Abstract
Borrelia burgdorferi , the agent of Lyme disease, spreads from the site of the tick bite to tissues such as heart, joints and the nervous system. Host glycosaminoglycans (GAGs), highly modified repeating disaccharides that are present on cell surfaces and in extracellular matrix, are common targets of microbial pathogens during tissue colonization. While several dermatan sulfate-binding B. burgdorferi adhesins have been identified, B. burgdorferi adhesins documented to promote spirochetal binding to heparan sulfate have not yet been identified. OspEF-related proteins (Erps), a large family of plasmid-encoded surface lipoproteins that are produced in the mammalian host, can be divided into the OspF-related, OspEF leader peptide (Elp), and OspE-related subfamilies. We show here that a member of the OspF-related subfamily, ErpG, binds to heparan sulfate, and when produced on the surface of an otherwise nonadherent B. burgdorferi strain, ErpG promotes heparan sulfate-mediated bacterial attachment to glial but not endothelial, synovial or respiratory epithelial cells. Six other OspF-related proteins were capable of binding heparan sulfate, whereas representative OspE-related and Elp proteins lacked this activity. These results indicate that OspF-related proteins are heparan sulfate-binding adhesins, at least one of which promotes bacterial attachment to glial cells.
Keywords: Lyme disease, glycosaminoglycan, Erp, adhesin
INTRODUCTION
Lyme disease, the most common arthropod-borne disease in the United States and Europe, is caused by the spirochete Borrelia burgdorferi sensu lato (Steere et al., 2004). Following a tick bite, this motile spirochete first causes a local skin infection, usually resulting in the characteristic rash “erythema migrans”, and may then spread to a variety of tissues including skin, joints, heart and nervous tissue (Radolf et al., 2012, Stanek et al., 2012). Indeed, the clinical manifestations of Lyme disease are quite variable and have been associated with the genotype of the infecting strain (Wang et al., 1999). Attachment to host tissues is a critical step in tissue colonization by many pathogens (Finlay et al., 1997), and B. burgdorferi can attach to a wide variety of mammalian cells, including lymphocytes, platelets, epithelial cells, endothelial cells, and neuroglia (Comstock et al., 1989, Garcia-Monco et al., 1989, Thomas et al., 1989, Szczepanski et al., 1990). Among host cell ligands recognized by this spirochete are integrins (Coburn et al., 1993), fibronectin (Probert et al., 1998, Brissette et al., 2009a, Moriarty et al., 2012, Gaultney et al., 2013), laminin (Brissette et al., 2009b, Verma et al., 2009), collagen (Zambrano et al., 2004, Zhi et al., 2015), and proteoglycans (Isaacs, 1994, Leong et al., 1995, Guo et al., 1998, Parveen et al., 2000, Parveen et al., 2003, Fischer et al., 2006).
Proteoglycans consist of a core protein and one or more covalently linked glycosaminoglycan (GAG) chains (Aquino et al., 2010). GAGs are long repeating disaccharides that can be divided into different classes, such as chondroitin 6-sulfate, dermatan sulfate, and heparan sulfate, depending upon the sugar composition, the glycosidic linkage between each monosaccharide, and the degrees and types of sulfation (Aquino et al., 2010). Upon adaptation to the mammalian host, B. burgdorferi exhibits enhanced dermatan sulfate- and heparin-binding activity (Parveen et al., 2003), and spirochetes deficient in some GAG-binding adhesins have been shown to be reduced in infectivity in a murine model, indicating a critical role for GAG-binding during mammalian infection (Seshu et al., 2006, Blevins et al., 2008, Shi et al., 2008, Weening et al., 2008). We recently showed that GAG-binding by the B. burgdorferi adhesin BBK32 specifically promotes joint colonization and disease (Lin et al., 2014b), so GAG-binding activities may play roles in colonization of specific tissues.
Although all infectious strains of Lyme disease spirochete are capable of binding to GAGs (Parveen et al., 1999), the specific subclasses of GAGs recognized vary among strains (Parveen et al., 1999). This variation appears to be due in part to allelic variation of one or more GAG-binding adhesins. The adhesin DbpA, which recognizes dermatan sulfate and the proteoglycan decorin, is allelically variable (Roberts et al., 1998), and DbpA sequence variation results in differences in binding to purified ligands in vitro (Benoit et al., 2011, Salo et al., 2011). In turn, GAG-binding variation among Lyme disease strains is associated with differences in cell type-specific attachment in vitro and tissue tropism in vivo. A characterization of GAG-binding specificity of six Lyme disease spirochete strains revealed that one group recognized primarily dermatan sulfate, another heparan sulfate, and a third both dermatan and heparan sulfate (Parveen et al., 1999). The GAG-binding preference of a particular strain is reflected in its cell-type binding specificity, because strains that recognized predominantly dermatan sulfate bound to cultured glial cells more efficiently than to endothelial cells, whereas strains that recognized heparan sulfate bound to both cultured glial and endothelial cells (Parveen et al., 1999). Finally, isogenic strains expressing different dbpA alleles display differences in tissue tropism, indicating that allelic variation of GAG-binding adhesins may contribute to strain-dependent differences in tissue tropism (Lin et al., 2014a).
Strain-specific GAG-binding variation might also reflect the production of different sets of GAG-binding adhesins, each with different GAG binding specificities. Several B. burgdorferi surface proteins, when in recombinant form, bind to different subclasses of GAGs (Parveen et al., 2000, Parveen et al., 2003, Fischer et al., 2006). A few B. burgdorferi surface proteins, when ectopically produced on the surface of a high passage, noninfectious and otherwise nonadherent B. burgdorferi strains, have been documented to promote spirochetal attachment to different subsets of GAGs, such as dermatan sulfate and/or chondroitin-6-sulfate (Fischer et al., 2003, Fischer et al., 2006, Benoit et al., 2011). In spite of the observation that infectious Lyme disease strains bind to heparan sulfate (Parveen et al., 1999), no such well-documented adhesin that confers heparan sulfate binding has been identified.
B. burgdorferi sensu lato strains carry 6 to 10 homologous 32-kb circular plasmids (termed cp32’s) encoding a family of surface lipoproteins termed OspEF-related (i.e. Erp) proteins (Akins et al., 1995, Marconi et al., 1996, Stevenson et al., 1996, Akins et al., 1999) (for review, see (Brissette et al., 2008)). The Erp proteins share a high degree of similarity in their respective promoter regions (previously given the term “upstream homology box” or “UHB” (Marconi et al., 1996)) and leader peptides. The production of many Erp proteins responds to signals that reflect the mammalian environment, such as higher temperature or growth in the dialysis membrane chambers implanted into rats (Akins et al., 1995, Stevenson et al., 1995, Babb et al., 2001, Hefty et al., 2001, Ojaimi et al., 2003). Erp proteins are produced during mammalian infection, although differences in their temporal expression patterns during infection suggest possible divergence of function (McDowell et al., 2001, Miller et al., 2006a, Miller et al., 2006b). Erp proteins can be divided into three subfamilies on the basis of homology of their respective mature peptide sequences: OspE-related and OspF-related proteins, and OspEF-leader peptide (Elp) proteins (Akins et al., 1999, Caimano et al., 2000). OspE-related proteins bind to host plasminogen and to mammalian complement regulator factor H and factor H-related proteins (Brissette et al., 2008, Kenedy et al., 2011, Kraiczy et al., 2013). ErpX, an Elp protein, has been shown to bind to the ECM component laminin in vitro (Brissette et al., 2009b). Finally, fragments of four OspF-related proteins (i.e. ErpK, ErpL, BBK2.10, and OspG) from B. burgdorferi strain N40-D10/E9, when artificially produced on the surface of filamentous bacteriophage, promoted phage localization to the joints, heart, or bladder, suggesting that these proteins play a role in adhesion (Antonara et al., 2007). Spirochetes deficient in ErpK or OspF showed reduced colonization activities at the joints, heart, and skin (Lin et al., 2012).
In this study, we showed that the OspF-related protein ErpG (i.e. the strain B31 homolog of OspG of strain N40-D10/E9) bound to purified heparan sulfate. When produced on the surface of an otherwise poorly adherent B. burgdorferi strain, ErpG promoted the attachment of spirochetes to heparan sulfate and to cultured glial but not endothelial, synovial or respiratory epithelial cells. We also showed that six other OspF-related proteins are capable of binding heparan sulfate, whereas representatives of OspE-related or Elp proteins lack this activity. These results suggest that OspF-related proteins are B. burgdorferi heparan sulfate-binding adhesins.
RESULTS
Recombinant ErpG binds to heparin and heparan sulfate
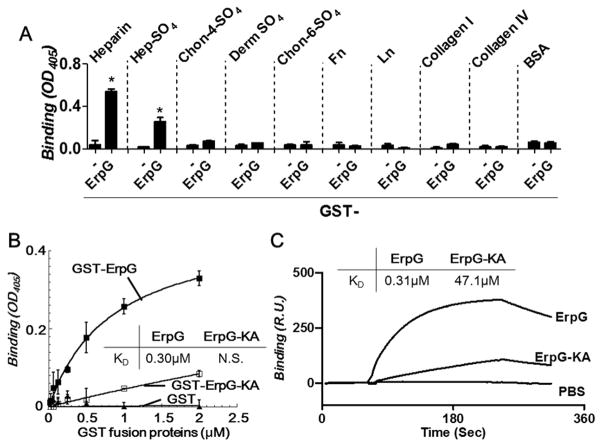
Artificial production of several strain N40-D10/E9 OspF-related proteins, including OspG, on the surface of phages was associated with the localization of recombinant phages to the murine joint, bladder, or heart after intravenous inoculation (Antonara et al., 2007). A glutathione-S-transferase (GST)-tagged recombinant version of strain B31 ErpG, a homolog of strain N40-D10/E9 OspG, was therefore tested for its ability to bind to ECM components such as fibronectin, laminin, vitronectin, collagen, elastin, or a variety of subclasses of GAGs. As expected, the control protein GST did not bind to any of the substrates tested (Fig. 1A and data not shown). In contrast, GST-ErpG bound to heparin and heparan sulfate, but not to BSA or to other ECM components, including chondroitin-4-sulfate, dermatan sulfate, or chondroitin-6-sulfate (Fig. 1A and data not shown). The heparan sulfate-binding affinity of ErpG was measured using quantitative ELISA and surface plasmon resonance (SPR). GST-ErpG bound to heparan sulfate in a dose dependent manner in a quantitative ELISA assay and with an affinity of 0.30±0.08μM (Fig. 1B; Table 1). The heparan sulfate-binding affinity of ErpG determined by SPR (0.31±0.01μM) was entirely consistent with the ELISA results (Fig. 1C; Table 1). Thus, ErpG is a GAG-binding protein that binds specifically to heparin and heparan sulfate.
Figure 1. Recombinant ErpG proteins bind to heparan sulfate.
(A) 2 μM recombinant GST-tagged ErpG or GST (as a negative control) was added to quadruplicate wells coated with heparin, heparan sulfate (Hep-SO4), chondroitin-4-sulfate (Chon-4-SO4), dermatan sulfate (Derm SO4), chondroitin-6-sulfate (Chon-6-SO4), fibronectin (Fn), laminin, (Ln), type I collagen (Collagen I), or type IV collagen (Collagen IV). Bound protein was measured by ELISA (see Experimental Procedures) and mean OD405 ± standard deviation was determined. Asterisks indicate that binding of GST-ErpG to heparin or heparan sulfate was statistically (p≤0.05 by Student’s t test) different than GST. Shown is a representative of three independently performed experiments. (B) The indicated concentrations of recombinant GST-ErpG, GST-ErpG-KA (i.e., the ErpG double alanine substitution at residues 195-196) or GST were added to quadruplicate wells coated with heparan sulfate, and protein binding was quantified by ELISA and mean OD405 ± standard deviation was determined. GST-ErpG bound to heparan sulfate significantly better than GST (p≤0.05 by Student’s t test). KD values obtained from the average of three independent experiments are shown in the inset. The binding of GST-ErpG-KA to heparan sulfate was not saturated (indicated as “N.S.”) and therefore the KD could not be determined. Shown is a representative of three independently performed experiments. (C) Varying concentrations (15.625 to 500 nM) of GST-tagged ErpG or ErpG-KA was flowed over a surface coated with 10 μg heparan sulfate. Binding was measured in response units (RU) by SPR (see Experimental Procedures). Shown is a representative of six experiments performed on three different occasions. KD values obtained from the average of three independent experiments are shown in the inset. (The kon, koff, and KD values are shown in Table 1.)
Table 1. Affinitiy of Erp proteins binding to heparan sulfate.
KD values were obtained from average of six experiments performed on three different occasions by SPR.
| Erp proteins | ELISA analysis | Surface Plasmon Resonance Analysis | ||
|---|---|---|---|---|
| KD (μM)b | KD (μM)a | kon (105s−1M−1) | koff (s−1) | |
| OspF family | ||||
| ErpG | 0.30±0.08 | 0.31±0.01 | 24.65±2.36 | 5.88±0.14 |
| ErpG-KA | n.s.a | 47.1±8.48b | 0.77±0.08b | 3.53±0.02b |
| ErpK | 0.42±0.07 | 0.44±0.03 | 26.10±2.36 | 11.70±0.18 |
| ErpL | 0.40±0.08 | 0.42±0.04 | 12.65±1.83 | 5.33±0.82 |
| ErpY | n.d.c | 2.31±0.12 | 4.53±1.06 | 6.90±0.94 |
| Erp25 | n.d.c | 1.72±0.60 | 5.02±0.77 | 8.64±0.17 |
| Erp27 | n.d.c | 2.52±0.33 | 3.09±0.27 | 7.64±0.33 |
| OspF | n.d.c | 8.17±0.35 | 6.83±0.13 | 5.51±0.86 |
| OspE family | ||||
| ErpP | n.d.c | n.b.d | n.b.d | n.b.d |
| Elp family | ||||
| ErpX | n.d.c | n.b.d | n.b.d | n.b.d |
All values represent the mean ± SEM of three experiments.
Not saturated
Not accurate due to weak binding
Not determined
No binding
Basic amino acids usually participate in recognition of GAGs, which are highly negatively charged (Hileman et al., 1998, Aquino et al., 2010). Three independent surface display phage clones that encode slightly different portions of N40-D10/E9 OspG were enriched for vascular binding in mice, and 22 basic amino acids were common to all three sequences (Antonara et al., 2007). Two adjacent lysine (K) residues (KK195”) among these residues were replaced with alanine residues in the homologous ErpG protein to generate a recombinant ErpG mutant that we term “ErpG-KA” (Fig. S1). Far-UV CD analysis indicated that the secondary structure of ErpG was unaltered by this double K to A substitution (Fig. S2), Although ErpG-KA retained partial heparan sulfate-binding activity, the affinity of this mutant, with a KD of ~47 μM, was reduced approximately 150-fold compared to wild type ErpG, indicating that the KK195 sequence is important for the heparan sulfate-binding activity of ErpG (Fig. 1B and C; Table 1).
Ectopic surface production of ErpG confers heparin- and heparan sulfate-binding activity on the otherwise non-adhesive strain B314
To test the GAG-binding activity of ErpG produced on the spirochetal outer membrane, this protein was ectopically produced in the non-infectious and non-adhesive B. burgdorferi strain B314 using the pBSV2-based shuttle vector pErpG (Table S1; (Sadziene et al., 1993, Stewart et al., 2001)). Although strain B314 carries plasmid cp32-3, which encodes erpG, it does not express this endogenous gene under the conditions used in this study (Fig. S3A and C). Strain B314/pErpG produced ErpG that was degraded upon addition of proteinase K to intact spirochetes, indicating that ErpG was exposed on the bacterial surface (Fig. S3A). To better quantify ErpG surface expression, strain B314/pErpG, or the control strain B314 harboring the parental vector pJF21, was stained with anti-ErpG and subjected to flow cytometric analysis (Fig. S3C). Analysis of the mean fluorescence index of stained bacteria, which reflects the relative amount of surface-exposed ErpG, indicated that B314/pErpG produced surface ErpG at a level indistinguishable from that produced by the infectious isolate B31, from which strain B314 was originally derived (Sadziene et al., 1993) (Fig. S3C).
To determine whether ErpG located on the cell surface promotes spirochete attachment to GAGs, [35S] methionine-labeled strain B314/pJF21 and B314/pErpG were tested for the ability to bind to wells coated with different species of GAG, or with BSA as a negative control. Strain B31 encodes multiple GAG-binding proteins (Guo et al., 1998, Parveen et al., 2000, Parveen et al., 2003, Fischer et al., 2006, Benoit et al., 2011, Russell et al., 2013) and bound to heparin, heparan sulfate and dermatan sulfate with high efficiency (i.e. >11% of inoculum bound, or >7.7-fold more efficiently than to wells coated with BSA; P<0.005; Fig. 2). In addition, ~5% of the strain B31 inoculum bound to chondroitin-6-sulfate, which was significantly (3.4-fold) above background levels (Fig. 2). Whereas strain B314 harboring the pJF21 vector did not bind to GAGs, B314/pErpG bound to immobilized heparin and heparan sulfate 3.6- to 4.4-fold more efficiently than to BSA (P<0.004) and indistinguishably from strain B31 (Fig. 2). Unlike strain B31, however, strain B31/pErpG did not bind dermatan sulfate or chondroitin-6-sulfate, indicating that ErpG specifically binds to heparin and heparan sulfate (Fig. 2).
Figure 2. B. burgdorferi producing ErpG binds to heparin and heparan sulfate.
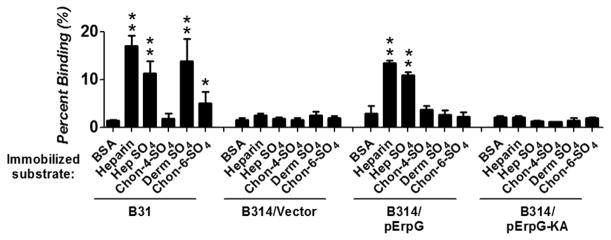
The percent of radiolabeled B. burgdorferi B31, B314/pErpG, B314/ErpG-KA, or B314/pJF21 (vector control) that bound to the indicated GAG, or to BSA as a negative control, was determined by scintillation counting (see Experimental Procedures). Each bar represents the mean of 12 independent determinations ± SEM. Significant differences in spirochetal binding relative to the BSA-coated wells was determined using the Student T test and are indicated (*,P-value of < 0.05; **, P-value of < 0.01).
ErpG-KA was produced on the surface of the strain B314 as efficiently as ErpG (Fig. S1), allowing us to determine if KK195 was required to promote spirochetal binding to GAGs. Strain B314/pErpG-KA bound to heparin or heparan sulfate no better than strain B314/pJF21 (vector control) and was 8- to 16-fold lower than binding by strain B314/pErpG (P<0.004; Fig. 2). Thus, the weak heparan sulfate-binding activity of recombinant ErpG-KA protein detected by ELISA and SPR (Fig. 1) is insufficient to support spirochetal binding to heparin or heparan sulfate.
Ectopic production of ErpG by strain B314 confers binding to cultured glial, but not endothelial, synovial or epithelial cells
To determine whether the heparan sulfate-binding activity of ErpG promotes the attachment of spirochetes to mammalian cells, we utilized two assays to measure the ability of ErpG to mediate attachment of strain B314 to a variety of cultured mammalian cells. First, we incubated bacteria with dispersed mammalian cells, then determined the percent of cells harboring bound bacteria by flow cytometry (Fig. 3, top panel). Strain B31, which has previously been shown to attach to C6 glial cells (Fischer et al., 2006) was able to attach to each of the cell lines with high efficiency (i.e. >25% of cells bound), whereas for all cell lines, between 5 and 11% of cells were bound to the control strain B314/pJF21 (Fig. 3, top panel). Ectopic production of ErpG failed to promote the attachment of strain B314 to SW982 synovial, SVEC endothelial, A549 epithelial, or CHO-K1 cells (Fig. 3, top panel). In contrast, ErpG-producing B314 bound to C6 cells 4-fold more efficiently than B314 bearing the empty vector (P< 0.05), and at a level equivalent to C6 cell binding by strain B31 (Fig. 3, top panel). Second, to assay binding to immobilized cells, the attachment of radiolabeled B. burgdorferi strains to cell monolayers was determined. Consistent with the suspension assays, the monolayer binding assays showed that both strains B31 and ErpG-producing B314 bound to C6 glial cells approximately 15-fold more efficiently than the negative control strain B314/pJF21(P<0.004) (Fig. 3, bottom panel). Whereas strain B31 was capable of binding to synovial, endothelial, A549 or CHO-K1 epithelial cells, B314/pErpG bound none of these cells (Fig. 3, bottom panel). Finally, attachment of B314 producing the double alanine substitution ErpG-KA bound to C6 glial cells no better than the control strain B314/pJF21, at a level 4-to 6-fold lower than B314/pErpG when measured by the two different assays (P<0.007; Fig. 3), thus correlating ErpG heparan sulfate-binding activity and ErpG-mediated spirochetal attachment to this cell line.
Figure 3. ErpG promotes spirochetal attachment to C6 glial cells.
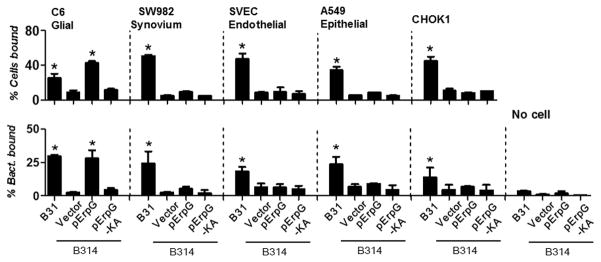
Top panel: Suspensions of the indicated cells were incubated with B. burgdorferi strains B31, B314/pErpG, B314/ErpG-KA, or B314/vector (negative control). Bound bacteria were stained with FITC-conjugated anti-B. burgdorferi antibody, and the percentage of cells bound to B. burgdorferi was measured by flow cytometry. Each bar represents the mean of 12 independent determinations± SEM. Binding efficiencies that are significantly different from that of strain B314/vector were determined by Student T test and are indicated (*, P<0.05). Bottom panel: The percentage of radiolabeled B. burgdorferi B31, B314/pErpG, B314/ErpG-KA, or B314/vector (negative control) that bound to the indicated cell monolayers was determined by scintillation counting (see Experimental Procedures). Each bar represents the mean of 12 independent determinations ± SEM. Significant differences in spirochetal binding relative to B314/vector were determined by Student T test and are indicated (*, P-value of < 0.05).
To functionally link GAG recognition with mammalian cell binding, we test whether pre-incubation of B. burgdorferi with purified GAGs prior to C6 glial cell infection resulted in diminished attachment. To first test the validity of this approach using a strain known to bind these substrates, we preincubated strain B31 with heparin, heparan sulfate or chondroitin-6-sulfate, each of which is recognized by this strain (Fig. 2). All three GAGs reduced attachment to C6 glial cells in both the cell suspension and monolayer assays (Fig. 4A and 4B). Although strain B31 binds dermatan sulfate, this GAG did not inhibit attachment of this strain to C6 cells in either assay (Fig. 4A and B), suggesting that dermatan sulfate is not a substrate for strain B31 on these cells. Preincubation of strain B31 with chondroitin-4-sulfate, which is not recognized by this B. burgdorferi strain, had no effect on cell binding.
Figure 4. Binding of ErpG-producing B. burgdorferi strain B314 to C6 glial cells is mediated by heparan sulfate.
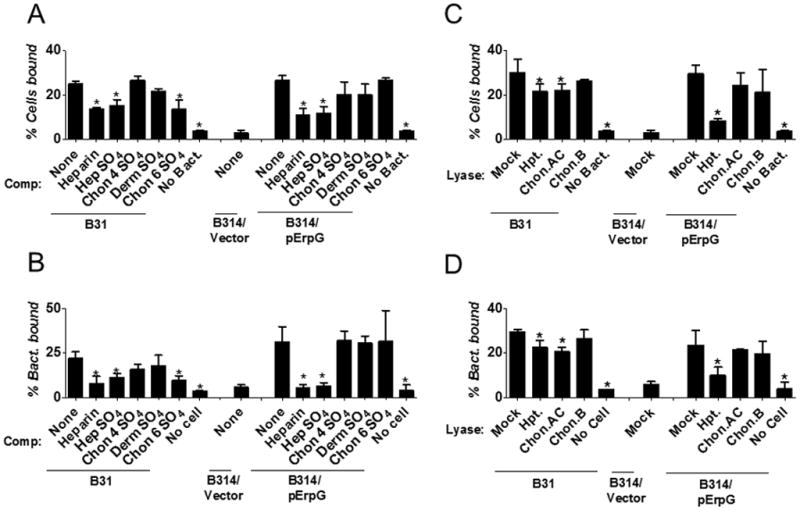
(A) Heparin, Hep-SO4, Chon-4-SO4, Derm SO4, or Chon-6-SO4 were added to the indicated B. burgdorferi strain to a final concentration of 6.25mg/ml prior to infection of C6 glial cells in suspension (see Experimental Procedures). PBS alone (“None”) was utilized as a negative control. The bacteria were stained with FITC-conjugated anti-B. burgdorferi antibody. The percentage of B. burgdorferi-bound cells was measured by flow cytometry. Each bar represents the mean of 12 independent determinations ± SEM. Statistically significant reductions in the percentage of cells bound by treated spirochetes compared to PBS-treated spirochetes are indicated (*, P-value <0.05). (B) Radiolabeled strains B31 and B314/pErpG were incubated with 2 mg/ml of the indicated GAG (or PBS alone as a negative control) for 30 min prior to infection of C6 glial cell monolayers, and the percentage of spirochetes stably bound was determined by scintillation counting. Each bar represents the mean of 12 independent determinations ± SEM. Significant reductions in spirochetal binding relative to spirochetes pre-incubated with PBS are indicated (*, P-value < 0.05). (C) The indicated B. burgdorferi strain was added to C6 glial cells in suspension that had been mock-treated (i.e. buffer alone) or treated with heparitinase (Hpt.) chondroitinase AC (Chon. AC), or chondroitinase B (Chon. B), and the percentage of B. burgdorferi-bound cells was measured as above. Note that the experiments in panels A and C were performed concurrently and binding of strain B314/Vector to mock-treated C6 glial cells in the absence of competitor served as a negative control for both experiments. Each bar represents the mean of 12 independent determinations ± SEM. Statistically significant reduction in the percentage of lyase-treated cells bound by spirochetes compared with PBS-treated cells is indicated (*, P-value <0.05). (D) The percentage of radiolabeled strains B31, B314/pErpG, and B314/pJF21 (“B314/vector”) (negative control) that stably bound to wells containing monolayers of C6 glial cells that had been mock-treated or treated with the indicated lyase was determined by scintillation counting. Note that the experiments in panels B and D were performed concurrently, and binding of strain B314/Vector to mock-treated C6 glial cells in the absence of competitor served as a negative control for both experiments. Each bar represents the mean of 12 independent determinations ± SEM. Significant reduction in spirochetal binding relative to PBS-treated cells is indicated (*, P-value < 0.05).
The role of ErpG, specifically, was then tested by preincubation of strain B314/pErpG with GAGs prior to glial cell binding. Purified chondroitin-4-sulfate, dermatan sulfate or chondroitin-6-sulfate, which are not recognized by this strain (Fig. 2), had no significant effect in either the cell suspension or monolayer assay. In contrast, preincubation with heparan sulfate or heparin diminished bacterial binding approximately two-fold (P<0.05; Fig. 4A and B; see Experimental Procedures).
To determine which cell-associated GAGs contributed to binding of strain B314/pErpG (or, as a control, strain B31), specific classes of GAGs on the surface of C6 glial cells were removed by digestion with lyases prior to binding. Treatment with hepartinase, which removes heparan sulfate, or chondroitinase AC, which removes chondroitin 4 sulfate and chondroitin 6 sulfate, decreased cell attachment of strain B31 in both the cell suspension and monolayer binding assays (Fig. 4C and D). Removal of dermatan sulfate with chondroitinase B did not (Fig. 4C and D), consistent with the finding that exogenous dermatan sulfate did not inhibit binding of B31 to C6 cells (Fig. 4A and B). Cell binding by B314/pErpG was not diminished in either assay by treatment of C6 glial cells with chondroitinase AC or chondroitinase B (Fig. 4C and D). In contrast, removal of heparan sulfate from C6 glial cells by heparitinase digestion led to a three- to four-fold reduction in binding by B314/pErpG in both the suspension and monolayer assay formats (P<0.05; Fig. 4C and D), indicating that heparan sulfate is required for maximal ErpG-mediated spirochetal attachment to C6 glial cells.
OspF-related Erp proteins bind heparin- and heparan sulfate
Surface display phages encoding not only OspG peptides, but also peptides of other OspF-related proteins of B. burgdorferi strain N40-D10/E9 such as BBK2.10, ErpK, and ErpL were localized to the joints or heart (Antonara et al., 2007). Furthermore, ErpK, fused either to filamentous phage or to maltose binding proteins (MBP), bound to respiratory epithelial cells in a dose-dependent manner (Antonara et al., 2007). To test whether strain B31 ErpK and ErpL, like ErpG, bind to heparin sulfate and heparin, GST-tagged recombinant versions of these proteins were tested for binding to different ECM components, including fibronectin, laminin, vitronectin, collagen, elastin, and a variety of subclasses of GAGs. As expected, GST did not bind to any of the substrates tested (Fig. S4A and data not shown). In contrast, GST-ErpK and GST-ErpL each bound to heparin- or heparan sulfate-coated wells at least 7-fold better (p<0.05) than to control (BSA-coated) wells (Fig. S4A). No other ECM components tested were recognized by either GST-ErpK or GST-ErpL (Fig. S4A and data not shown).
To determine whether ErpK and ErpL bound to heparan sulfate with affinities similar to that of ErpG, we utilized quantitative ELISA to assess their binding. GST-ErpK and GST-ErpL each bound to heparan sulfate in a dose-dependent manner with KD values of 0.42 μM and 0.40 μM, respectively (Table 1 and Fig. S4B, left panel). KD values obtained using SPR were highly similar (Table 1, Fig. 5 and Fig. S4B, right panel). Thus, both ErpK and ErpL bind to heparan sulfate with affinities roughly equivalent to that of GST-ErpG (i.e. ~0,31 μM).
Figure 5. OspF-related proteins bind to heparan sulfate.

A rooted phylogram of the OspF-related proteins (in red font) encoded by the B. burgdorferi strains B31 (ErpL, ErpY, ErpK, ErpG), B. burgdorferi N40-D10/E9 (Erp25 and Erp27), and B. burgdorferi 297 (OspF) was prepared using complete protein sequences and MEGA version 5.2 (Tamura et al. Mol. Biol. Evol. 2011). Strain B31 ErpX (in blue font) and ErpP (in green font) were included as representatives of OspEF-like leader peptides and OspE-related proteins, respectively. The KD for heparan sulfate (Hep-SO4)-binding of each Erp protein determined by SPR (Experimental Procedures) is shown. Alignment of the putative GAG-binding sites is shown on the right, with the red asterisks indicating the KK195 sequence that is required for full heparan sulfate binding.
Interestingly, the dual lysine sequence KK195 of ErpG, which is critical for its heparan sulfate-binding activity, is conserved among all OspF-related proteins. The sequences flanking these residues are also well conserved (Fig. 5). In contrast, the sequences of this region in OspE-related and Elp proteins are divergent from OspF-related proteins (Fig. 5). To test whether heparan sulfate binding might be a common property of OspF-related proteins, we measured the heparan sulfate-binding activity of four other OspF-related proteins, i.e. ErpY, Erp25, Erp27, and OspF by SPR. Each of these OspF-related proteins bound to heparan sulfate, albeit with somewhat lower affinity than ErpK, ErpL, or ErpG (Table 1 and Fig. 5). To determine if this ligand-binding activity is specific for OspF-related proteins, we also tested heparan sulfate binding by ErpX and ErpP, representatives of the OspE-related and Elp subfamilies, respectively. Neither ErpP nor ErpX, were able to bind heparan sulfate (Table 1 and Fig. 5), indicating that the ability to bind heparan sulfate is a common and specific property of OspF-related proteins.
DISCUSSION
Microbial attachment to mammalian cells is an important step in colonization, and several subclasses of GAGs, including chondroitin-6-sulfate, dermatan sulfate and heparan sulfate, have been demonstrated to participate in the attachment of the Lyme disease spirochete to host cells (Leong et al., 1998, Parveen et al., 1999, Fischer et al., 2003, Parveen et al., 2003). The B. burgdorferi adhesins DbpA, DbpB and BBK32 recognize dermatan sulfate or chondroitin-6-sulfate, (Parveen et al., 2000, Fischer et al., 2003, Parveen et al., 2003, Fischer et al., 2006, Benoit et al., 2011), and each of these adhesins promotes colonization of the mammalian host. However, no well-documented B. burgdorferi heparan sulfate-binding adhesin has previously been identified.
Members of the OspF family of B. burgdorferi proteins were identified as potential adhesins because surface display of fragments of these proteins derived from strain N40-D10/E9 resulted in localization of phage to the joints, heart, and bladder of mice after i.v. injection (Antonara et al., 2007). In this study, we showed that ErpG, the strain B31 homolog of the OspF family member OspG of strain N40-D10/E9, binds to heparan sulfate with sub-micromolar affinity. When ectopically produced on the surface B. burgdorferi, ErpG promoted spirochetal attachment to purified heparan sulfate and to C6 glial cells. In addition, several other OspF-related proteins, such as ErpL and ErpK, were found to also promote the localization of surface display phage to specific mouse tissues (Antonara et al., 2007), and we found that recombinant derivatives of strain B31 ErpL and ErpK, as well as four other OspF-related proteins, i.e. OspF, ErpY, Erp25 and Erp27, recognized heparan sulfate. This binding is not a property of all Erp subfamilies, because recombinant derivatives of ErpX and ErpP, members of the Elp and OspE-related proteins, respectively (Marconi et al., 1996, Stevenson et al., 1996, Akins et al., 1999), failed to bind this GAG. Recombinant ErpX has been shown to bind the mammalian extracellular matrix protein laminin (Brissette et al., 2008) and OspE-related proteins bind to plasminogen, factor H, and factor H-related proteins (Kraiczy et al., 2013). Therefore, the three Erp subfamilies have distinct ligand-binding properties and likely distinct functions.
A dual lysine sequence, KK195, is conserved in all OspF family members but lacking in members of the Elp and OspE-related proteins. Clusters of basic amino acids of GAG-binding proteins are typically critical for activity (Cardin et al., 1989), and dual alanine substitution of KK195 abrogated high affinity heparan sulfate binding by ErpG, as well as its ability to promote attachment to C6 glial cells. It is notable, however, that four ErpK and one ErpL peptides that promoted the localization of surface display phage to various tissues in mice lack this (and nearby) sequences, indicating that other ErpK/ErpL sequences are capable of promoting localization of surface display phage to murine tissues (Antonara et al., 2007). All selected phage clones encoded at least one region containing a cluster of basic amino acids (Antonara et al., 2007), but it is unknown whether these alternate binding sequences promote binding to heparan sulfate (or other host ligands).
B. burgdorferi strains that produce BBK32 or DbpA mutants specifically deficient in dermatan sulfate binding display colonization defects (Fortune et al., 2014, Lin et al., 2014a, Lin et al., 2014b), raising the possibility that heparan sulfate binding by OspF-related proteins may promote colonization. B. burgdorferi strains with transposon insertions in erpK or erpY displayed reduced colonization at the joints, heart, and skin (Lin et al., 2012), although no analyses have yet been performed on targeted mutations of either of these genes. Moreover, the GAG-binding specificity of Lyme disease spirochetes has the capacity to influence which tissues are most efficiently colonized during mammalian infection. For example, a B. burgdorferi B31 mutant that specifically lacks the dermatan sulfate-binding activity of the adhesin BBK32 retains wild type capacity to colonize the skin but is partially defective for joint colonization (Lin et al., 2014b). We found here that ErpG promoted binding to C6 rat glial cells but not to four other cell lines tested. Although it is possible that differences such as the parental mammalian species (rat, mouse, hamster or human) of the cell lines influence ErpG recognition, our results suggest that this adhesin may promote colonization of a specific (e.g. nervous) tissue during mammalian infection. Finally, that variant alleles of DbpA that display different GAG-binding properties also differ in their ability to promote colonization of the heart or joint (Lin et al., 2014a) raises the possibility that the collective diversity of the OspF-related subfamily may provide diversity in colonization function. Strain B31 5A3 and B31 M1-16 each encode four OspF family members whose temporal expression patterns during mammalian infection are not identical (McDowell et al., 2001, Miller et al., 2006a), and the OspF-related proteins tested here bound to heparan sulfate with affinities that varied 26-fold. Whether the differences among OspF-related proteins in temporal expression or heparan sulfate recognition correspond to differences in in vivo function will require a careful genetic analysis of OspF family mutants. This study, by defining a specific host ligand for these proteins, as well as identifying residues important for ligand binding, provides a foundation for such future studies.
EXPERIMENTAL PROCEDURES
Bacterial strains and growth conditions
The Borrelia and E. coli strains used in this study are described in Table S1. Escherichia coli strains DH5α, BL21 and derivatives were grown in Luria-Bertani (BD Bioscience, Franklin lakes, NJ) broth or agar, supplemented with kanamycin (50 μg/ml) or ampicillin (100 μg/ml) where appropriate. All B. burgdorferi strains were grown in BSK-H complete medium supplemented with kanamycin (200 μg/ml).
Generation of recombinant Erp proteins and ErpG antisera
To generate recombinant Glutathione S-transferase (GST)-tagged Erp proteins, the open reading frames of erpK, erpL, erpG, erpY, erpP, and erpX from B. burgdorferi strains B31, erp25 and erp27 from B. burgdorferi strain N40-D10/E9, and ospF from B. burgdorferi strain 297 were amplified using primers described in Table S2 and inserted into pGEX4T2 (GE Healthcare, Piscataway, NJ) as previously described (Benoit et al., 2011) (see Table S1). The amplified erp open reading frames were engineered to lack the putative signal sequence and to encode a restriction site at the 5′ ends and a stop codon followed by a second restriction site at the 3′ end. PCR products were sequentially digested with the appropriate restriction enzymes and then inserted into pGEX4T2 digested with the same enzymes. The resulting plasmids were transformed into E. coli strain BL21 and the plasmid inserts were sequenced (Tufts core sequencing facility). The GST-tagged Erp proteins were produced and purified by GST bead affinity chromatography according to the manufacturer’s instructions (GE Healthcare, Piscataway, NJ). To generate antisera against strain B31 ErpG, four-week-old BALB/c mice were immunized with recombinant ErpG, similar to the immunization protocol described previously (Benoit et al. Infect. Immun. 2011).,
Quantification of attachment to extracellular matrix molecules
One hundred μL of 10 μg/mL of heparin, heparan sulfate, chondroitin-4-sulfate, dermatan sulfate, chondroitin-6-sulfate, human plasma fibronectin, human plasma vitronectin, laminin, elastin, type I collagen, type IV collagen or BSA (negative control) (Sigma-Aldrich, St. Louis, MO) was coated onto microtiter plate wells as described previously (Benoit et al. Infect. Immun. 2011). One hundred microliters of increasing concentrations (0.03125, 0.0625, 0.125, 0.25, 0.5, 1, 2 μM) of GST-tagged ErpK, ErpL, or ErpG, or the GST negative control were then added to the wells. To detect the binding of GST-tagged proteins, goat anti-GST tag (Sigma-Aldrich, St. Louis, MO; 1:200) and HRP-conjugated anti-goat IgG (Sigma-Aldrich, St. Louis, MO; 1:1000) were used as primary and secondary antibodies, respectively. The plates were washed three times with PBST (0.05% Tween20 in PBS buffer), and 100 μL of tetramethyl benzidine (TMB) solution (KPL, Gaithersburg, MD) were added to each well and incubated for five minutes. The reaction was stopped by adding 100 μL of 0.5% hydro sulfuric acid to each well. Plates were read at 405 nm using a Synergy HT ELISA plate reader (BioTek, Winooski, VT). To determine the dissociation constant (KD), the data were fitted by the following equation using KaleidaGraph software (Version 4.1.1 Synergy Software, Reading, PA).
| (Equation 1) |
Surface Plasmon Resonance (SPR)
Interactions of Erp proteins with heparan sulfate were analyzed by a SPR technique using a Biacore 3000 (GE Healthcare, Piscataway, NJ). Ten μg of biotinylated heparan sulfate was conjugated to an SA chip (GE Healthcare, Piscataway, NJ). A control flow cell was injected with PBS buffer without heparan sulfate. To quantify heparan sulfate binding, ten μL of increasing concentrations (0, 15.625, 31.25, 62.5, 125, 250, 500 nM) of ErpK, ErpL, ErpG, ErpY, Erp25, Erp27, OspF, ErpP, ErpX, or ErpG-KA were injected into the control cell and the cell with immobilized heparan sulfate at 10 μL/min, 25°C. To obtain the kinetic parameters of the interaction, sensogram data were fitted by means of BIAevaluation software version 3.0 (GE Healthcare, Piscataway, NJ), using the one step biomolecular association reaction model (1:1 Langmuir model), resulting in the optimum mathematical fit with the lowest Chi square values.
Shuttle Plasmid construction
To generate plasmids encoding erpG, the gene was first PCR amplified with the addition of a SalI site and a BamH1 site at the 5′ and 3′ ends, respectively, using the primers listed in Table S2. Amplified DNA was inserted into the TA cloning vector pCR2.1-TOPO (Invitrogen, Houston, TX) (see Table S2), to generate plasmid pCR2.1-erpG. The plasmid was then digested with SalI and BamHI to release the erpG, which was then inserted into the SalI and BamHI sites of plasmid pJF21 (Table S1). The resulting ErpG-producing plasmid was transformed into E. coli strain BL21 and the plasmid insert sequence was confirmed by DNA sequencing (Tufts core sequencing facility).
Generation of an ErpG-KA-producing shuttle vector
Site-directed, ligase-independent mutagenesis (SLIM) was used to replace lysines-194 and -195 of ErpG with alanines as described previously (Chiu et al., 2004). Briefly, pGEX4T2-ErpG and pJF21-ErpG were used as templates to PCR amplify pGEX4T2-ErpG-KA and pJF21-ErpG-KA, using the primers shown in Table S2. The PCR products were treated with DpnI in 20 mM MgCl2, 20 mM Tris, pH 8.0 and 5 mM DTT for 60 min at 37°C, and then the reaction was terminated by adding 30 μL of H-buffer (300 mM NaCl, 50 mM Tris, pH 9.0, 20 mM EDTA, pH 8.0). DNA was then denatured at 99°C for 3 min, then hybridized at 65°C for 5 min followed by 30°C for 15 min. Subsequently, 20 μL of the hybridization product was transformed into E. coli XL1-blue by electroporation (Eppendorf, Hauppauge, NY), and plasmid inserts were sequenced (Tufts core sequencing facility).
Plasmid transformation into B. burgdorferi B314
Electrocompetent B. burgdorferi strain B314 was transformed separately with 80 μg shuttle plasmid encoding erpG or erpG-KA (table S1), then cultured in BSK II medium at 37°C for 24 hours as previously described (Margolis et al., 1995). Aliquots of the culture were mixed with 1.8% analytical grade agarose (BioRad; Hercules, CA) and plated on a solidified BSK II/agarose layer in sterile 100 × 20 mm tissue culture dishes (Corning Incorporated, Corning, NY). Plates were incubated at 37°C in 5% CO2 for two weeks. Kanamycin-resistant colonies of B. burgdorferi carrying plasmids encoding erpG or erpG-KA were selected and expanded at 37°C in liquid BSK II medium containing kanamycin. To verify the presence of the kanamycin-resistance gene in transformants, PCR was carried out using kan-specific primers (Table S2).
Surface localization of ErpG detected by Proteinase K treatment, SDS-PAGE, and Western immunoblotting
To test whether plasmid-encoded ErpG and ErpG-KA produced in B. burgdorferi B314 were localized to the spirochete surface, spirochetes were treated with proteinase K, lysed, fractionated by SDS-PAGE, and immunoblotted with α-ErpG (1:5,000) and the α-FlaB monoclonal antibody CB1 (1:10,000; a gift from J. Benach, Stony Brook University; Stony Brook, NY) as the primary antibodies, and goat anti-mouse IgG/horseradish peroxidase (HRP) as the secondary antibody (1:10,000). The blot was developed using the ECL system (GE, Piscataway, NJ), as described previously (Benoit et al., 2011).
Surface localization of ErpG detected by Flow Cytometry
1 × 108 B. burgdorferi cells were washed thrice with HBSC buffer containing DB (25 mM Hepes acid, 150 mM sodium chloride, 1 mM MnCl2, 1 mM MgCl2, 0.25 mM CaCl2, 0.1% dextrose, and 0.2% BSA, final concentration) then resuspended into 500 μL of the same buffer. To generate permeablized spirochetes, the bacteria were suspended in 100% methanol and incubated for one hour, then washed thrice by HBSC DB buffer. The mouse antisera raised against ErpG (1:250x), or mouse anti-FlaB (negative control) (1:250x), was used as a primary antibody, and Alexa488-conjugated goat anti-mouse IgG (Invitrogen) (1:250x) was used as a secondary antibody. Subsequently, binding was fixed with 300 μL of formalin (0.1%). Surface production of ErpG or FlaB (as a negative control) was measured by flow cytometry using a Becton-Dickinson FACSCalibur (BD Bioscience, Franklin Lakes, NJ). All flow cytometry experiments were performed within two days of collection of B. burgdorferi samples. Spirochetes in the suspension were distinguished on the basis of their distinct light scattering properties in a Becton Dickinson FACSCalibur™ flow cytometer equipped with a 15 mW, 488 nm air-cooled argon laser, a standard three-color filter arrangement, and CELLQuest™ Software (BD Bioscience, Franklin Lakes, NJ). The mean fluorescence index (MFI) of each sample, representing the surface production of the indicated proteins, was obtained from FlowJo software (Three star Inc, Ashland, OR).
Binding of radiolabeled B. burgdorferi to purified GAG and to mammalian cells
Binding of B. burgdorferi to purified GAG or to mammalian cells was determined as previously described (Benoit et al., 2011). Briefly, prior to the addition of spirochetes, 100μL of 2.5 mg/mL of purified heparin, heparan sulfate, chondroitin-4-sulfate, dermatan sulfate, chondroitin-6-sulfate, or BSA (negative control) was added to break-apart microtiter plate wells (ThermoScientific, Pittsburgh PA) for 16 hours at 4°C. For the cell-binding assay, 1 × 106 mammalian cells were incubated on the break-apart microtiter plate wells overnight at 37°C. Spirochetes were then radiolabeled with [35S] methionine, and 1 × 108 radiolabeled bacteria were suspended in BSK-H medium and incubated at room temperature for 2 hours. The radiolabeled spirochetes were then diluted 1:3 into GHS buffer (10mM glucose, 10mM HEPES. 50mM NaCl at pH 7.0) and added to quadruplicate wells with the immobilized cells or GAGs at 106 spirochetes/wells. To enhance spirochete-cell contact, the plates were centrifuged at 1,000 rpm for 5 minutes and then rocked at room temperature for 1 hour. Unbound bacteria were removed by washing with PBS containing 0.2% BSA. Plates were air-dried, and percent binding was determined by liquid scintillation counting.
B. burgdorferi binding to mammalian cells in suspension
Mammalian cells were washed twice and resuspended in HBSC buffer containing DB. Spirochetes were washed thrice and resuspended in HBSC buffer containing DB, and 20 μL of a 2 × 108 spirochetes/mL suspension were added to 20 μL of a 5 × 106 cells/mL suspension and incubated for 1 hour at 37°C. 10 μL of a 1:240 dilution of Alexa488-conjugated mouse antisera raised against B. burgdorferi (Abcam, Cambridge MA) was added to the spirochete-cell mixture, followed by fixation with 300 μL of formalin (0.1%). The percentage of the cells bound by B. burgdorferi was measured by flow cytometry using a Becton-Dickinson FACSCalibur and analyzed by FlowJo software as described above.
Inhibition of binding with exogenous GAGs
Spirochetes were prepared as described above and incubated for 30 min at room temperature in BSK-H supplemented with 6.25 mg/ml of GAGs. After incubation, spirochetes were diluted 1:3 in GHS buffer (10mM glucose, 10mM Hepes, and 50mM NaCl at pH 7.0) before incubating with C6 glial cells as described above.
Enzymatic removal of specific classes of GAGs
C6 glial cells were incubated for 2 hours with 0.5 U of heparitinase, chondroitinase AC, or chondroitinase B (Sigma)/ml at 37°C in RPMI supplemented with 1% BSA, 10−2 U of aprotinin/mL, and 165 μg of PMSF/mL. Cell monolayers were then washed with PBS, and spirochetes were incubated with the pretreated C6 glial cells, as described above.
Circular dichroism (CD) spectroscopy
CD analysis was performed on a Jasco 810 spectropolarimeter (Jasco Analytical Instrument, Easton, MD) under N2. CD spectra were measured at RT (25°C) in a 1 mm path length quartz cell. Spectra of ErpG (10 μM) and ErpG-KA (10 μM) were recorded in Tris buffer at 25°C, and three far-UV CD spectra were recorded from 190 to 250 nm for far-UV CD in 1 nm increments. The background spectrum of buffer without protein was subtracted from the protein spectra. CD spectra were initially analyzed using the Spectra Manager Program software. Analysis of spectra to extrapolate secondary structures was performed by Dichroweb (http://www.cryst.bbk.ac.uk/cdweb/html/home), using the K2D and Selcon 3 analysis programs (Lin et al., 2009).
Statistical analysis
Significant differences between samples were determined using the one-way ANOVA test following logarithmic transformation of the data. P-values were determined for each sample. A P-value < 0.05 (*) was considered to be significant, and a P-value < 0.01 (**) was considered to be extremely significant.
Supplementary Material
Acknowledgments
We thank Vivian Benoit, for valuable technical advice, Marcia Osburne for critical reading of the manuscript, Jorge Benach for α-FlaB CB1 monoclonal antibody, James Baleja for help with circular dichroism, Allen Parmelee and Stephen Kwok for assisting with flow cytometry, and Michael Waring of the Ragon Institute Imaging Core Facility for providing access to and assistance with SPR instrumentation. This work was supported by NIH R01AI37601 (to J.M.L.) and R01AI093104 (to J.C and J.M.L.), and American Heart Association Postdoctoral Fellowship 12POST11660031 (to Y.L.).
Footnotes
The authors declare no competing financial interests.
References
- Akins DR, Caimano MJ, Yang X, Cerna F, Norgard MV, Radolf JD. Molecular and evolutionary analysis of Borrelia burgdorferi 297 circular plasmid-encoded lipoproteins with OspE- and OspF-like leader peptides. Infection and Immunity. 1999;67:1526–1532. doi: 10.1128/iai.67.3.1526-1532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akins DR, Porcella SF, Popova TG, Shevchenko D, Baker SI, Li M, et al. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homologue. Molecular Microbiology. 1995;18:507–520. doi: 10.1111/j.1365-2958.1995.mmi_18030507.x. [DOI] [PubMed] [Google Scholar]
- Antonara S, Chafel RM, LaFrance M, Coburn J. Borrelia burgdorferi adhesins identified using in vivo phage display. Molecular Microbiology. 2007;66:262–276. doi: 10.1111/j.1365-2958.2007.05924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquino RS, Lee ES, Park PW. Diverse functions of glycosaminoglycans in infectious diseases. Progress in Molecular Biology and Translational Science. 2010;93:373–394. doi: 10.1016/S1877-1173(10)93016-0. [DOI] [PubMed] [Google Scholar]
- Babb K, El-Hage N, Miller JC, Carroll JA, Stevenson B. Distinct regulatory pathways control expression of Borrelia burgdorferi infection-associated OspC and Erp surface proteins. Infection and Immunity. 2001;69:4146–4153. doi: 10.1128/IAI.69.6.4146-4153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour AG. Isolation and cultivation of Lyme disease spirochetes. The Yale Journal of Biology and Medicine. 1984;57:521–525. [PMC free article] [PubMed] [Google Scholar]
- Benoit VM, Fischer JR, Lin YP, Parveen N, Leong JM. Allelic variation of the Lyme disease spirochete adhesin DbpA influences spirochetal binding to decorin, dermatan sulfate, and mammalian cells. Infection and Immunity. 2011;79:3501–3509. doi: 10.1128/IAI.00163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins JS, Hagman KE, Norgard MV. Assessment of decorin-binding protein A to the infectivity of Borrelia burgdorferi in the murine models of needle and tick infection. BMC Microbiol. 2008;8:82. doi: 10.1186/1471-2180-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissette CA, Bykowski T, Cooley AE, Bowman A, Stevenson B. Borrelia burgdorferi RevA antigen binds host fibronectin. Infection and Immunity. 2009a;77:2802–2812. doi: 10.1128/IAI.00227-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissette CA, Cooley AE, Burns LH, Riley SP, Verma A, Woodman ME, et al. Lyme borreliosis spirochete Erp proteins, their known host ligands, and potential roles in mammalian infection. International Journal of Medical Microbiology: IJMM. 2008;298(Suppl 1):257–267. doi: 10.1016/j.ijmm.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissette CA, Verma A, Bowman A, Cooley AE, Stevenson B. The Borrelia burgdorferi outer-surface protein ErpX binds mammalian laminin. Microbiology. 2009b;155:863–872. doi: 10.1099/mic.0.024604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Yang X, Popova TG, Clawson ML, Akins DR, Norgard MV, Radolf JD. Molecular and evolutionary characterization of the cp32/18 family of supercoiled plasmids in Borrelia burgdorferi 297. Infection and Immunity. 2000;68:1574–1586. doi: 10.1128/iai.68.3.1574-1586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardin AD, Weintraub HJ. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis. 1989;9:21–32. doi: 10.1161/01.atv.9.1.21. [DOI] [PubMed] [Google Scholar]
- Chiu J, March PE, Lee R, Tillett D. Site-directed, Ligase-Independent Mutagenesis (SLIM): a single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Research. 2004;32:e174. doi: 10.1093/nar/gnh172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn J, Leong JM, Erban JK. Integrin alpha IIb beta 3 mediates binding of the Lyme disease agent Borrelia burgdorferi to human platelets. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:7059–7063. doi: 10.1073/pnas.90.15.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comstock LE, Thomas DD. Penetration of endothelial cell monolayers by Borrelia burgdorferi. Infection and Immunity. 1989;57:1626–1628. doi: 10.1128/iai.57.5.1626-1628.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BB, Falkow S. Common themes in microbial pathogenicity revisited. Microbiology and molecular biology reviews: MMBR. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer JR, LeBlanc KT, Leong JM. Fibronectin binding protein BBK32 of the Lyme disease spirochete promotes bacterial attachment to glycosaminoglycans. Infection and Immunity. 2006;74:435–441. doi: 10.1128/IAI.74.1.435-441.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer JR, Parveen N, Magoun L, Leong JM. Decorin-binding proteins A and B confer distinct mammalian cell type-specific attachment by Borrelia burgdorferi, the Lyme disease spirochete. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:7307–7312. doi: 10.1073/pnas.1231043100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune DE, Lin YP, Deka RK, Groshong AM, Moore BP, Hagman KE, et al. Identification of Lysine Residues in the Borrelia burgdorferi DbpA Adhesin Required for Murine Infection. Infection and Immunity. 2014;82:3186–3198. doi: 10.1128/IAI.02036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Monco JC, Fernandez-Villar B, Benach JL. Adherence of the Lyme disease spirochete to glial cells and cells of glial origin. The Journal of Infectious Diseases. 1989;160:497–506. doi: 10.1093/infdis/160.3.497. [DOI] [PubMed] [Google Scholar]
- Gaultney RA, Gonzalez T, Floden AM, Brissette CA. BB0347, from the lyme disease spirochete Borrelia burgdorferi, is surface exposed and interacts with the CS1 heparin-binding domain of human fibronectin. PloS One. 2013;8:e75643. doi: 10.1371/journal.pone.0075643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo BP, Brown EL, Dorward DW, Rosenberg LC, Hook M. Decorin-binding adhesins from Borrelia burgdorferi. Molecular Microbiology. 1998;30:711–723. doi: 10.1046/j.1365-2958.1998.01103.x. [DOI] [PubMed] [Google Scholar]
- Hefty PS, Jolliff SE, Caimano MJ, Wikel SK, Radolf JD, Akins DR. Regulation of OspE-related, OspF-related, and Elp lipoproteins of Borrelia burgdorferi strain 297 by mammalian host-specific signals. Infection and Immunity. 2001;69:3618–3627. doi: 10.1128/IAI.69.6.3618-3627.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hileman RE, Fromm JR, Weiler JM, Linhardt RJ. Glycosaminoglycan-protein interactions: definition of consensus sites in glycosaminoglycan binding proteins. BioEssays: news and reviews in molecular, cellular and developmental biology. 1998;20:156–167. doi: 10.1002/(SICI)1521-1878(199802)20:2<156::AID-BIES8>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Isaacs RD. Borrelia burgdorferi bind to epithelial cell proteoglycans. The Journal of Clinical Investigation. 1994;93:809–819. doi: 10.1172/JCI117035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenedy MR, Akins DR. The OspE-related proteins inhibit complement deposition and enhance serum resistance of Borrelia burgdorferi, the Lyme disease spirochete. Infection and Immunity. 2011;79:1451–1457. doi: 10.1128/IAI.01274-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraiczy P, Stevenson B. Complement regulator-acquiring surface proteins of Borrelia burgdorferi: Structure, function and regulation of gene expression. Ticks and Tick-borne diseases. 2013;4:26–34. doi: 10.1016/j.ttbdis.2012.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong JM, Morrissey PE, Ortega-Barria E, Pereira ME, Coburn J. Hemagglutination and proteoglycan binding by the Lyme disease spirochete, Borrelia burgdorferi. Infection and Immunity. 1995;63:874–883. doi: 10.1128/iai.63.3.874-883.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong JM, Wang H, Magoun L, Field JA, Morrissey PE, Robbins D, et al. Different classes of proteoglycans contribute to the attachment of Borrelia burgdorferi to cultured endothelial and brain cells. Infection and Immunity. 1998;66:994–999. doi: 10.1128/iai.66.3.994-999.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Gao L, Zhang C, Odeh E, Jacobs MB, Coutte L, et al. Analysis of an ordered, comprehensive STM mutant library in infectious Borrelia burgdorferi: insights into the genes required for mouse infectivity. PloS One. 2012;7:e47532. doi: 10.1371/journal.pone.0047532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YP, Benoit V, Yang X, Martinez-Herranz R, Pal U, Leong JM. Strain-specific variation of the decorin-binding adhesin DbpA influences the tissue tropism of the lyme disease spirochete. PLoS Pathogens. 2014a;10:e1004238. doi: 10.1371/journal.ppat.1004238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YP, Chen Q, Ritchie JA, Dufour NP, Fischer JR, Coburn J, Leong JM. Glycosaminoglycan binding by Borrelia burgdorferi adhesin BBK32 specifically and uniquely promotes joint colonization. Cellular microbiology. 2014b doi: 10.1111/cmi.12407. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YP, Greenwood A, Nicholson LK, Sharma Y, McDonough SP, Chang YF. Fibronectin binds to and induces conformational change in a disordered region of leptospiral immunoglobulin-like protein B. The Journal of Biological Chemistry. 2009;284:23547–23557. doi: 10.1074/jbc.M109.031369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marconi RT, Sung SY, Hughes CA, Carlyon JA. Molecular and evolutionary analyses of a variable series of genes in Borrelia burgdorferi that are related to ospE and ospF, constitute a gene family, and share a common upstream homology box. Journal of Bacteriology. 1996;178:5615–5626. doi: 10.1128/jb.178.19.5615-5626.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis N, Samuels DS. Proteins binding to the promoter region of the operon encoding the major outer surface proteins OspA and OspB of Borrelia burgdorferi. Molecular biology reports. 1995;21:159–164. doi: 10.1007/BF00997238. [DOI] [PubMed] [Google Scholar]
- McDowell JV, Sung SY, Price G, Marconi RT. Demonstration of the genetic stability and temporal expression of select members of the lyme disease spirochete OspF protein family during infection in mice. Infection and Immunity. 2001;69:4831–4838. doi: 10.1128/IAI.69.8.4831-4838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Stevenson B. Borrelia burgdorferi erp genes are expressed at different levels within tissues of chronically infected mammalian hosts. International Journal of Medical Microbiology: IJMM. 2006a;296(Suppl 40):185–194. doi: 10.1016/j.ijmm.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Miller JC, von Lackum K, Woodman ME, Stevenson B. Detection of Borrelia burgdorferi gene expression during mammalian infection using transcriptional fusions that produce green fluorescent protein. Microbial Pathogenesis. 2006b;41:43–47. doi: 10.1016/j.micpath.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Moriarty TJ, Shi M, Lin YP, Ebady R, Zhou H, Odisho T, et al. Vascular binding of a pathogen under shear force through mechanistically distinct sequential interactions with host macromolecules. Molecular Microbiology. 2012;86:1116–1131. doi: 10.1111/mmi.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojaimi C, Brooks C, Casjens S, Rosa P, Elias A, Barbour A, et al. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infection and Immunity. 2003;71:1689–1705. doi: 10.1128/IAI.71.4.1689-1705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parveen N, Caimano M, Radolf JD, Leong JM. Adaptation of the Lyme disease spirochaete to the mammalian host environment results in enhanced glycosaminoglycan and host cell binding. Molecular Microbiology. 2003;47:1433–1444. doi: 10.1046/j.1365-2958.2003.03388.x. [DOI] [PubMed] [Google Scholar]
- Parveen N, Leong JM. Identification of a candidate glycosaminoglycan-binding adhesin of the Lyme disease spirochete Borrelia burgdorferi. Molecular Microbiology. 2000;35:1220–1234. doi: 10.1046/j.1365-2958.2000.01792.x. [DOI] [PubMed] [Google Scholar]
- Parveen N, Robbins D, Leong JM. Strain variation in glycosaminoglycan recognition influences cell-type-specific binding by lyme disease spirochetes. Infection and Immunity. 1999;67:1743–1749. doi: 10.1128/iai.67.4.1743-1749.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probert WS, Johnson BJ. Identification of a 47 kDa fibronectin-binding protein expressed by Borrelia burgdorferi isolate B31. Molecular Microbiology. 1998;30:1003–1015. doi: 10.1046/j.1365-2958.1998.01127.x. [DOI] [PubMed] [Google Scholar]
- Radolf JD, Caimano MJ, Stevenson B, Hu LT. Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nature reviews Microbiology. 2012;10:87–99. doi: 10.1038/nrmicro2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts WC, Mullikin BA, Lathigra R, Hanson MS. Molecular analysis of sequence heterogeneity among genes encoding decorin binding proteins A and B of Borrelia burgdorferi sensu lato. Infection and Immunity. 1998;66:5275–5285. doi: 10.1128/iai.66.11.5275-5285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell TM, Johnson BJ. Lyme disease spirochaetes possess an aggrecan-binding protease with aggrecanase activity. Molecular Microbiology. 2013;90:228–240. doi: 10.1111/mmi.12276. [DOI] [PubMed] [Google Scholar]
- Sadziene A, Wilske B, Ferdows MS, Barbour AG. The cryptic ospC gene of Borrelia burgdorferi B31 is located on a circular plasmid. Infection and Immunity. 1993;61:2192–2195. doi: 10.1128/iai.61.5.2192-2195.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo J, Loimaranta V, Lahdenne P, Viljanen MK, Hytonen J. Decorin binding by DbpA and B of Borrelia garinii, Borrelia afzelii, and Borrelia burgdorferi sensu Stricto. The Journal of Infectious Diseases. 2011;204:65–73. doi: 10.1093/infdis/jir207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshu J, Esteve-Gassent MD, Labandeira-Rey M, Kim JH, Trzeciakowski JP, Hook M, Skare JT. Inactivation of the fibronectin-binding adhesin gene bbk32 significantly attenuates the infectivity potential of Borrelia burgdorferi. Molecular Microbiology. 2006;59:1591–1601. doi: 10.1111/j.1365-2958.2005.05042.x. [DOI] [PubMed] [Google Scholar]
- Shi Y, Xu Q, McShan K, Liang FT. Both decorin-binding proteins A and B are critical for the overall virulence of Borrelia burgdorferi. Infection and Immunity. 2008;76:1239–1246. doi: 10.1128/IAI.00897-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek G, Wormser GP, Gray J, Strle F. Lyme borreliosis. Lancet. 2012;379:461–473. doi: 10.1016/S0140-6736(11)60103-7. [DOI] [PubMed] [Google Scholar]
- Steere AC, Coburn J, Glickstein L. The emergence of Lyme disease. The Journal of Clinical Investigation. 2004;113:1093–1101. doi: 10.1172/JCI21681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B, Schwan TG, Rosa PA. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infection and Immunity. 1995;63:4535–4539. doi: 10.1128/iai.63.11.4535-4539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B, Tilly K, Rosa PA. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. Journal of Bacteriology. 1996;178:3508–3516. doi: 10.1128/jb.178.12.3508-3516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PE, Thalken R, Bono JL, Rosa P. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Molecular Microbiology. 2001;39:714–721. doi: 10.1046/j.1365-2958.2001.02256.x. [DOI] [PubMed] [Google Scholar]
- Szczepanski A, Furie MB, Benach JL, Lane BP, Fleit HB. Interaction between Borrelia burgdorferi and endothelium in vitro. The Journal of Clinical Investigation. 1990;85:1637–1647. doi: 10.1172/JCI114615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DD, Comstock LE. Interaction of Lyme disease spirochetes with cultured eucaryotic cells. Infection and Immunity. 1989;57:1324–1326. doi: 10.1128/iai.57.4.1324-1326.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma A, Brissette CA, Bowman A, Stevenson B. Borrelia burgdorferi BmpA is a laminin-binding protein. Infection and Immunity. 2009;77:4940–4946. doi: 10.1128/IAI.01420-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, van Dam AP, Schwartz I, Dankert J. Molecular typing of Borrelia burgdorferi sensu lato: taxonomic, epidemiological, and clinical implications. Clin Microbiol Rev. 1999;12:633–653. doi: 10.1128/cmr.12.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weening EH, Parveen N, Trzeciakowski JP, Leong JM, Hook M, Skare JT. Borrelia burgdorferi lacking DbpBA exhibits an early survival defect during experimental infection. Infection and Immunity. 2008;76:5694–5705. doi: 10.1128/IAI.00690-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrano MC, Beklemisheva AA, Bryksin AV, Newman SA, Cabello FC. Borrelia burgdorferi binds to, invades, and colonizes native type I collagen lattices. Infection and Immunity. 2004;72:3138–3146. doi: 10.1128/IAI.72.6.3138-3146.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi H, Weening EH, Barbu EM, Hyde JA, Hook M, Skare JT. The BBA33 lipoprotein binds collagen and impacts Borrelia burgdorferi pathogenesis. Molecular Microbiology. 2015 doi: 10.1111/mmi.12921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.