Abstract
Introduction
Melanoma origin has always been a debated subject, as well as the role of adjacent melanocytic nevi. Epidemiological and histopathological studies point to melanomas arising either de novo or from a nevus.
Methods
Sixty-one melanomas found in association with a preexisting nevus were microdissected, after careful selection of cell subpopulations and submitted to Sanger sequencing of the BRAF, NRAS, C-KIT, PPP6C, STK19 and RAC1 genes. Each gene was evaluated twice in all samples by sequencing or by sequencing and another confirmation method, allele-specific fluorescent polymerase chain reaction (PCR) and capillary electrophoresis detection, or by SNaPshot Analysis. Only mutations confirmed via two different molecular methods or twice by sequencing were considered positive.
Results
The majority of cases presented concordance of mutational status between melanoma and the associated nevus for all 6 genes (40/60; 66.7%). Nine cases presented concomitant BRAF and NRAS mutations, including one case, in which both the melanoma and the adjacent nevus harbored V600E and Q61K double mutations. In two cases, both melanoma and associated nevus, located on acral sites were BRAF mutated, including an acral lentiginous melanoma.
Conclusions
This is the largest nevus-associated melanoma series molecularly evaluated to our knowledge. The majority of melanomas and adjacent nevi in our sample share the same mutational profile, corroborating the theory that the adjacent nevus and melanoma are clonally related and that melanoma originated within a nevus.
Keywords: melanoma, nevus, BRAF, NRAS, V600E
Introduction
Melanoma origin has always been a much debated subject, as well as the precursor role of pigmented lesions. Considering epidemiological and histological studies, melanoma may arise in association with a pre-existing nevus or de novo, without any associated lesion 1. It is controversial whether nevus-associated melanoma and de novo melanomas have a different prognosis 2–6, but a prior nevus-associated melanoma seems to be associated with a 9-fold increased risk of presenting another nevus-associated melanoma 7. According to histopathological studies, the incidence of nevus-associated melanoma may vary from 20–50% of all melanoma cases 2,5,8–18. Some authors suggest that benign melanocytic lesions could correspond to precursor lesions in melanoma genesis 19–23. Although the contiguity of nevus cells with melanoma, may provide some clues as to the etiopathogenesis of malignant melanoma, direct evidence of a causal relationship between the two is still lacking. The extent of genetic alterations has been shown to increase in parallel with the transitions from benign nevi to dysplastic nevi (DN) to melanoma, supporting multistep tumorigenesis 24. Further evidence that nevi could correspond to initial steps in melanomagenesis are the facts that nevi and melanoma share genetic alterations and BRAF mutated melanomas are more likely to be associated with multiple melanocytic nevi than BRAF-wild type melanomas 25,26.
Although molecular studies of melanoma and nevus present in contiguity suggested that melanoma and nevus cells could be clonally related 22,27–30, there is still no consensus for the nevus-to-melanoma progression model 31. While previous studies 22,27–32 differentiated nevus and melanoma cells using morphologic criteria only, it is important to point out that melanoma diagnosis may vary even among experts 33–35 and distinction of contiguous nevus cells and melanoma cells can be doubtful, especially in nevoid melanomas 35,36 and melanomas with adjacent dysplastic nevus 37,38. Thus possible selection bias of cell populations in the previous studies cannot be ruled out when considering clonality between melanoma and adjacent nevus. A combination of morphologic and immunohistochemistry markers would be the most reliable diagnostic method for melanoma 39. Therefore we sought to evaluate the presence of mutations in genes from well known melanomagenesis pathways, such as BRAF, NRAS, c-KIT, as well as new candidates as driver genes: RAC1, PPP6C and STK19 40, after careful selection of melanoma and nevus cell subpopulations in the largest series of nevus-associated melanomas, to our knowledge.
Materials and Methods
Cases of melanoma found in histological association with a nevus were evaluated. All Formalin-fixed, paraffin-embedded (FFPE) samples studied were collected from the archives of a dermatopathology referral center in São Paulo-Brazil and the Hospital Clínic of Barcelona-Spain. The clinico-pathological data was extracted from the histopathological history. New slides stained with hematoxylin and eosin (HE) were independently evaluated by experienced dermatopathologists (N.M., M.M.S.S.E. and L.A.) and a dermatologist (D.S.). The nevus cytology criteria used for differentiation from melanoma cells were described elsewhere 41–43. Sixty-one cases (34 pairs from Brazil and 27 from Spain), which presented concordance for the diagnosis of nevus-associated melanoma by at least two of the evaluators and fulfilled the following criteria: undisputed distinction between nevus and melanoma and sufficient biological material from both melanoma and nevus cells for further analysis, were included.
All study participants gave their written informed consent to participate and the study was approved by the research ethic committees of the Federal University of São Paulo/UNIFESP-EPM (number 188.713 of 24/01/2013) and the Hospital Clínic of Barcelona.
Immunohistochemistry
Available tumor tissue from 47 cases was stained using HMB-45 (Cell Marque, NL) and/or Ki-67 (Spring Bioscience, USA), Melan-A (Dako, DEN) as recommended by the manufacturer and slides were evaluated by two observers (D.S. and M.M.S.S.E.) in order to properly differentiate melanoma cells from nevus cells.
Laser Microdissection and DNA extraction
Contiguous nevus-melanoma specimens from FFPE archival tissue were sectioned (5 μm thick) on polyethylene naphthalate membranes (PEN Membrane slides®, LEICA, US) mounted on regular glass slides, according to manufacturer’s instructions, and stained with HE. Melanoma and nevus cells were microdissected using LEICA microdissector (LMD6500®/ LEICA Microsystems) according to previous marking on correspondent HE slides. DNA was extracted using the QIAmp DNA Micro kit (QIAGEN® Sample & Assay Technologies, Spain).
Mutational screening of BRAF, NRAS, C-KIT, PPP6C, STK19 and RAC1 by Sanger sequencing
BRAF (exons 11 and 15), NRAS (exons 2 and 3), c-KIT (exon 11), PPP6C (exon 7), RAC1 (exon 2) and STK19 (exon 2) genes were amplified using nested PCR with 40–60ng of DNA for each reaction. PCR conditions were: denaturation at 95°C 5 min, 10 cycles (94°C 40sec, 60°C–55°C 40 sec, 72°C 1 min), followed by 25 cycles (94°C 40sec, 57°C 40 sec, 72°C 1 min) and extension at 72°C (10 min). The second PCR mixture was prepared using 1μl from the first PCR reaction. The nested PCR conditions were the same, except that the annealing temperature was 57°C. Nested primers were designed surrounding the exons (200–300bp long) with standard M13 forward (M13F: 5′ TGTAAAACGACGGCCAGT 3′) and M13 reverse (M13R: 5′CAGGAAACAGCTATGACC 3′) tails being included on the internal primers to standardize the capillary sequencing. Nucleotide sequences of nested primers are reported in Tables 1a and 1b. The PCR products were cleaned up using 6μL of Exo-SAP-IT (Applied Biosystems, CA, USA) and amplification was confirmed by 2% agarose gel electrophoresis. The entire amplicons were sequenced using both primers M13RF and M13R with BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, CA, USA) on an ABI PRISM 3130xl automatic sequencer (Applied Biosystems, CA, USA) and analyzed using the SeqPilot 4.0.1 software (JSI Medical Systems).
Table 1a.
Primers used for the amplification of BRAF, NRAS, C-KIT, PPP6C, RAC1 and STK19 genes.
| Gene | Primer | Exon | Primer Sequence | Length of product (bp) |
|---|---|---|---|---|
| BRAF | Forward | 11 | 5′ TTTCTTTTTCTGTTTGGCTTG | 200 |
| BRAF | Reverse | 11 | 5′ ACTTGTCACAATGTCACCACA | 200 |
| BRAF | Forward | 15 | 5′ TGCTTGCTCTGATAGGAAAA | 204 |
| BRAF | Reverse | 15 | 5′ TCAGTGGAAAAATAGCCTCA | 204 |
| NRAS | Forward | 2 | 5′ CGCCAATTAACCCTGATTAC | 201 |
| NRAS | Reverse | 2 | 5′ AGAGACAGGATCAGGTCAGC | 201 |
| NRAS | Forward | 3 | 5′ CCCCTTACCCTCCACACC | 245 |
| NRAS | Reverse | 3 | 5′ AACACAAAGATCATCCTTTCAGA | 245 |
| C-KIT | Forward | 11 | 5′ TGTTCTCTCTCCAGAGTGCTCTAA | 291 |
| C-KIT | Reverse | 11 | 5′ AAACAAAGGAAGCCACTGGA | 291 |
| PPP6C | Forward | 7 | 5′ AAACTCATCTGCAGAGCACA | 271 |
| PPP6C | Reverse | 7 | 5′ AAGAAGAGGGCAGAAAAATG | 271 |
| RAC1 | Forward | 2 | 5′ TGTGATGTATATGCCTTGATTTT | 254 |
| RAC1 | Reverse | 2 | 5′ AGCAAAACAAATGGTCAAAG | 254 |
| STK19 | Forward | 2 | 5′ GACAAGTTGACGCTCCTTTC | 270 |
| STK19 | Reverse | 2 | 5′ AGAGGATCCGACTCCACAG | 270 |
Table 1b.
Primers used in nested PCR.
| Gene | Primer | Exon | Primer sequence | Length of product (bp) |
|---|---|---|---|---|
| BRAF | Forward | 11 | 5′ TGTAAAACGACGGCCAGTTTTCTTTTTCTGTTTGGCTTG | 222 |
| BRAF | Reverse | 11 | 5′ CAGGAAACAGCTATGACCACTTGTCACAATGTCACCACA | 222 |
| BRAF | Forward | 15 | 5′ TGTAAAACGACGGCCAGTTGCTTGCTCTGATAGGAAAA | 217 |
| BRAF | Reverse | 15 | 5′ CAGGAAACAGCTATGACCTCAGTGGAAAAATAGCCTCA | 217 |
| C-KIT | Forward | 11 | 5′ TGTAAAACGACGGCCAGTTGTTCTCTCTCCAGAGTGCTCTAA | 270 |
| C-KIT | Reverse | 11 | 5′ CAGGAAACAGCTATGACCAAACAAAGGAAGCCACTGGA | 270 |
| NRAS | Forward | 2 | 5′ TGTAAAACGACGGCCAGTACCCTGATTACTGGTTTCCA | 253 |
| NRAS | Reverse | 2 | 5′ CAGGAAACAGCTATGACCGATCAGGTCAGCGGGCTA | 253 |
| NRAS | Forward | 3 | 5′ TGTAAAACGACGGCCAGTCTTACCCTCCACACCCCC | 274 |
| NRAS | Reverse | 3 | 5′ CAGGAAACAGCTATGACCCAAAGATCATCCTTTCAGAGAA | 274 |
| PPP6C | Forward | 7 | 5′ TGTAAAACGACGGCCAGTCATCAACTAGTGCACGAAGG | 210 |
| PPP6C | Reverse | 7 | 5′ CAGGAAACAGCTATGACCCGTTCTGGGAGGAATAACAC | 210 |
| RAC1 | Forward | 2 | 5′ TGTAAAACGACGGCCAGTTTTAGAGCTGTAGGTAAAACTTGC | 148 |
| RAC1 | Reverse | 2 | 5′ CAGGAAACAGCTATGACCGCAAAACAAATGGTCAAAGA | 148 |
| STK19 | Forward | 2 | 5′ TGTAAAACGACGGCCAGTAAGTTGACGCTCCTTTCGT | 192 |
| STK19 | Reverse | 2 | 5′ CAGGAAACAGCTATGACCGGATCAGGTGATGCCTCTT | 192 |
Letters in bold correspond to the M13 tails sequence included on the nested primers.
BRAF p.V600E Mutational Analysis
The c.1799T>A (p.V600E) mutation was determined using allele-specific fluorescent PCR and capillary electrophoresis detection. Genomic DNA samples were amplified using primers described elsewhere 44. PCR conditions were: denaturation at 95°C for 5 min; 35 cycles (94°C for 30 sec, 56°C for 40 sec, 72°C for 40 sec); extension at 72°C for 10 min. Amplicons were detected using capillary electrophoresis on ABI PRISM 3130xl and analyzed using GeneMapper Software (Applied Biosystems/Life Technologies, Grand Island, NY).
SNaPshot Analysis
The SNaPshot® Multiplex System was performed to confirm the results obtained by sequencing in the BRAF, NRAS and c-KIT genes45. We performed the multiplex PCR and extension PCR under the same conditions, using previously described primers 45.
Results
Table 2 summarizes the clinical characteristics for the 61paired cases (melanomas and their associated nevi) and their complete molecular screening results. Each gene was evaluated twice in all samples either by two different molecular methods or by Sanger sequencing twice and only confirmed mutations were considered as positive. The DNA extraction of nevus number 6 failed; therefore this pair was excluded from the analysis.
Table 2.
Clinical characteristics and molecular screening results of melanomas and associated nevi
| CASE | BRAF | NRAS | CKIT | PPP6C | STK19 | RAC1 | LOCAL | SUBTYPE | BRESLOW | CLARK | type of nevus |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | WT A,B,* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | HEAD | N/S | 0.7 | III | N/A |
| 2 | V600E A,B,* | WT A,B | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | N/S | 1.25 | III | N/A |
| 3 | V600E A,B,* | WT A,B | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | N/S | IS | I | N/A |
| 4 | WT A,B,* | WTA,#,Q61KB,# | WT A,B | WT A,B | WT A,B | WT A,B | HEAD | SSM | 0.45 | III | N/A |
| 5 | WT A,B,* | WT A,B | WT A,B | WT A,B | WT A,B | WT A,B | EXT | N/S | IS | I | N/A |
| 6 | N/AA,V600EB,* | N/AA,WTB | N/AA,WTB | N/AA,WTB | N/AA,WTB | N/AA,WTB | EXT | NM | 2.5 | III | junctional |
| 7 | V600EA,WTB | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | N/S | IS | I | N/A |
| 8 | WT A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | N/S | IS | I | N/A |
| 9 | WT A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | N/S | IS | I | junctional |
| 10 | WT A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | N/A | SSM | 0.8 | III | N/A |
| 11 | WT A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | N/S | IS | I | compound |
| 12 | WT A,B* | WT A,B | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | 1.15 | III | N/A |
| 13 | WT A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | N/S | IS | I | compound |
| 14 | WTA,V600EB* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | N/A | SSM | 0.4 | II | junctional |
| 15 | V600E A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | 0.4 | II | N/A |
| 16 | V600E A,B* | Q61KA,#, WTB,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | 0.5 | II | compound |
| 17 | V600E A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | EXT | SSM | 0.35 | II | N/A |
| 18 | WT A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | 0.75 | III | compound |
| 19 | V600E A,B* | WT A,B | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | N/S | IS | I | intradermal |
| 20 | WT A,B* | WT A,B | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | 0.7 | III | compound |
| 21 | WT A,B* | WT A,B | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | N/S | IS | I | compound |
| 22 | WT A,B* | WT A,B | WT A,B | WT A,B | WT A,B | WT A,B | EXT | N/S | IS | I | junctional |
| 23 | V600E A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | EXT | N/S | IS | I | congenital |
| 24 | V600E A,B* | WTA,#,Q61KB,# | WT A,B | WT A,B | WT A,B | WT A,B | EXT | N/S | IS | I | junctional |
| 25 | V600E A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | N/A | SSM | 0.45 | II | junctional |
| 26 | V600EA,WTB | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | 0.5 | II | compound |
| 27 | V600EA*,WTB* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | EXT | N/S | IS | I | N/A |
| 28 | WTA*,V600EB* | WT A,B | WT A,B | WT A,B | WT A,B | WT A,B | N/A | N/S | IS | I | N/A |
| 29 | V600EA*,WTB* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | IS | I | N/A |
| 30 | WTA*,V600EB* | WTA,#,Q61KB,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | 0.5 | II | N/A |
| 31 | WT A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | IS | I | compound |
| 32 | WTA*,V600EB* | WTA,#,Q61KB,# | WT A,B | WT A,B | WT A,B | WT A,B | EXT | SSM | IS | I | N/A |
| 33 | WT A,B* | Q61K A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | N/S | IS | I | compound |
| 34 | V600E A,B* | WT A,B | WT A,B | WT A,B | WT A,B | WT A,B | EXT | SSM | 0.45 | II | compound |
| 35 | V600E A,B* | Q61K A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | EXT | SSM | 0.5 | III | N/A |
| 36 | WT A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | N/A | N/A | N/A | N/A | N/A |
| 37 | V600E A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | 0.52 | III | N/A |
| 38 | V600EA*,WTB* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | 0.6 | III | N/A |
| 39 | V600E A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | IS | I | N/A |
| 40 | WT A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | 1.01 | IV | N/A |
| 41 | V600E A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | 0.8 | III | congenital |
| 42 | V600EA*,WTB* | WTA,#,Q61KB,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | 0.7 | III | N/A |
| 43 | WT A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | 0.5 | III | N/A |
| 44 | V600EA*,WTB* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | N/A | SSM | 0.7 | IV | N/A |
| 45 | WT A,B* | WTA,#,Q61KB,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | IS | I | congenital |
| 46 | WT A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | EXT | SSM | 0.3 | II | N/A |
| 47 | V600EA*,WTB* | Q70H/Q61K A,#, Q61KB,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | 0.9 | III | N/A |
| 48 | WTA*,V600EB* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | 0.8 | III | congenital |
| 49 | WT A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | EXT | SSM | 1.2 | IV | intradermal |
| 50 | V600E A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | ACRAL | ACRAL LENTIGINOUS | 1.3 | IV | intradermal |
| 51 | WT A,B* | WT A,B | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | 1.2 | IV | N/A |
| 52 | WT A,B* | Q61K A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | 0.5 | II | N/A |
| 53 | V600E A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | N/A | SSM | 0.85 | III | N/A |
| 54 | V600E A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | 0.6 | III | congenital |
| 55 | V600E A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | EXT | SSM | 1 | III | N/A |
| 56 | V600E A,B* | Q61KA,#, WTB,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | 1.4 | III | N/A |
| 57 | V600E A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | N/A | SSM | 0.55 | I | N/A |
| 58 | WT A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | EXT | SSM | 0.5 | II | N/A |
| 59 | V600E A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | EXT | N/A | N/A | N/A | N/A |
| 60 | V600E A,B* | WT A,B,# | WT A,B | WT A,B | WT A,B | WT A,B | TRUNK | SSM | 1.2 | IV | congenital |
| 61 | V600E A,B* | D33D/Q61KA,#, WTB,# | WT A,B | WT A,B | WT A,B | WT A,B | ACRAL | N/S | IS | I | N/A |
N/A: not available; N/S: not specified
Nevus result;
Melanoma result; EXT: extremities; SSM: superficial spreading melanoma; IS: in situ melanoma;
Result determined by Sanger sequencing and V600E allele-specific fluorescent PCR;
Result determined by Sanger sequencing and SNaPshot method.
Clinical sample characteristics (Table 2)
Most melanomas were located on trunk and extremities (34/61 and 15/61 respectively). No mucosal melanomas were found. Most melanomas were superficial spreading melanomas (SSM) (39/61), one nodular melanoma, one acral lentiginous melanoma (ALM) and 18 were incipient melanomas which did not fulfill all the criteria for SSM. Two cases had incomplete pathological information.
Six associated nevi (10.0%) corresponded to congenital subtype, while 6 were junctional, 10 compound and 3 intradermal common nevi and in 36 cases, no subtype could be specified. Only one of these latter showed signs of dysplasia (1/36). Twenty-one melanomas were in situ melanomas, and most invasive melanomas were thin melanomas, with a mean Breslow thickness of 0.75mm.
Frequency of BRAF mutations in melanomas and associated nevi
BRAF exons 11 and 15 were evaluated in 60 paired cases. No mutation was detected in exon 11. In exon 15, we detected the V600E (c.1799T>A) mutation in 46.6% (28/60) of melanomas and in 51.7% (31/60) of nevi. Regarding BRAF mutational status, most paired cases were concordant (78.3%; 47/60; Fig. 1). In 38.3% of cases a V600E-mutated melanoma had a preexisting V600E-mutated nevus (23/60). Among the discordant cases, a V600E-mutated melanoma had an adjacent BRAF-WT nevus in 8.3% of cases (5/60), while in 13% a V600E-mutated nevus was associated with a BRAF-WT melanoma (8/60).
Figure 1.
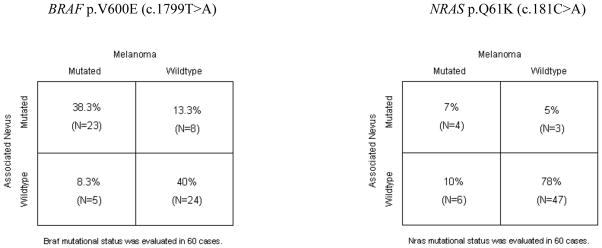
Comparison of mutational status between melanomas and associated nevi, adapted with the numbers of the present study from Tschandl et al.
The V600E mutation was confirmed in all cases using allele-specific fluorescent PCR. With this technique we could also estimate the percentage of mutated DNA present in each sample. Focusing on the concordant BRAF V600E mutated cases (23 pairs), we observed the same percentage of V600E mutation in 39% of these cases (9/23). However, 11 melanomas showed a higher percentage of V600E mutation than their associated nevi (48%; 11/23) and 3 melanomas showed a lower percentage of V600E mutation in comparison with their associated nevi (13%; 3/23).
Frequency of NRAS mutations in melanomas and associated nevi
NRAS exons 2 and 3 were evaluated in 60 paired cases. No mutation was detected in exon 2, but in exon 3 we observed the Q61K (c.181C>A) mutation in 21.7% (13/60) of cases (Fig. 1). Regarding NRAS mutational status, 85% of cases were concordant, being 78% (47/60) WT and 7% mutated (4/60). However, in 10% of cases we detected a mutated melanoma adjacent to a WT nevus (6/60) and the remaining 5% were wild-type melanomas associated with a mutated nevus (3/60). Additionally, we observed a double mutation in NRAS exon 3 in two nevi (Table 2): in one case, melanoma and its associated nevus were both Q61K mutated but the nevus also harbored the Q70H (c.210A>T) variation. In the other case, an NRAS wild-type melanoma was associated with a Q61K mutated nevus that also harbored a D33D (c.99T>C) synonymous variation. These two nucleotide variations have not been previously reported.
Frequency of C-KIT, PPP6C, STK19 and RAC1 mutations in melanomas and associated nevi
No mutations were detected. All paired cases were wild-type for these genes.
Discussion
Driver mutations, being either the activation of an oncogene or the loss of a tumor suppressor allele, could trigger the establishment of a benign lesion. Thereafter the senescence program would control the tumor expansion, and another mutation would be necessary for tumor progression 30,46,47. Molecular analysis of melanomas and so called “precursor lesions” separately, so far point to alterations in critical genes of the MAPK and the PI3-Kinase-Akt pathways, as well as in genes involved in cell cycle regulation, as molecular pathways involved in melanoma genesis in the majority of cases 48,49. Indeed, most cases in this study show mutations in either BRAF or NRAS (67.2%; 41/61). The presence of BRAF V600E mutation in 31 of the studied nevi is in concordance with the hypothesis that this mutation may be an early event in melanocytic tumorigenesis, even though alone it is not sufficient for progression to melanoma50–55. Most of the associated nevi included in this study were common melanocytic nevi. Since there is controversy regarding the diagnosis of dysplastic nevi 56,57, and since we only included cases in which distinction between melanoma and nevus cells was concordant among the observers, it may have led to a selection bias toward the inclusion of nevi with less dysplasia. A limitation observed by other authors 31. Unfortunately, since most of the nevi observed in our study were not dysplastic, even though the majority of our cases depicted the same mutational status between melanoma and nevus, no conclusion regarding the Clark multi-step melanoma progression can be made.
In our study the majority of cases (78.3%; 47/60; Table 2; Fig. 1) presented the same BRAF mutational status for both melanoma and nevus (mut/mut; WT/WT), being mutated in 48% (23/47) of concordant pairs, similar to previous studies 27,29,31,58. Considering these latter, the percentage of V600E mutation was similar in melanoma and nevus in 39% (9/23) of cases, while in 48% (11/23) the melanomas harbored a higher percentage of mutant cells compared to the adjacent nevus. The results support the hypothesis that the presence of V600E BRAF mutation confers an advantage for melanoma progression.
In a significant proportion of our cases (21.7%; 13/60) we observed that either the melanoma or the nevus was mutated while the counterpart was WT. It has been suggested that BRAF mutations are fully clonal in melanocytic nevi, so that it is expected that melanomas deriving from these nevi would be BRAF mutated as well 59. Therefore cases in which only the nevus was mutated favors the possibility of the absence of a clonal relationship between these two lesions. In cases where the BRAF mutation was only present in the melanoma, the possibility of the melanoma arising in the nevus cannot be excluded, since the melanoma may have later acquired the mutation or mutant parts of the associated nevus may have been overgrown by the melanoma and therefore may not have been accessible for analysis.
Recently Kakavand et al., after obtaining 100% concordance of BRAF mutational status between melanomas and associated nevi, suggested that a higher percentage of discordant melanomas and associated nevi obtained by Sanger sequencing, might be related to the technical difficulty of micro-dissecting out melanoma and admixed nevus cells. This could eventually explain why in case 47, both melanoma and nevus were Q61K mutated, while only the nevus was V600E mutated. Nevertheless cases with only the melanoma BRAF and NRAS mutated (cases 30 and 32) and the presence of a nevus with mutation in BRAF while the melanoma harbored mutation in NRAS (case 42), are more difficult to explain by technical differences among mutation detection techniques, and could highlight the fact that in some cases the contiguity of melanoma and nevus correspond to mere fortuitous collision.
Although it has been referred that BRAF V600 and NRAS Q61R 60 are among the most recurrent nucleotide substitutions in melanoma, all NRAS mutations observed in this study were Q61K, being this mutation described in melanomas and congenital nevi 61. Only one case in previous study 27–31 on melanoma and associated nevus described BRAF and NRAS mutations in the same lesion, while we found 9 cases with concomitant BRAF V600E and NRAS Q61K mutations. A case, in which both melanoma and associated nevus harbored V600E and Q61K (case 35, Table 2; Fig. 2) and case 24, in which melanoma and nevus were V600E mutated, and only the melanoma acquired a Q61K mutation, further favors the possibility of melanoma and nevus cells being clonally related. BRAF and NRAS mutations are said to be mutually exclusive 49,62–66. Mutation of both BRAF and RAS, may not provide an additional significant growth/survival advantage (epistatic theory) 67 or is not compatible with cell survival (synthetic lethality theory) 68,69. Nevertheless there have been a few reports of primary melanomas harboring both mutations 70–73, with the hypothesis that these melanomas represent a mosaic with both mutations being mutually exclusive at a single cell level 70. This mosaic explanation may explain the recurrence of melanomas after BRAF inhibition 74. The presence of the Q61K mutation in both melanoma and nevus suggest that this mutation alone may not be sufficient for melanoma induction 75.
Figure 2.
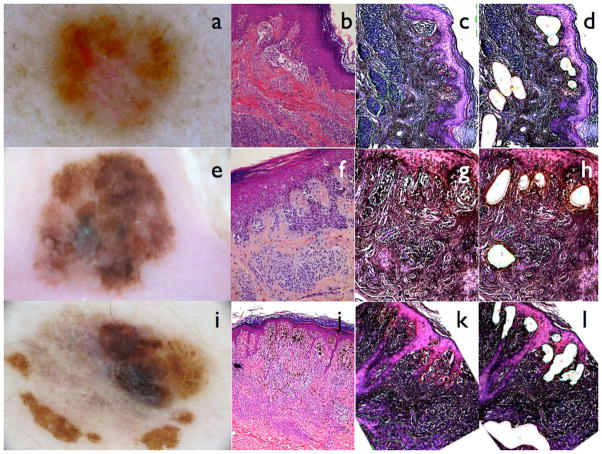
Melanomas and associated nevi: Dermoscopy (a), Haematoxylin-eosin (b) and laser microdissection (c) (d) of case 35, melanoma and associated nevus both mutated for BRAF V600E and NRAS Q61K. Dermoscopy (e), HE (f) and laser microdissection (g)(h) of case 50, acral lentiginous melanoma and adjacent nevus both BRAF V600E mutated. Dermoscopy (i), HE (j) and laser microdissection (k)(l) of case 53, superficial spreading melanoma and adjacent nevus both BRAF V600E mutated. (c) and (k) Delimitation of melanoma cells in red and naevus cells in green. (g) Delimitation of melanoma cells in green and naevus cells in red. (d), (h) and (l) show the empty areas of melanoma and nevus cells after laser dissection.
This is the first molecular study to evaluate the presence of cKIT among melanoma and associated nevus 22,27–32. KIT has been related to acral, mucosal and chronic sun exposure melanomas 53. We did not observe mutations in cKIT, not even among the Brazilian cases, even though solar exposure in Brazil is high. Interestingly, 2 cases located on acral sites were V600E mutated for both melanomas and nevi (Cases 50 and 61). One was an ALM (case 50) and the other was an in situ melanoma with no specified subtype (case 61). In the latter the nevus also harbored mutations in NRAS (D33D and Q61K), not present in the melanoma. Even though it was suggested that most acral melanomas would correspond to de novo melanomas 47, recently Lacruz et al described two cases of multiple BRAF-mutated acral melanomas. One of the cases presented a subungueal melanoma and a nevus-associated melanoma on the toe, both BRAF mutated. The findings of our study corroborate the idea that nevus-associated melanomas located on acral sites would not be related to cKIT as de novo acral melanomas, but instead to BRAF, as nevus-associated melanomas from other sites, such as trunk 76.
Only two cases from our sample were located on the head, one wild-type (Case 1), while in the other, only the melanoma harbored a Q61K mutation (Case 4). Both cases were BRAF-WT, concordant with the fact that BRAF is less frequent in high sun exposure areas, when compared to intermittent sun exposure areas 77. It has been suggested that there may be more than one pathway to melanoma 19,78. Roughly melanomas would be classified: 1) CSD melanomas (Chronic sun damage) related to chronic sun exposure, KIT or NRAS mutations and radial growth phase comprised of melanocytes arranged as solitary units with poor lateral circumscription; 2) non-CSD melanomas related to intermittent sun exposure, located on trunk and proximal extremities, association with nevus either directly adjacent or elsewhere, greater frequency of BRAF mutations and larger, slightly pigmented melanocytes arranged primarily in nests and that display upward intraepidermal scatter in their radial growth phase; 3) Acral melanomas with about 20% of mutations in KIT and 4) mucosal melanomas. The findings of the present study on 61 nevus-associated melanomas, that the majority of mutations observed affected BRAF, most cases were located on the trunk and proximal extremities and of the SSMM subtype and also the two acral nevus-associated melanomas were BRAF mutated, corroborate the idea that melanomas may have different origins and that most nevus-associated melanomas may be classified amongst the non-CSD subtype.
Recently a list of genes with a statistically significant functional mutation burden was described after whole-genome sequencing. Among the most important were six well-known cancer genes (BRAF, NRAS, PTEN, TP53, p16INK4a and MAP2K1) and five new candidates for driver mutations, SNX31, TACC1, PPP6C, RAC1 and STK19 40,79, the latter three harboring recurrent mutations. Despite the high rate of amplification in our study, we didn’t find any mutations in the candidate driver genes (RAC1, STK19, PPP6C) evaluated. Mutations in RAC1, including the P29S mutation (UV-signature activating mutation)40, and in PPP6C are more frequently observed in sun-exposed melanomas. Even though our sample was mostly comprised of chronic or intermittent sun exposed melanomas, no mutation in these genes was detected.
Considering altogether the molecular results for the 6 genes evaluated (BRAF, NRAS, c-KIT, RAC1, PPP6C and STK19), the majority of cases in our study presented concordance of mutational status between melanoma and nevus (40/60; 66.7%).
When evaluating the hypothesis of clonality between melanoma and nevus, the correct sampling of cells is vital, especially in nevoid melanomas and melanomas adjacent to dysplastic nevi. We carefully selected melanoma and nevus cells allying morphologic criteria and immunohistochemistry, since none of these criteria is 100% specific in the diagnosis of melanoma 39 and have included cases where the distinction between melanoma and nevus was concordant among the observers. Therefore, after minimizing the selection bias for the subpopulation of melanoma and adjacent nevus cells, the majority of melanomas and adjacent nevus share the same mutational profile regarding the most relevant genetic hallmarks described in melanoma genesis, corroborating the theory that these lesions may be clonally related and that melanoma most probably originated within the nevus. Nevertheless, at least for some cases, the possibility of a fortuitous collision event cannot be ruled out 18. Even though we tested a broad panel of genes, the most frequently mutated genes in nevus-associated melanoma remain related to the MAPK pathway.
What is known about this topic?
Epidemiological and histopathological studies point to melanomas arising either de novo or associated with a nevus, but the role of the adjacent benign lesion remains to be elucidated.
What does this study add?
The majority of melanomas and adjacent nevi share the same mutational profile, corroborating the theory that melanoma and the adjacent nevus are clonally related and that melanoma originated within a nevus
Acknowledgments
Funding sources: The research at Federal University of São Paulo is funded by Grants from FAPESP (Fundação de Auxílio à Pesquisa do Estado de São Paulo número 2012/15238-0). The research at the Melanoma Unit in Barcelona is partially funded by Grants from Fondo de Investigaciones Sanitarias (12/00840) from the Health Institute Carlos III Spain, by the AGAUR 2014_SGR_603 of the Catalan Government, Spain; by the European Commission under the 6th Framework Programme, Contract nr: LSHC-CT-2006-018702 (GenoMEL) and by the National Cancer Institute (NCI) of the US National Institute of Health (NIH) (CA83115).
We thank all melanoma patients for participating in this study. We thank Helena Kruyer for the English editing of the manuscript.
Footnotes
Conflict of interest
The authors declare no conflicts of interest.
Author contribution
Dr(s)Shitara, Tell-Martí, Badenas, Enokihara, Alós, Larque, Michalany, Carrera, Malvehy, Bagatin, and Puig had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: Dr(s) Puig. Acquisition of data: Dr(s)Shitara, Carrera, Malvehy, Alós, Larque, Michalany, Enokihara. Analysis and interpretation of data: Dr(s)Shitara, Tell-Martí, Badenas, Puig-Butille and Puig. Drafting of the manuscript: Dr(s)Shitara, Tell-Martí, Bagatin and Puig. Critical revision of the manuscript for important intellectual content: Dr(s) Shitara, Tell-Martí, Badenas, Enokihara, Alós, Michalany, Carrera, Malvehy, Bagatin, Puig-Butille and Puig. Statistical analysis: Dr Puig. Obtained funding: Dr(s) Puig, Shitara and Bagatin. Administrative, technical or material support: Dr(s) Carrera, Malvehy, Puig and Bagatin. Study supervision: Dr(s) Puig and Bagatin.
References
- 1.Thomas NE, Groben P. Invasive superficial spreading melanomas arising from clinically normal skin. J Am Acad Dermatol. 2004;51:466–70. doi: 10.1016/j.jaad.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 2.Purdue MP, From L, Armstrong BK, et al. Etiologic and other factors predicting nevus-associated cutaneous malignant melanoma. Cancer Epidemiol Biomarkers Prev. 2005;14:2015–22. doi: 10.1158/1055-9965.EPI-05-0097. [DOI] [PubMed] [Google Scholar]
- 3.Lin WM, Luo S, Muzikansky A, et al. Outcome of patients with de novo versus nevus-associated melanoma. J Am Acad Dermatol. 2015;72:54–8. doi: 10.1016/j.jaad.2014.09.028. [DOI] [PubMed] [Google Scholar]
- 4.Weatherhead SC, Haniffa M, Lawrence CM. Melanomas arising from naevi and de novo melanomas--does origin matter? Br J Dermatol. 2007;156:72–6. doi: 10.1111/j.1365-2133.2006.07570.x. [DOI] [PubMed] [Google Scholar]
- 5.Kaddu S, Smolle J, Zenahlik P, et al. Melanoma with benign melanocytic naevus components: reappraisal of clinicopathological features and prognosis. Melanoma Res. 2002;12:271–8. doi: 10.1097/00008390-200206000-00011. [DOI] [PubMed] [Google Scholar]
- 6.Shitara D, Nascimento MM, Puig S, et al. Nevus-associated melanomas: clinicopathologic features. Am J Clin Pathol. 2014;142:485–91. doi: 10.1309/AJCP4L5CJGKTJVDD. [DOI] [PubMed] [Google Scholar]
- 7.Echeverria B, Botella-Estrada R, Serra-Guillen C, et al. Increased risk of developing a second primary cutaneous nevus-associated melanoma in patients previously diagnosed with the disease. Actas Dermosifiliogr. 2010;101:710–6. [PubMed] [Google Scholar]
- 8.Togawa Y, Nakamura Y, Kamada N, et al. Melanoma in association with acquired melanocytic nevus in Japan: a review of cases in the literature. Int J Dermatol. 2010;49:1362–7. doi: 10.1111/j.1365-4632.2010.04602.x. [DOI] [PubMed] [Google Scholar]
- 9.Stolz W, Schmoeckel C, Landthaler M, et al. Association of early malignant melanoma with nevocytic nevi. Cancer. 1989;63:550–5. doi: 10.1002/1097-0142(19890201)63:3<550::aid-cncr2820630325>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 10.Skender-Kalnenas TM, English DR, Heenan PJ. Benign melanocytic lesions: risk markers or precursors of cutaneous melanoma? J Am Acad Dermatol. 1995;33:1000–7. doi: 10.1016/0190-9622(95)90294-5. [DOI] [PubMed] [Google Scholar]
- 11.Tannous ZS, Mihm MC, Jr, Sober AJ, et al. Congenital melanocytic nevi: clinical and histopathologic features, risk of melanoma, and clinical management. J Am Acad Dermatol. 2005;52:197–203. doi: 10.1016/j.jaad.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 12.Watt AJ, Kotsis SV, Chung KC. Risk of melanoma arising in large congenital melanocytic nevi: a systematic review. Plast Reconstr Surg. 2004;113:1968–74. doi: 10.1097/01.prs.0000122209.10277.2a. [DOI] [PubMed] [Google Scholar]
- 13.Zaal LH, Mooi WJ, Sillevis Smitt JH, et al. Classification of congenital melanocytic naevi and malignant transformation: a review of the literature. Br J Plast Surg. 2004;57:707–19. doi: 10.1016/j.bjps.2004.04.022. [DOI] [PubMed] [Google Scholar]
- 14.Kinsler VA, Chong WK, Aylett SE, et al. Complications of congenital melanocytic naevi in children: analysis of 16 years’ experience and clinical practice. Br J Dermatol. 2008;159:907–14. doi: 10.1111/j.1365-2133.2008.08775.x. [DOI] [PubMed] [Google Scholar]
- 15.Bett BJ. Large or multiple congenital melanocytic nevi: occurrence of cutaneous melanoma in 1008 persons. J Am Acad Dermatol. 2005;52:793–7. doi: 10.1016/j.jaad.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 16.Wise SR, Capra G, Martin P, et al. Malignant melanoma transformation within a nevus of Ito. J Am Acad Dermatol. 2010;62:869–74. doi: 10.1016/j.jaad.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 17.Smolle J, Kaddu S, Kerl H. Non-random spatial association of melanoma and naevi--a morphometric analysis. Melanoma Res. 1999;9:407–12. doi: 10.1097/00008390-199908000-00011. [DOI] [PubMed] [Google Scholar]
- 18.Massi D, Carli P, Franchi A, et al. Naevus-associated melanomas: cause or chance? Melanoma Res. 1999;9:85–91. doi: 10.1097/00008390-199902000-00011. [DOI] [PubMed] [Google Scholar]
- 19.Whiteman DC, Pavan WJ, Bastian BC. The melanomas: a synthesis of epidemiological, clinical, histopathological, genetic, and biological aspects, supporting distinct subtypes, causal pathways, and cells of origin. Pigment Cell Melanoma Res. 2011;24:879–97. doi: 10.1111/j.1755-148X.2011.00880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsao H, Bevona C, Goggins W, et al. The transformation rate of moles (melanocytic nevi) into cutaneous melanoma: a population-based estimate. Arch Dermatol. 2003;139:282–8. doi: 10.1001/archderm.139.3.282. [DOI] [PubMed] [Google Scholar]
- 21.Hussein MR. Melanocytic dysplastic naevi occupy the middle ground between benign melanocytic naevi and cutaneous malignant melanomas: emerging clues. J Clin Pathol. 2005;58:453–6. doi: 10.1136/jcp.2004.019422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bogdan I, Smolle J, Kerl H, et al. Melanoma ex naevo: a study of the associated naevus. Melanoma Res. 2003;13:213–7. doi: 10.1097/01.cmr.0000056226.78713.99. [DOI] [PubMed] [Google Scholar]
- 23.Bevona C, Goggins W, Quinn T, et al. Cutaneous melanomas associated with nevi. Arch Dermatol. 2003;139:1620–4. doi: 10.1001/archderm.139.12.1620. discussion 4. [DOI] [PubMed] [Google Scholar]
- 24.Hussein MR. Genetic pathways to melanoma tumorigenesis. J Clin Pathol. 2004;57:797–801. doi: 10.1136/jcp.2003.015800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maldonado JL, Fridlyand J, Patel H, et al. Determinants of BRAF mutations in primary melanomas. J Natl Cancer Inst. 2003;95:1878–90. doi: 10.1093/jnci/djg123. [DOI] [PubMed] [Google Scholar]
- 26.Thomas NE, Edmiston SN, Alexander A, et al. Number of nevi and early-life ambient UV exposure are associated with BRAF-mutant melanoma. Cancer Epidemiol Biomarkers Prev. 2007;16:991–7. doi: 10.1158/1055-9965.EPI-06-1038. [DOI] [PubMed] [Google Scholar]
- 27.Dadzie OE, Yang S, Emley A, et al. RAS and RAF mutations in banal melanocytic aggregates contiguous with primary cutaneous melanoma: clues to melanomagenesis. Br J Dermatol. 2009;160:368–75. doi: 10.1111/j.1365-2133.2008.08887.x. [DOI] [PubMed] [Google Scholar]
- 28.Demunter A, Stas M, Degreef H, et al. Analysis of N- and K-ras mutations in the distinctive tumor progression phases of melanoma. J Invest Dermatol. 2001;117:1483–9. doi: 10.1046/j.0022-202x.2001.01601.x. [DOI] [PubMed] [Google Scholar]
- 29.Yazdi AS, Palmedo G, Flaig MJ, et al. Mutations of the BRAF gene in benign and malignant melanocytic lesions. J Invest Dermatol. 2003;121:1160–2. doi: 10.1046/j.1523-1747.2003.12559.x. [DOI] [PubMed] [Google Scholar]
- 30.Vredeveld LC, Possik PA, Smit MA, et al. Abrogation of BRAFV600E-induced senescence by PI3K pathway activation contributes to melanomagenesis. Genes Dev. 2012;26:1055–69. doi: 10.1101/gad.187252.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tschandl P, Berghoff AS, Preusser M, et al. NRAS and BRAF mutations in melanoma-associated nevi and uninvolved nevi. PLoS One. 2013;8:e69639. doi: 10.1371/journal.pone.0069639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winnepenninckx V, van den Oord JJ. p16INK4A expression in malignant melanomas with or without a contiguous naevus remnant: a clue to their divergent pathogenesis? Melanoma Res. 2004;14:321–2. doi: 10.1097/01.cmr.0000134855.12474.f3. [DOI] [PubMed] [Google Scholar]
- 33.Edwards SL, Blessing K. Problematic pigmented lesions: approach to diagnosis. J Clin Pathol. 2000;53:409–18. doi: 10.1136/jcp.53.6.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shoo BA, Sagebiel RW, Kashani-Sabet M. Discordance in the histopathologic diagnosis of melanoma at a melanoma referral center. J Am Acad Dermatol. 2010;62:751–6. doi: 10.1016/j.jaad.2009.09.043. [DOI] [PubMed] [Google Scholar]
- 35.McKee PH. Clues to the diagnosis of atypical melanocytic lesions. Histopathology. 2010;56:100–11. doi: 10.1111/j.1365-2559.2009.03451.x. [DOI] [PubMed] [Google Scholar]
- 36.Safa G, Fromentoux S, Darrieux L, et al. Nodal Melanoma Metastasis under Infliximab Therapy in a Patient with Nevoid Melanoma First Misdiagnosed as Benign Nevus: A Potentially Dangerous Diagnostic Pitfall in the Era of Biologic Therapies. Case Rep Dermatol. 2013;5:290–4. doi: 10.1159/000355670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cerroni L, Kerl H. Tutorial on melanocytic lesions. Am J Dermatopathol. 2001;23:237–41. doi: 10.1097/00000372-200106000-00016. [DOI] [PubMed] [Google Scholar]
- 38.McNutt NS. “Triggered trap”: nevoid malignant melanoma. Semin Diagn Pathol. 1998;15:203–9. [PubMed] [Google Scholar]
- 39.Chudnovsky Y, Khavari PA, Adams AE. Melanoma genetics and the development of rational therapeutics. J Clin Invest. 2005;115:813–24. doi: 10.1172/JCI24808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hodis E, Watson IR, Kryukov GV, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Veronese LA, Corrente JE, Marques MEA. Melanoma maligno cutàneo: sistema de pontos (scoring system) para auxílio no diagnóstico histopatologico. J Bras Patol Med Lab. 2006;42:375–83. [Google Scholar]
- 42.Veronese LA, Corrente JE, Marques MEA. Critérios histopatológicos para diagnóstico de melanoma maligno cutâneo: análise comparativa de sua frequência em lesões benignas e melanomas de pequena espessura (<2mm) J Bras Patol Med Lab. 2007;43:363–8. [Google Scholar]
- 43.Barnhill R. Textbook of Dermatopathology. United Statesof America: McGraw-Hill; 1998. [Google Scholar]
- 44.Giudice S, Benassi L, Bertazzoni G, et al. Biological evaluation of MR36, a novel non-polyglutamatable thymidylate synthase inhibitor that blocks cell cycle progression in melanoma cell lines. Invest New Drugs. 2012;30:1484–92. doi: 10.1007/s10637-011-9733-2. [DOI] [PubMed] [Google Scholar]
- 45.Ponti G, Luppi G, Losi L, et al. p16 immunohistochemistry of multiple primary melanomas as screening to identify Familial Melanoma Syndrome. Int J Dermatol. 2012;51:488–92. doi: 10.1111/j.1365-4632.2010.04496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Michaloglou C, Vredeveld LC, Mooi WJ, et al. BRAF(E600) in benign and malignant human tumours. Oncogene. 2008;27:877–95. doi: 10.1038/sj.onc.1210704. [DOI] [PubMed] [Google Scholar]
- 47.Takata M, Murata H, Saida T. Molecular pathogenesis of malignant melanoma: a different perspective from the studies of melanocytic nevus and acral melanoma. Pigment Cell Melanoma Res. 2010;23:64–71. doi: 10.1111/j.1755-148X.2009.00645.x. [DOI] [PubMed] [Google Scholar]
- 48.Dutton-Regester K, Hayward NK. Reviewing the somatic genetics of melanoma: from current to future analytical approaches. Pigment Cell Melanoma Res. 2012;25:144–54. doi: 10.1111/j.1755-148X.2012.00975.x. [DOI] [PubMed] [Google Scholar]
- 49.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 50.Hocker TL, Singh MK, Tsao H. Melanoma genetics and therapeutic approaches in the 21st century: moving from the benchside to the bedside. J Invest Dermatol. 2008;128:2575–95. doi: 10.1038/jid.2008.226. [DOI] [PubMed] [Google Scholar]
- 51.Singh M, Lin J, Hocker TL, et al. Genetics of melanoma tumorigenesis. Br J Dermatol. 2008;158:15–21. doi: 10.1111/j.1365-2133.2007.08316.x. [DOI] [PubMed] [Google Scholar]
- 52.Pollock PM, Harper UL, Hansen KS, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 53.Nikolaou VA, Stratigos AJ, Flaherty KT, et al. Melanoma: new insights and new therapies. J Invest Dermatol. 2012;132:854–63. doi: 10.1038/jid.2011.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ross AL, Sanchez MI, Grichnik JM. Molecular nevogenesis. Dermatol Res Pract. 2011;2011:463184. doi: 10.1155/2011/463184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dessars B, De Raeve LE, Morandini R, et al. Genotypic and gene expression studies in congenital melanocytic nevi: insight into initial steps of melanotumorigenesis. J Invest Dermatol. 2009;129:139–47. doi: 10.1038/jid.2008.203. [DOI] [PubMed] [Google Scholar]
- 56.Ackerman AB. What naevus is dysplastic, a syndrome and the commonest precursor of malignant melanoma? A riddle and an answer. Histopathology. 1988;13:241–56. doi: 10.1111/j.1365-2559.1988.tb02036.x. [DOI] [PubMed] [Google Scholar]
- 57.Clark WH, Jr, Reimer RR, Greene M, et al. Origin of familial malignant melanomas from heritable melanocytic lesions ‘The B-K mole syndrome’. Arch Dermatol. 1978;114:732–8. [PubMed] [Google Scholar]
- 58.Kakavand H, Crainic O, Lum T, et al. Concordant BRAFV600E mutation status in primary melanomas and associated naevi: implications for mutation testing of primary melanomas. Pathology. 2014;46:193–8. doi: 10.1097/PAT.0000000000000077. [DOI] [PubMed] [Google Scholar]
- 59.Yeh I, von Deimling A, Bastian BC. Clonal BRAF mutations in melanocytic nevi and initiating role of BRAF in melanocytic neoplasia. J Natl Cancer Inst. 2013;105:917–9. doi: 10.1093/jnci/djt119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang FW, Hodis E, Xu MJ, et al. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–9. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kinsler VA, Thomas AC, Ishida M, et al. Multiple congenital melanocytic nevi and neurocutaneous melanosis are caused by postzygotic mutations in codon 61 of NRAS. J Invest Dermatol. 2013;133:2229–36. doi: 10.1038/jid.2013.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chattopadhyay C, Ellerhorst JA, Ekmekcioglu S, et al. Association of activated c-Met with NRAS-mutated human melanomas. Int J Cancer. 2012;131:E56–65. doi: 10.1002/ijc.26487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goydos JS, Mann B, Kim HJ, et al. Detection of B-RAF and N-RAS mutations in human melanoma. J Am Coll Surg. 2005;200:362–70. doi: 10.1016/j.jamcollsurg.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 64.VanBrocklin MW, Verhaegen M, Soengas MS, et al. Mitogen-activated protein kinase inhibition induces translocation of Bmf to promote apoptosis in melanoma. Cancer Res. 2009;69:1985–94. doi: 10.1158/0008-5472.CAN-08-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–47. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 66.Colombino M, Lissia A, Capone M, et al. Heterogeneous distribution of BRAF/NRAS mutations among Italian patients with advanced melanoma. J Transl Med. 2013;11:202. doi: 10.1186/1479-5876-11-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rajagopalan H, Bardelli A, Lengauer C, et al. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 68.Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–98. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 69.Petti C, Molla A, Vegetti C, et al. Coexpression of NRASQ61R and BRAFV600E in human melanoma cells activates senescence and increases susceptibility to cell-mediated cytotoxicity. Cancer Res. 2006;66:6503–11. doi: 10.1158/0008-5472.CAN-05-4671. [DOI] [PubMed] [Google Scholar]
- 70.Sensi M, Nicolini G, Petti C, et al. Mutually exclusive NRASQ61R and BRAFV600E mutations at the single-cell level in the same human melanoma. Oncogene. 2006;25:3357–64. doi: 10.1038/sj.onc.1209379. [DOI] [PubMed] [Google Scholar]
- 71.Akslen LA, Angelini S, Straume O, et al. BRAF and NRAS mutations are frequent in nodular melanoma but are not associated with tumor cell proliferation or patient survival. J Invest Dermatol. 2005;125:312–7. doi: 10.1111/j.0022-202X.2005.23788.x. [DOI] [PubMed] [Google Scholar]
- 72.Kumar R, Angelini S, Hemminki K. Activating BRAF and N-Ras mutations in sporadic primary melanomas: an inverse association with allelic loss on chromosome 9. Oncogene. 2003;22:9217–24. doi: 10.1038/sj.onc.1206909. [DOI] [PubMed] [Google Scholar]
- 73.Goel VK, Lazar AJ, Warneke CL, et al. Examination of mutations in BRAF, NRAS, and PTEN in primary cutaneous melanoma. J Invest Dermatol. 2006;126:154–60. doi: 10.1038/sj.jid.5700026. [DOI] [PubMed] [Google Scholar]
- 74.Nazarian R, Shi H, Wang Q, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ensembl.org. [last access 18 May 2015];Variation gene table. 2014 2014 Mutation ID: COSM580. Available at Catalogue of Somatic mutations in cancer ( www.sanger.ac.uk) [Google Scholar]
- 76.Lacruz G, Cardenas I, Carrera C, et al. Multiple primary acral melanomas in two young caucasian patients. Dermatology. 2014;228:307–10. doi: 10.1159/000362207. [DOI] [PubMed] [Google Scholar]
- 77.Berger MF, Garraway LA. Applications of genomics in melanoma oncogene discovery. Hematol Oncol Clin North Am. 2009;23:397–414. vii. doi: 10.1016/j.hoc.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Whiteman DC, Watt P, Purdie DM, et al. Melanocytic nevi, solar keratoses, and divergent pathways to cutaneous melanoma. J Natl Cancer Inst. 2003;95 :806–12. doi: 10.1093/jnci/95.11.806. [DOI] [PubMed] [Google Scholar]
- 79.Krauthammer M, Kong Y, Ha BH, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–14. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]