primarily affects variable NL contacts
We considered the possibility that competition between LADs could explain why some LADs contact the NL in only a fraction of all cells: such LADs may have lower NL binding affinities and fail to compete with stronger LADs in some of the cells. We wondered whether this balance could be altered by changing the total amount of genomic DNA in the nucleus. To test this, we took advantage of the fact that KBM7 cells spontaneously form diploid cells at low frequency (Kotecki et al., 1999). Such diploid cells should be genetically identical to the haploid cells, except for their ploidy. We derived a clonal diploid line from clone #14, with normal cell cycle behaviour (Figure S2G), and generated a total of 51 single-cell Dam-LmnB1 contact maps.
Comparison of the single-cell maps from haploid and diploid cells is not straightforward, because our current DamID method cannot determine whether the sequence reads from diploid cells are derived from one homolog or both. We therefore constructed “pseudo-diploid” reference maps by combining equal numbers of sequence reads from pairs of single haploid cells, and then merged the reads and processed the data as above, as if they were derived from a single cell. Hence, these pseudo-diploid maps are simulated maps in which two homologs of each chromosome are present, but there cannot be any biological effect of increased ploidy (because the homologs were in different cells), while technical skews due to the inability to discriminate the two homologs should be identical to those in diploid cells.
Although the overall pattern of NL contacts was similar between pseudo-diploid and diploid cells, many loci showed decreased CFs in diploid cells compared to haploid cells (Figure 3E), while increased CFs were rare. Strikingly, these changes occurred preferentially in genomic segments that have intermediate CFs in pseudo-diploid (and thus haploid) cells. This result indicates that diploidization leads to preferential loss of NL interactions of LADs that contact the NL less robustly in haploid cells. We obtained similar results with 32 single cells from a diploid clonal line derived from clone #5.5 (Figure S2H). The loss of NL contacts in diploid cells suggests increased competition for NL contacts. This may be caused by a reduced surface:volume ratio and an increase in the average distance of loci to the NL, which are expected to accompany the increase in nuclear volume due to the doubled DNA content. However, other mechanisms cannot be ruled out.
Single-cell maps point to a multivalent mechanism of LAD – NL interactions
We wondered what biophysical principle could explain the differences in apparent NL affinity between genomic loci. The domain pattern of NL contacts suggest that these interactions are not mediated by focal attachments, but rather by multivalent interactions within each LAD. Because interactions with higher valency typically have a higher avidity, we predicted that long LADs interact more stably with the NL than short LADs. To analyze this, we defined LADs here as continuous stretches of 100 kb segments with CF>1% across the 118 cells. This yielded a total of 1,358 LADs. Strikingly, the mean CF within each LAD shows a clear positive correlation with LAD length (Spearman’s ρ = 0.81; P < 2.2*10−16), reaching a plateau for sizes larger than ~6–8Mb (Figure 4A). This supports a model of multivalent local genome – NL interactions.
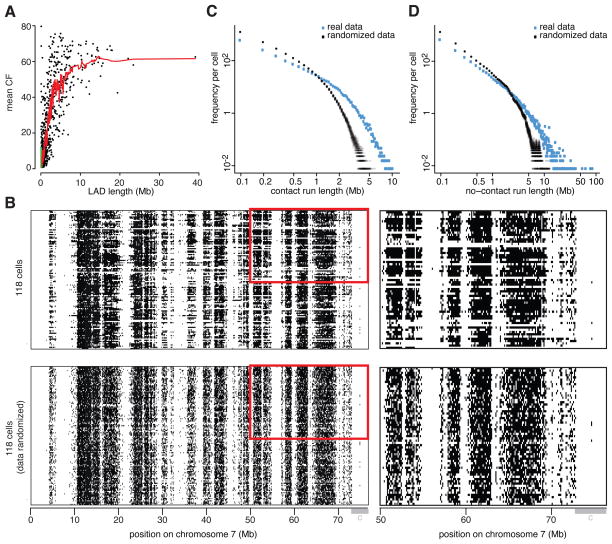
Figure 4. Evidence for multivalent NL interactions.
(A) Correlation between the mean CF and the length of LADs. Here, LADs are defined as continuous regions in which all 100 kb segments have CF>1% across all 118 clone #14 cells. Curve shows running mean with bin size 15. (B). Binarized NL contact map of the p-arm of chr7 in 118 cells (top panels), and the same data after a random shuffling procedure that keeps CFs and the number of contacts per cell the same, to simulate the complete absence of coordination between neighboring segments (bottom panels). Grey bar marked “c” indicates centromeric region. Right-hand panels are magnified views of the regions outlined by red boxes in the left-hand panels. (C) Distribution of genome-wide NL contact run lengths in 118 single-cell datasets (blue) compared to 100 sets of randomized data (grey). (D) Genome-wide length distribution of no-contact runs in real (blue) and 100 sets of randomized (grey) data. No-contact runs present in all 118 cells (i.e., regions that never contact the NL) were excluded in this analysis. See also Figure S3.
This multivalency model predicts that there are continuous stretches of contact in individual LADs in single cells. Indeed, such runs of contact could be frequently observed (black horizontal streaks in Figure 4B, top panel). For comparison, we randomly shuffled the contact data to simulate a “random button” model, in which each segment maintains exactly the same CF as in the real data, but contacts the NL independently of its neighboring segments (Figure 4B, bottom panel). This pattern has a much more fine-grained appearance than the original contact maps, with fewer long runs of contact. Quantitative analysis showed that long contact runs, particularly above ~1.5Mb, occur more frequently in the real data than in the simulated random button model (Figure 4C). This supports the multivalent interaction model.
Long-range coordination of NL contacts within chromosomes
Interestingly, we also observed long runs of no-contact on many single chromosomes (white horizontal streaks in Figure 4B, top panel). Particularly in the range >5Mb such runs occur more frequently than may be expected by random chance (Figure 4D). We interpret these long runs of no-contact as large chromosomal regions (often including multiple LADs) that completely dissociate from the NL in an incidental manner. These long no-contact runs are not generally due to loss of large chromosomal regions during sample processing, because our Dam-only single cell maps have more homogeneous methylation patterns (Figure 2C), and lack long runs that are completely devoid of mappable sequence reads (Figure S3A).
This coordinated detachment of neighboring LADs from the NL prompted us to systematically search for evidence of coordination of NL contacts, by calculating pair-wise NL contact correlations for all 100 kb segments within each chromosome, based on the binary NL contact maps of the 118 single cells. The resulting correlation matrices, which indicate how pairs of loci may coordinately attach and detach from the NL in single cells, showed remarkable plaid-like patterns on all chromosomes (Figure 5A). Along the diagonal, squares of consistently positive correlations represent units of coordinated NL contacts, which tend to be one to several Mb in size. Most of these units exhibit additional off-diagonal correlations with other units, sometimes with striking specificity (Figure 5A, examples marked with arrows and boxes). Such coordinated units can be tens of megabases apart, although the frequency decays gradually with distance (Figure 5B). Randomized contact maps yield much lower average correlation coefficients over all distances, demonstrating that the prevalence of positive correlations is not due to random chance (Figure 5B). Intra-chromosomal coordination is on average higher than inter-chromosomal coordination, and the latter is not higher than may be expected by random chance (Figure S4A). These results point to extensive coordination of NL contacts within chromosomes, often over long genomic distances.
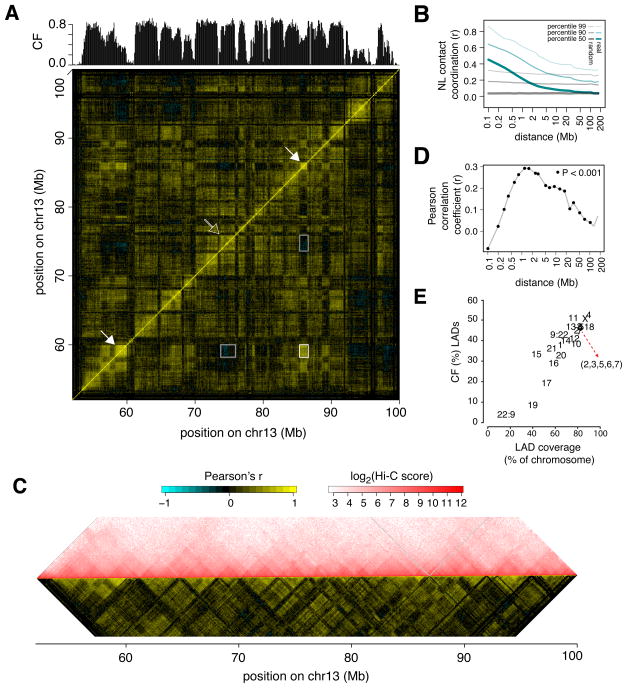
Figure 5. Intra-chromosomal coordination of NL contacts over a wide distance range.
(A) NL contact correlation matrix for part of chr13 in clone #14, showing the Pearson correlation of binary NL contacts across the 118 cells for all possible pairs of genomic segments. Color key is shown in panel (C). White arrows mark two example regions located ~26Mb apart that exhibit coordinated contacts (white box); open arrow indicates a region that is not positively correlated with these regions (grey boxes). (B) Genome-wide coordinated NL contacts as a function of linear distance. Median, 90th and 99th percentile Pearson’s r values are shown for real (cyan) and randomized (grey) binary NL contact data from 118 single cells. (C) Comparison of NL contact coordination matrix to Hi-C interaction matrix. The matrix in (A) was turned clock-wise by 45 degrees and only a section below the diagonal is shown; the corresponding part of the Hi-C interaction matrix (white-red color scale) is juxtaposed to facilitate comparison. Grey lines mark a repetitive region. (D) Genome-wide correlation between NL contact coordination and Hi-C interactions, plotted as function of linear distance along chromosomes. Dots mark distances at which the correlation is significantly different from 0 (P<0.001). (E) Positive correlation (Spearman’s ρ = 0.85, P = 8.96*10−7) between the fraction of each chromosome covered by LADs and the average CF in these LADs. See also Figure S4.
Coordination of NL contacts is partially linked to physical proximity detected by Hi-C
The plaid-like correlation patterns reminded us of patterns commonly observed with Hi-C (Lieberman-Aiden et al., 2009; Dixon et al., 2012), a technology that maps proximity of genomic sequences in nuclear space (Dekker et al., 2013). An intriguing possibility was therefore that regions with coordinated NL contacts could be in spatial proximity to one another. To investigate this, we generated Hi-C maps from KBM7 clone #14 cells. The resulting Hi-C interaction matrices appeared partially similar to the NL contact correlation matrices (Figure 5C).
To quantify the similarity we calculated the correlation between the degree of NL contact coordination and the Hi-C interaction frequency as a function of genomic distance (Figure 5D). The results show that Hi-C interactions correlate moderately but significantly with the degree of NL contact coordination. This correlation is most prominent at ~1–2Mb distance and declines gradually over longer distances, but is still statistically significant (P<0.001) at ~100 Mb. Somewhat surprisingly, this positive correlation appears absent among directly neighboring 100 kb segments. One possibility is that pairs of adjacent 100 kb segments have very high Hi-C interaction frequencies due to their physical linkage, regardless of any coordination of NL contacts; indeed our analyses show that finer-scale features of Hi-C do not always correspond to coordination of NL contacts (see below). Together, these data indicate that over a broad range of linear distances, coordinated NL contacts have a tendency to be linked to close proximity in nuclear space.
Previous Hi-C studies have revealed multi-scale compartmentalization of chromatin. At the highest level, megabase-sized domains are segregated into two main compartments that can be identified by eigenvector decomposition of the Hi-C matrix (Lieberman-Aiden et al., 2009). The CF pattern shows a remarkably tight correlation with this compartment score (Figure S4B; genome-wide Spearman’s ρ = −0.88, weighted average of by-chromosome correlations), indicating that the two main Hi-C compartments largely correspond to NL-interacting and internally located chromatin.
At a finer scale, the genome is partitioned into topologically associated domains (TADs), which are discrete domains that have many intra-domain Hi-C interactions but relatively few interactions with neighboring TADs (Dixon et al., 2012; Nora et al., 2012). We computed TAD boundary positions and compared them to the NL contact coordination patterns. We found examples where TAD boundaries coincide with borders of units of coordinated NL contacts, but also cases where they do not coincide (Figure S4B). We discovered that only TAD boundaries that coincide with strong transitions in CF values also mark the edges of of units of NL contact coordination, while TAD boundaries located in regions with relatively uniform CF values typically do not (Figure S4C). This is not due to a general difference in TAD boundary strength, because the average TAD boundary scores were highly similar (Figure S4C, bottom panels). We conclude that TADs overlap only partially with units of coordinated NL contacts.
Chromosomal CF patterns suggest long-range cooperativity
The prevalence of intra-chromosomally coordinated NL contacts raised the possibility that multiple LADs across a chromosome associate with the NL in a cooperative manner. In support of this, we noticed a strong correlation between the average CF of all LADs along each chromosome and the overall density of LADs on the same chromosome (Figure 5E; Spearman’s rho=0.85, P = 9*10−7). This is for example illustrated by the stark contrast between chr18 and chr19: the former has a very high density of LADs, of which many have consistent NL contacts, while the latter has only a few LADs that infrequently contact the NL (Figure 3A). The tight correlation between chromosomal density of LADs and their average CF is suggestive of chromosome-wide cooperative LAD–NL interactions.
NL contacts often involve embedding of DNA in the NL
To gain more insight into the nature of NL contacts at the scale of single LADs, we visualized these contacts by Ground State Depletion (GSD) super-resolution fluorescence microscopy (Folling et al., 2008) in combination with the m6A-Tracer method (Kind et al., 2013). We used a previously established clonal cell line (derived from HT1080 fibrosarcoma cells) that expresses inducible Dam-LmnB1 together with a GFP-tagged m6A-binding protein; this system allows for direct visualization of DNA that is, or has been, in contact with the NL (Kind et al., 2013). We labeled the NL in the same cells with an antibody against LmnB1 and used GSD microscopy to obtain super-resolution two-color images.
Twenty hours after Dam-LmnB1 induction the m6A-Tracer signal exhibits a striking speckled pattern that is mostly confined to a zone of ~1μm underneath the NL (Figure 6A, B). The signal consists of clusters with diameters in the range of approximately 50–300nm. A similar pattern is seen 10 hours after Dam-LmnB1 induction (Figure S5A), although the signals are more sparse than after 20 hours. Many of the m6A-Tracer signals do not touch the NL directly, indicating that they represent loci that contacted the NL in the recent past and subsequently moved over a short distance towards the nuclear interior, as noted previously (Kind et al., 2013). Nevertheless, close association of m6A-Tracer signals with LmnB1 staining is frequently observed (Figure 6A).
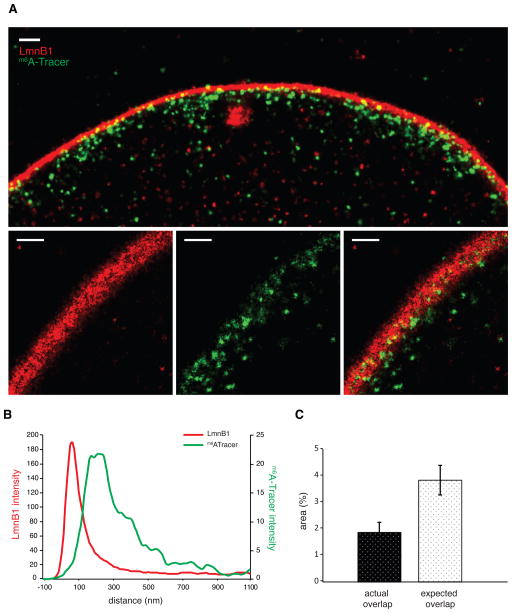
Figure 6. LAD–NL interactions involve partial embedding of chromatin in the NL.
(A) GSD microscopy image sections perpendicular (top panel) and oblique (bottom panels) to the NL. Red: LmnB1; green: m6A-Tracer signal 20 hours after induction of Dam-LmnB1. Large red structure in the center of the top panel may be an invagination of the NL. Scale bars represent 500nm. (B) Average pixel intensity of m6A-Tracer and LmnB1 signals as a function of the distance to the center of the NL. Average of 4 images. (C) Quantification of the overlap of m6A-Tracer and LaminB1 signals within the confines of the NL, compared to random overlap. Average of 4 nuclei. Error bars indicate s.d. See also Figure S5.
It is noteworthy that the edge of the LmnB1 signal at the NL is less sharply defined at the nucleoplasmic side than at the inner nuclear membrane side (Figure 6A, B). This could indicate that lamin filaments extend to varying degrees into the nuclear interior, creating a somewhat fuzzy surface. Interestingly, the m6A-Tracer signals that abut the NL often appear partially embedded in this LmnB1 meshwork (Fig 6A, top panel). Oblique sections show that m6A-Tracer signals tend to occupy small pockets in the Lamin B1 signals (Figure 6A, bottom panels). Indeed, quantitative analysis of oblique sections showed that, within the confines of the NL, the overlap between m6A-Tracer clusters and LmnB1 is less than expected by chance (Figure 6C). This embedding is not caused by thickening of the NL due to expression of Dam-LmnB1, because the thickness of the NL is similar to that of cells in which Dam-LmnB1 is not induced (Figure S5B). Together, these results show that contacts of LADs with the NL often involves embedding of chromatin in the relatively fuzzy nucleoplasmic surface of the NL.
Links between NL contacts, single-cell gene expression and chromatin state
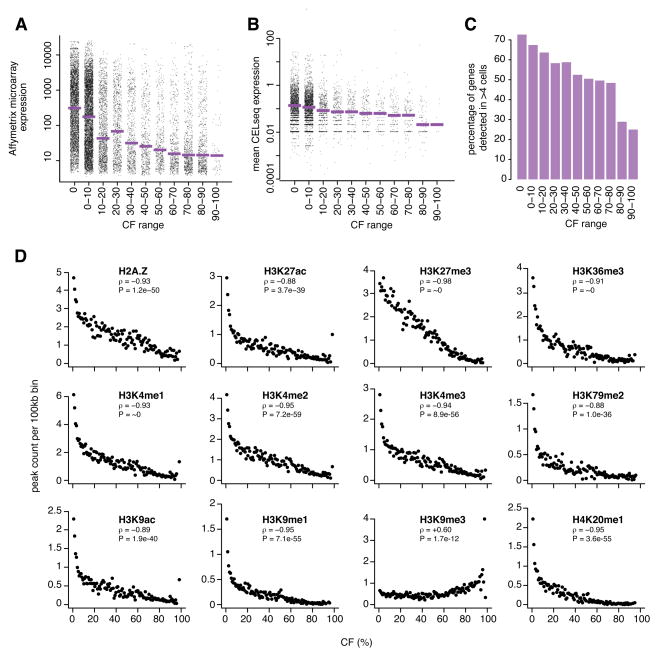
Because most NL interactions have been linked to gene repression (Finlan et al., 2008; Guelen et al., 2008; Reddy et al., 2008; Peric-Hupkes et al., 2010), we asked how transcriptional activity of genes is linked to their CF. Analysis of publicly available gene expression profiles from pools of KBM7 cells revealed that with increasing CFs the distribution of gene expression levels shifts gradually towards lower values (Figure 7A). To investigate this further, we employed a modified CEL-seq method (Grun et al., 2014) to generate genome-wide mRNA expression profiles from 96 single KBM7 cells. These data show that the average mRNA expression level, as well as the fraction of genes with detectable mRNA, decline with increasing CF values (Figure 7B, C), which is in agreement with the cell pool expression analysis. Thus, in general, the more stably a gene is associated with the NL, the less active it tends to be. We considered that genes with mid-range CFs (i.e., genes that associate with the NL in only a subset of cells) might exhibit more cell-to-cell variation in gene expression. Analysis of the CEL-seq data did not uncover such a relationship (Figure S6A-B), although we note that technical noise in CEL-seq data may may obscure such biological noise, particularly because NL-associated genes are generally expressed at very low levels.
Figure 7. Links of CF to gene expression and chromatin composition in single cells.
(A) Gene expression in pools of KBM7 cells (n=7 independent clones) as a function of CF. Dots represent genes; purple bars show median values. (B) Gene expression level in single clone #14 cells (mean of 96 single cells) as a function of CF. Dots represent genes; two genes with expression levels >100 are not shown. Purple bars show median values. Only genes with detectable expression in at least one cell are included. (C) Fraction of genes with detectable expression in at least 5 out of 96 cells, as function of CF. Genes not detected in any of the 96 cells are not counted. (D) Links of CF to epigenome mapping data from K562 cells (Consortium, 2012). See also Figure S6.
Finally, we investigated whether CFs are linked to the presence of specific histone modifications. While no genome-wide maps of these modifications are available for KBM7 cells, extensive datasets are available from K562 cells (Consortium, 2012), which is another myeloid leukemia cell line. These data reveal clear negative correlations between CFs and 10 out of 11 tested histone marks as well as the histone variant H2A.Z (Figure 7D), most of which have previously been linked to active transcription. Only H3K9me3, which is generally linked to gene repression (Consortium, 2012), is positively correlated with CF values (Spearman’s rho = 0.60, P = 1.7*10−12). This is consistent with the previously reported role of H3K9 methylation in NL tethering (Towbin et al., 2012; Bian et al., 2013). In contrast, H3K27me3, which is also linked to gene repression, shows a strong negative correlation with CFs, suggesting that it does not play a role in NL tethering in KBM7 cells.
DISCUSSION
Single-cell protein – DNA interaction mapping by DamID
Here we demonstrate that a modified DamID protocol can be used to map protein–DNA contacts genome-wide in single cells. At present, the resolution of the resulting maps is approximately 100 kb, which is suited to study proteins that form large domains along the genome (Bickmore and van Steensel, 2013). We expect that the resolution may be further improved by optimization of the DamID protocol, and by deeper sequencing of samples so that more unique reads are recovered. It will be of interest to integrate single-cell DamID mapping with the recently reported single-cell Hi-C approach (Nagano et al., 2013).
Most chromosomes have a ‘backbone’ of consistent NL interactions
One interesting outcome of this study is that about 15% of the genome contacts the NL in most of the cells (CF>80%). We propose that these consistently interacting loci together form a ‘backbone’ that may help to shape the overall chromosome architecture. Their strong overlap with constitutive LADs suggest a common backbone function in many cell types. The extremely low gene density within these loci suggests that they may have evolved to play a structural role. Not all cLADs have high CFs, but because virtually all chromosomes have multiple cLADs the backbone may be robust due to redundancy. The precise distribution of loci with stable NL contacts may be critical; this is suggested by the previously reported strong evolutionary conservation of the boundaries of cLADs (Meuleman et al., 2013). It will be of interest to investigate the consequences of deleting such stable contact sites from chromosomes.
Intra-chromosomal coordination of variable NL contacts and spatial proximity
Scattered between these stable contact sites are many loci that associate with the NL in only a subset of cells. Such loci may be subject to a balance between mechanisms that tether them to the NL and mechanisms that sequester them in the nuclear interior. These variable loci, which together cover about one third of the genome, exhibit a complex pattern of coordinated NL contacts within chromosomes. This domain-like pattern presumably reflects the overall chromosomal architecture. Indeed, our Hi-C analysis shows that loci with intra-chromosomally coordinated NL contacts tend to be in close proximity in nuclear space, particularly in the 0.5–5Mb distance range. Physical interactions between these loci may facilitate the coordination of their NL contacts. However, it is also possible that loci with coordinated NL contacts are more often in spatial proximity of one another simply because they are located in the same nuclear compartment.
Multivalent interactions and embedding of chromatin in the NL
The non-random long runs of NL contacts observed in our single-cell maps strongly suggest a multivalent mode of interaction. Considering that both the NL and chromosomes consist of polymer structures, such a mechanism is quite plausible. The Mb range over which we find this mechanism to be active appears at odds with a report claiming that a short 400bp repetitive sequence was sufficient to target a locus to the NL (Zullo et al., 2012). One possibility is that this sequence was integrated as a long tandem repeat, which often happens in stable transfections. Another study identified three independent “peripheral targeting regions” in a human LAD, one of which could be narrowed down to 6.3kb (Bian et al., 2013). However, this element was unable to target a free plasmid to the NL, while in genomic context its ability to interact with the NL was strongly dependent on the integration site, indicating that this element requires support from other elements.
H3K9me3 is strongly correlated with CF values, and several studies have found that di-and tri-methylation of H3K9 can promote NL interactions (Towbin et al., 2012; Bian et al., 2013; Kind et al., 2013). These histone modifications tend to cover large genomic regions (Bickmore and van Steensel, 2013) and are thus probable candidates to be involved in multivalent chromatin–NL contacts.
The largest m6A-Tracer clusters (~300 nm in size) that we observed by super-resolution microscopy may contain ~100–250kb of DNA, a rough estimate that we infer from previous polymer modeling of FISH distance measurements in mammalian nuclei (Mateos-Langerak et al., 2009; Giorgetti et al., 2014). However, most m6A-Tracer clusters are substantially smaller. We therefore suggest that a typical long (>1Mb) contact run as observed in the single-cell maps may consist of a string of such clusters, each typically shorter than 100 kb, that contact the NL in a multivalent manner. Future improvement of the resolution of single-cell DamID mapping may enable the identification of individual m6A-Tracer clusters at the sequence level.
EXPERIMENTAL PROCEDURES
Detailed methods are described in the Extended Experimental Procedures.
Single cell DamID
The protocol for the detection of Dam methylation in single-cell DamID is similar to that of conventional DamID (Vogel et al., 2007) and uses largely the same reagents. Key differences are (i) use of clonal cell lines with controlled expression of Dam-LmnB1; (ii) use of the Fucci system and flow sorting to collect single cells at the G1/S transition; (iii) the solution used to lyse the cells; (iv) performance of all enzymatic steps to detect Dam methylation in a single well of a 96-well plate, by sequential addition of reagents without any purification of the DNA; (v) 4–6 additional PCR cycles; (vi) use of Illumina sequencing instead of microarrays as readout. Multiplexing of samples was done with indexing primers as listed in Table S2. A detailed description is provided in the Extended Experimental Procedures, which also documents the processing of sequencing reads.
Hi-C, gene expression analysis and CEL-seq
Hi-C data from clone #14 cells were generated in duplicate and processed as described (Belton et al., 2012; Lajoie et al., 2015). Gene expression profiles from pools of KBM7 cells were obtained from Gene Expression Omnibus (GEO) accession GSE56465. CEL-seq was done essentially as described (Grun et al., 2014).
GSD microscopy
Super-resolution microscopy was performed with a Leica SR GSD microscope (Leica Microsystems, Wetzlar, Germany) with a Sumo Stage (#11888963) for drift free imaging. Images were collected with an EMCCD Andor iXon camera (Andor Technology, Belfast, UK) and an oil immersion objective (PL Apo 160X, NA 1.46). Lasers used are 405 nm/30 mW (back-pumping only), 488 nm/300 mW and 647 nm/500 mW. Between 10,000 to 50,000 frames were collected at 100 Hz for each SR image. Data were analyzed with the Image J ThunderStorm analysis module (Ovesny et al., 2014).
Data availability
Data are available from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/), accession numbers GSE69423 (single-cell and conventional DamID), GSE69841 (Hi-C), and GSE68596 (CEL-seq). Processed single-cell DamID data (OE scores) are available as Supplementary Datasets S1 – S5.
Supplementary Material
Tab-delimited file. First three columns indicate the coordinates of the genomic segments; all other columns represent OE values of individual cells. NA, no data available.
Tab-delimited file. First three columns indicate the coordinates of the genomic segments; all other columns represent OE values of individual cells. NA, no data available.
Tab-delimited file. First three columns indicate the coordinates of the genomic segments; all other columns represent OE values of individual cells. NA, no data available.
Tab-delimited file. First three columns indicate the coordinates of the genomic segments; all other columns represent OE values of individual cells. NA, no data available.
Tab-delimited file. First three columns indicate the coordinates of the genomic segments; all other columns represent OE values of individual cells. NA, no data available.
Figure S1, Related to Figure 1: Characteristics of the KBM7 #14 Dam-LmnB1 clonal line. (A) Quantitative PCR analysis of m6A accumulation without (grey bars) or with (black bars) 15 hour Dam-LmnB1 induction. All cells were FACS-sorted in the beginning of S-phase based on the expression of both red and green Fucci markers (hence such cells appear as yellow). The y-axis indicates the fraction of m6A at four specific GATC sites located either outside (iLAD1 and iLAD2) or within a LAD (LAD1 and LAD2). m6A levels were determined by resistance to DpnII digestion. (B) PCR amplification product smears resulting from 25 amplification cycles of 0, 1, 5, 25 or 125 cells without (left image) or with (right image) 15 hour Dam-LmnB1 induction. Images are different parts from the same agarose gel. The graphs below show background-corrected average pixel intensities of the smears in the corresponding gel lanes. (C) Flow cytometry profiles of clone #14. (i) DNA content profile of unsorted cells. (ii) Fucci marker profile: red marks cells in G1 phase, green marks cells in S/G2 phase, yellow marks cells in early S phase. The yellow population was used for single-cell DamID mapping. (iii) DNA content profiles of gated cell populations indicated in (ii). (iv) Same as (iii), but normalized for easier comparison of the DNA content. Note that the DNA content of the “yellow” population is just slightly higher than the G1 (red) population and much lower than the G2 peak (green most right peak), thus the great majority of the cells in the “yellow” fraction are in early S phase. (D) Immunofluorescent labeling of LmnB1 (red) without (left) or with (right) Shield1 induction for 15 hours, showing that the expression of Dam-LmnB1 does not cause alterations in the nuclear distribution of LmnB1. (E) Sequences of the adaptor and primer oligonucleotides used for single-cell DamID, and the resulting 51nt sequence read structure after PCR amplification. NNNN: random nucleotide tail added at the 5′ end of the primer; nnn...: nucleotide sequence of the amplified genomic DNA fragment. (F) Typical electrophoresis gel image after the PCR amplification step, showing 48 samples. Included are two 20-cell positive controls, two negative controls with no cells, and 44 single-cell samples. (G) Overview of the 51bp reads obtained from 118 single-cell Dam-LmnB1 samples. The raw reads (1.2 * 106 on average) are first filtered on the correct adaptor structure (68%), then uniquely aligned to the human genome (40%), removing those reads that do not start with the expected GATC motif (37%). Finally all duplicate reads are removed, resulting in a list of unique GATC-flanked mappable reads (13%). (H) Histogram of the distribution of uniquely mappable read numbers across the 118 single-cell Dam-LmnB1 samples.
Figure S2, Related to Figure 3: OE value distribution and characterization of additional KBM7 clones. (A) Distribution of OE values in 118 clone #14 single-cell datasets. The vertical black dotted line at OE=1 indicates the cut-off used in this study to define NL contacts. Values above the horizontal red dotted line were clipped. (B) Scatterplot comparing the average OE values to CF. Green line depicts linear regression fit; Spearman’s ρ = 0.98. (C) Scatterplot comparing single-cell #14 data subsampled to the one-but-lowest read count (104k) to CFs obtained with the original data. Orange line depicts the diagonal (X = Y); Spearman’s ρ = 0.99. (D) Scatterplot comparing the estimated CFs for all 100kb segments in clones #14 and #5.5. Green line depicts linear regression fit; Spearman’s ρ = 0.94. (E) Comparison of CF maps of clone #14 (dark blue) and clone #5.5 (light blue) for a subset of chromosomes. (F) Quantitative PCR analysis of m6A accumulation 20 hours after Dam-LmnB1 induction in clone #14 (left) or clone #5.5 (right). The y-axis indicates the fraction of m6A at a GATC site within a LAD (LAD2). (G) Flow cytometry profiles of clone #5.5. (i) DNA content profile of unsorted cells. (ii) Fucci marker profile: red marks cells in G1 phase, green marks cells in S/G2 phase, green plus red (yellow) marks cells in early S phase. The yellow population was used for single-cell DamID mapping. (iii) DNA content profiles of gated cell populations indicated in (ii). (iv) Same as (iii), but normalized for easier comparison of the DNA content. Note that the DNA content of the “yellow” population is just slightly higher than the G1 (red) population and much lower than the G2 peak (green most right peak), thus the great majority of the cells in the “yellow” fraction are in early S phase. (H) Comparison of CFs in clone #5.5 diploid cells and pseudo-diploid cells. The latter are simulated by combining equal numbers of sequence reads from pairs of clone #5.5 haploid cells. Note that the effect of diploidization is similar to that observed for clone #14 (Figure 3E).
Figure S3, Related to Figure 4: Stretches of no-contact are not due to loss of DNA during sample processing. (A) Length distribution of genome-wide NL no-contact runs (OE<1) in clone #14 (red) compared to length distribution of regions lacking mappable sequence reads (OE=0) in the Dam-only (green) clone. Note that the continuous regions without any reads are much shorter than the Dam-LmnB1 no-contact runs, indicating that the no-contact runs reflect detachment of extended regions from the NL and are not due to loss of long DNA stretches during sample processing.
Figure S4, Related to Figure 5: Additional analyses of NL contact coordination. (A) Average Pearson correlation of binary NL contacts across 118 clone #14 cells, for all possible pairs of 100 kb segments within (red) or between (blue) chromosomes. Vertical axis: experimental data; horizontal axis: after randomization of NL contacts (average of 100 randomizations). Note that within-chromosome correlations are higher than may be expected by random chance, while this is not the case for between-chromosome correlations. (B) Comparison of NL contact coordination to Hi-C data and derivative parameters. Top panels: four examples of NL contact coordination (upper-right triangles) compared to Hi-C interactions (lower-left triangles). Lower three panels show CF, compartment score based on Hi-C data, and directionality score based on Hi-C data for the respective genomic regions. (C) Average NL contact correlation maps around TAD boundaries, stratified by change in CF (ΔCF) across the boundaries. Correlation maps are oriented such that higher CF regions are in the upper left quadrant (i.e., ΔCF decreases over the TAD boundary). A profile of average CFs around each class of boundaries is shown below each map. These boundaries were then assigned a ΔCF by considering the difference in CF for the 300kb upstream and downstream of the TAD boundary. At this 100kb resolution, this shows that LAD-internal TAD boundaries do not break these correlations, unless they coincide with high ΔCF (i.e. they tend to be at LAD boundaries).
Figure S5, Related to Figure 6: Additional super-resolution microscopy analyses. (A) GSD microscopy image section perpendicular to the NL 10 hours after induction of Dam-LmnB1. Red: LmnB1; green: m6A-Tracer signal. Some invaginations of the NL are visible in the nuclear interior. Scale bar, 500nm. (B) GSD image quantification of the width (nm) of the nuclear lamina 0, 10 and 20 hours after induction of Dam-LmnB1. The width of the nuclear lamina is measured based on immunofluorescence labeling of LmnB1. Bars represent the average of the measurements of 4 (0 hrs), 3 (10 hrs) and 5 (20 hrs) cells (± s.d.).
Figure S6, Related to Figure 7: Cell-to-cell variation in gene expression is not detectably linked to CF. (A) For all genes, we calculated cell-to-cell variation of transcript abundance across the 96 cells as a Coefficient of Variance (CV = s.d./mean). This CV is inversely correlated to the average mRNA expression, presumably reflecting Poissonian sampling noise. (B) Because average expression levels are also correlated with CF values (Figure 7B), we analyzed the correlation between expression CV and CF conditional on average expression levels in order to remove average expression level as a confounding factor. The result shows that for any group of genes with similar expression levels the CV is largely independent of CF values.
Table S1. Gene ontology analysis of genes located in segments with CF>80% – Related to Figure 3.
Table S2. List of indexing primers used for multiplexing of single--cell DamID samples – Related to Experimental Procedures. Positions of the Illumina sequencing primer index sequence (cyan) are highlighted in the first sequence entry.
Acknowledgments
We thank the NKI Genomics Core Facility for technical assistance, Thijn Brummelkamp for help with KBM7 cells, Erik Wernersson for FISH image analysis, and members of the BvS lab for helpful comments. This study was supported by a Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO) ALW-VENI fellowship (JK); NHMRC Early Career Fellowship (CAdG); NWO ZonMW-TOP and European Research Council (ERC) Advanced Grant AdG 293662-CHROMATINPRINCIPLES (BvS); STW grant 12150 (KJ); ERC Advanced Grant AdG 294325-GeneNoiseControl and NWO VICI Award (SSD, MB, AvO); National Institute of Health R01 GM114190 (LM, MI, GF); and National Human Genome Research Institute R01 HG003143 (JD, LM, MI, GF).
Footnotes
AUTHOR CONTRIBUTIONS.
JK: conceived & designed study; single-cell DamID development & experiments; data exploration; paper writing. LP and SSdV made equal contributions. LP: data processing pipeline; data analysis. SSdV: clonal cell line establishment, single-cell DamID, control experiments and experimental design. LN: superresolution microscopy; SSD: CEL-seq experiments & data analysis; MB: FISH experiments & data analysis. YZ: Hi-C experiments; BL: Hi-C data processing; CAdG: unpublished LAD atlas; data processing support; MA: constructs, experimental support. GF: data analysis; MI: data analysis; LM: advisor of GF and MI, data analysis, intellectual input; KJ: advisor of LN, superresolution microscopy experimental design and image analysis; JD: advisor of YZ and BL; intellectual input. AvO: advisor of SSD and MB; intellectual input; BvS: conceived & designed study; data analysis; project coordination; paper writing.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bau D, Sanyal A, Lajoie BR, Capriotti E, Byron M, Lawrence JB, Dekker J, Marti-Renom MA. The three-dimensional folding of the alpha-globin gene domain reveals formation of chromatin globules. Nat Struct Mol Biol. 2011;18:107–114. doi: 10.1038/nsmb.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belton JM, McCord RP, Gibcus JH, Naumova N, Zhan Y, Dekker J. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods. 2012;58:268–276. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Q, Khanna N, Alvikas J, Belmont AS. beta-Globin cis-elements determine differential nuclear targeting through epigenetic modifications. J Cell Biol. 2013;203:767–783. doi: 10.1083/jcb.201305027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickmore WA. The spatial organization of the human genome. Annu Rev Genomics Hum Genet. 2013;14:67–84. doi: 10.1146/annurev-genom-091212-153515. [DOI] [PubMed] [Google Scholar]
- Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Muller S, Eils R, Cremer C, Speicher MR, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckstummer T, Banning C, Hainzl P, Schobesberger R, Kerzendorfer C, Pauler FM, Chen D, Them N, Schischlik F, Rebsamen M, et al. A reversible gene trap collection empowers haploid genetics in human cells. Nat Methods. 2013;10:965–971. doi: 10.1038/nmeth.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G, Misteli T. Functional implications of genome topology. Nat Struct Mol Biol. 2013;20:290–299. doi: 10.1038/nsmb.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer M, von Hase J, Volm T, Brero A, Kreth G, Walter J, Fischer C, Solovei I, Cremer C, Cremer T. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 2001;9:541–567. doi: 10.1023/a:1012495201697. [DOI] [PubMed] [Google Scholar]
- Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folling J, Bossi M, Bock H, Medda R, Wurm CA, Hein B, Jakobs S, Eggeling C, Hell SW. Fluorescence nanoscopy by ground-state depletion and single-molecule return. Nat Methods. 2008;5:943–945. doi: 10.1038/nmeth.1257. [DOI] [PubMed] [Google Scholar]
- Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013;49:773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti L, Galupa R, Nora EP, Piolot T, Lam F, Dekker J, Tiana G, Heard E. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell. 2014;157:950–963. doi: 10.1016/j.cell.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun D, Kester L, van Oudenaarden A. Validation of noise models for single-cell transcriptomics. Nat Methods. 2014;11:637–640. doi: 10.1038/nmeth.2930. [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol. 2012;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- Kotecki M, Reddy PS, Cochran BH. Isolation and characterization of a near-haploid human cell line. Exp Cell Res. 1999;252:273–280. doi: 10.1006/excr.1999.4656. [DOI] [PubMed] [Google Scholar]
- Lajoie BR, Dekker J, Kaplan N. The Hitchhiker’s guide to Hi-C analysis: practical guidelines. Methods. 2015;72:65–75. doi: 10.1016/j.ymeth.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis L, Borden J. Reproducible compartmentalization of individual chromosome domains in human CNS cells revealed by in situ hybridization and three-dimensional reconstruction. Chromosoma. 1988;96:397–410. doi: 10.1007/BF00303033. [DOI] [PubMed] [Google Scholar]
- Marshall WF, Dernburg AF, Harmon B, Agard DA, Sedat JW. Specific interactions of chromatin with the nuclear envelope: positional determination within the nucleus in Drosophila melanogaster. Mol Biol Cell. 1996;7:825–842. doi: 10.1091/mbc.7.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos-Langerak J, Bohn M, de Leeuw W, Giromus O, Manders EM, Verschure PJ, Indemans MH, Gierman HJ, Heermann DW, van Driel R, et al. Spatially confined folding of chromatin in the interphase nucleus. Proc Natl Acad Sci U S A. 2009;106:3812–3817. doi: 10.1073/pnas.0809501106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuleman W, Peric-Hupkes D, Kind J, Beaudry JB, Pagie L, Kellis M, Reinders M, Wessels L, van Steensel B. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2013;23:270–280. doi: 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs RL, Press RI. Centromere autoantigens are associated with the nucleolus. Exp Cell Res. 1992;200:339–350. doi: 10.1016/0014-4827(92)90181-7. [DOI] [PubMed] [Google Scholar]
- Ovesny M, Krizek P, Borkovec J, Svindrych Z, Hagen GM. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014;30:2389–2390. doi: 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Therizols P, Illingworth RS, Courilleau C, Boyle S, Wood AJ, Bickmore WA. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science. 2014;346:1238–1242. doi: 10.1126/science.1259587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin BD, Gonzalez-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Dekker J. Genomics tools for unraveling chromosome architecture. Nat Biotechnol. 2010;28:1089–1095. doi: 10.1038/nbt.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel MJ, Peric-Hupkes D, van Steensel B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat Protoc. 2007;2:1467–1478. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- Zullo JM, Demarco IA, Pique-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Bernstein BE, Pritchard JK, Reddy KL, et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–1487. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tab-delimited file. First three columns indicate the coordinates of the genomic segments; all other columns represent OE values of individual cells. NA, no data available.
Tab-delimited file. First three columns indicate the coordinates of the genomic segments; all other columns represent OE values of individual cells. NA, no data available.
Tab-delimited file. First three columns indicate the coordinates of the genomic segments; all other columns represent OE values of individual cells. NA, no data available.
Tab-delimited file. First three columns indicate the coordinates of the genomic segments; all other columns represent OE values of individual cells. NA, no data available.
Tab-delimited file. First three columns indicate the coordinates of the genomic segments; all other columns represent OE values of individual cells. NA, no data available.
Figure S1, Related to Figure 1: Characteristics of the KBM7 #14 Dam-LmnB1 clonal line. (A) Quantitative PCR analysis of m6A accumulation without (grey bars) or with (black bars) 15 hour Dam-LmnB1 induction. All cells were FACS-sorted in the beginning of S-phase based on the expression of both red and green Fucci markers (hence such cells appear as yellow). The y-axis indicates the fraction of m6A at four specific GATC sites located either outside (iLAD1 and iLAD2) or within a LAD (LAD1 and LAD2). m6A levels were determined by resistance to DpnII digestion. (B) PCR amplification product smears resulting from 25 amplification cycles of 0, 1, 5, 25 or 125 cells without (left image) or with (right image) 15 hour Dam-LmnB1 induction. Images are different parts from the same agarose gel. The graphs below show background-corrected average pixel intensities of the smears in the corresponding gel lanes. (C) Flow cytometry profiles of clone #14. (i) DNA content profile of unsorted cells. (ii) Fucci marker profile: red marks cells in G1 phase, green marks cells in S/G2 phase, yellow marks cells in early S phase. The yellow population was used for single-cell DamID mapping. (iii) DNA content profiles of gated cell populations indicated in (ii). (iv) Same as (iii), but normalized for easier comparison of the DNA content. Note that the DNA content of the “yellow” population is just slightly higher than the G1 (red) population and much lower than the G2 peak (green most right peak), thus the great majority of the cells in the “yellow” fraction are in early S phase. (D) Immunofluorescent labeling of LmnB1 (red) without (left) or with (right) Shield1 induction for 15 hours, showing that the expression of Dam-LmnB1 does not cause alterations in the nuclear distribution of LmnB1. (E) Sequences of the adaptor and primer oligonucleotides used for single-cell DamID, and the resulting 51nt sequence read structure after PCR amplification. NNNN: random nucleotide tail added at the 5′ end of the primer; nnn...: nucleotide sequence of the amplified genomic DNA fragment. (F) Typical electrophoresis gel image after the PCR amplification step, showing 48 samples. Included are two 20-cell positive controls, two negative controls with no cells, and 44 single-cell samples. (G) Overview of the 51bp reads obtained from 118 single-cell Dam-LmnB1 samples. The raw reads (1.2 * 106 on average) are first filtered on the correct adaptor structure (68%), then uniquely aligned to the human genome (40%), removing those reads that do not start with the expected GATC motif (37%). Finally all duplicate reads are removed, resulting in a list of unique GATC-flanked mappable reads (13%). (H) Histogram of the distribution of uniquely mappable read numbers across the 118 single-cell Dam-LmnB1 samples.
Figure S2, Related to Figure 3: OE value distribution and characterization of additional KBM7 clones. (A) Distribution of OE values in 118 clone #14 single-cell datasets. The vertical black dotted line at OE=1 indicates the cut-off used in this study to define NL contacts. Values above the horizontal red dotted line were clipped. (B) Scatterplot comparing the average OE values to CF. Green line depicts linear regression fit; Spearman’s ρ = 0.98. (C) Scatterplot comparing single-cell #14 data subsampled to the one-but-lowest read count (104k) to CFs obtained with the original data. Orange line depicts the diagonal (X = Y); Spearman’s ρ = 0.99. (D) Scatterplot comparing the estimated CFs for all 100kb segments in clones #14 and #5.5. Green line depicts linear regression fit; Spearman’s ρ = 0.94. (E) Comparison of CF maps of clone #14 (dark blue) and clone #5.5 (light blue) for a subset of chromosomes. (F) Quantitative PCR analysis of m6A accumulation 20 hours after Dam-LmnB1 induction in clone #14 (left) or clone #5.5 (right). The y-axis indicates the fraction of m6A at a GATC site within a LAD (LAD2). (G) Flow cytometry profiles of clone #5.5. (i) DNA content profile of unsorted cells. (ii) Fucci marker profile: red marks cells in G1 phase, green marks cells in S/G2 phase, green plus red (yellow) marks cells in early S phase. The yellow population was used for single-cell DamID mapping. (iii) DNA content profiles of gated cell populations indicated in (ii). (iv) Same as (iii), but normalized for easier comparison of the DNA content. Note that the DNA content of the “yellow” population is just slightly higher than the G1 (red) population and much lower than the G2 peak (green most right peak), thus the great majority of the cells in the “yellow” fraction are in early S phase. (H) Comparison of CFs in clone #5.5 diploid cells and pseudo-diploid cells. The latter are simulated by combining equal numbers of sequence reads from pairs of clone #5.5 haploid cells. Note that the effect of diploidization is similar to that observed for clone #14 (Figure 3E).
Figure S3, Related to Figure 4: Stretches of no-contact are not due to loss of DNA during sample processing. (A) Length distribution of genome-wide NL no-contact runs (OE<1) in clone #14 (red) compared to length distribution of regions lacking mappable sequence reads (OE=0) in the Dam-only (green) clone. Note that the continuous regions without any reads are much shorter than the Dam-LmnB1 no-contact runs, indicating that the no-contact runs reflect detachment of extended regions from the NL and are not due to loss of long DNA stretches during sample processing.
Figure S4, Related to Figure 5: Additional analyses of NL contact coordination. (A) Average Pearson correlation of binary NL contacts across 118 clone #14 cells, for all possible pairs of 100 kb segments within (red) or between (blue) chromosomes. Vertical axis: experimental data; horizontal axis: after randomization of NL contacts (average of 100 randomizations). Note that within-chromosome correlations are higher than may be expected by random chance, while this is not the case for between-chromosome correlations. (B) Comparison of NL contact coordination to Hi-C data and derivative parameters. Top panels: four examples of NL contact coordination (upper-right triangles) compared to Hi-C interactions (lower-left triangles). Lower three panels show CF, compartment score based on Hi-C data, and directionality score based on Hi-C data for the respective genomic regions. (C) Average NL contact correlation maps around TAD boundaries, stratified by change in CF (ΔCF) across the boundaries. Correlation maps are oriented such that higher CF regions are in the upper left quadrant (i.e., ΔCF decreases over the TAD boundary). A profile of average CFs around each class of boundaries is shown below each map. These boundaries were then assigned a ΔCF by considering the difference in CF for the 300kb upstream and downstream of the TAD boundary. At this 100kb resolution, this shows that LAD-internal TAD boundaries do not break these correlations, unless they coincide with high ΔCF (i.e. they tend to be at LAD boundaries).
Figure S5, Related to Figure 6: Additional super-resolution microscopy analyses. (A) GSD microscopy image section perpendicular to the NL 10 hours after induction of Dam-LmnB1. Red: LmnB1; green: m6A-Tracer signal. Some invaginations of the NL are visible in the nuclear interior. Scale bar, 500nm. (B) GSD image quantification of the width (nm) of the nuclear lamina 0, 10 and 20 hours after induction of Dam-LmnB1. The width of the nuclear lamina is measured based on immunofluorescence labeling of LmnB1. Bars represent the average of the measurements of 4 (0 hrs), 3 (10 hrs) and 5 (20 hrs) cells (± s.d.).
Figure S6, Related to Figure 7: Cell-to-cell variation in gene expression is not detectably linked to CF. (A) For all genes, we calculated cell-to-cell variation of transcript abundance across the 96 cells as a Coefficient of Variance (CV = s.d./mean). This CV is inversely correlated to the average mRNA expression, presumably reflecting Poissonian sampling noise. (B) Because average expression levels are also correlated with CF values (Figure 7B), we analyzed the correlation between expression CV and CF conditional on average expression levels in order to remove average expression level as a confounding factor. The result shows that for any group of genes with similar expression levels the CV is largely independent of CF values.
Table S1. Gene ontology analysis of genes located in segments with CF>80% – Related to Figure 3.
Table S2. List of indexing primers used for multiplexing of single--cell DamID samples – Related to Experimental Procedures. Positions of the Illumina sequencing primer index sequence (cyan) are highlighted in the first sequence entry.
Data Availability Statement
Data are available from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/), accession numbers GSE69423 (single-cell and conventional DamID), GSE69841 (Hi-C), and GSE68596 (CEL-seq). Processed single-cell DamID data (OE scores) are available as Supplementary Datasets S1 – S5.