Abstract
To study how miR-34a acts as a tumor suppressor in inhibiting the invasion and metastasis of the gastric cancer cells. First, real-time polymerase chain reaction (PCR) and western blot analysis were used to analyze the expression of miR-34a and Tgif2 in gastric cancer tissues and the adjacent normal tissues. Next, gastric cancer cells were transfected with miR-34a mimic and Tgif2 siRNA, respectively. After transfection, real-time PCR and western blot analysis were used to detect the relative Tgif2 expression level. Cell proliferation was monitored by the colorimetric water-soluble tetrazolium salt and apoptosis analysis was performed with Annexin-V-FITC Apoptosis Detection Kit I. The expression of miR-34a in the adjacent non-tumor tissues was higher than that in gastric cancer tissues, but Tgif2 was opposite. In gastric cancer cells transfected with miR-34a mimic/Tgif siRNA, Tgif2 expression was remarkably down-regulated. Cells transfected with miR-34a mimic/Tgif2 siRNA grew more slowly than the control groups. The percentage of apoptotic cells in gastric cancer cells transfected with miR-34a mimic/Tgif siRNA was much higher compared to the controls. Therefore, we concluded that miR-34a could inhibit tumor invasion and metastasis in gastric cancer by targeting Tgif2 and may be a novel therapeutic candidate for gastric cancer.
Keywords: Gastric cancer cells, MiR-34a, Tgif2, cell proliferation, apoptosis
Introduction
Gastric cancer ranks as the second leading cause of cancer death worldwide, probably accounting for about 10% of newly diagnosed cancers although the incidence and mortality rates have generally declined during the past decades [1]. The outcome of gastric cancer patients mainly resulted from widespread metastasis. Metastasis is a complex, multi-step process whereby cancer cells migrate from the primary neoplasm to a distant location [2]. It occurs through a specific series of steps, starting with local invasion, followed by entrance of cancer cells into the bloodstream (intravasation), survival in the circulation, exit from blood vessels (extravasation), initiation, and maintenance of micrometastases at distant sites and finally, vascularization of the resulting tumors [3].
MicroRNAs (miRNAs) are a newly discovered class of small non-coding RNAs encoded by the genomes of a wide range of multicellular organisms [4]. They have been predicted to play a role in regulating human protein-coding genes expression [5]. They are involved in numerous bioprocesses of an organism, such as development, differentiation, apoptosis and proliferation [6]. Recently, many reports have showed that altered miRNA level resulted from mutation or aberrant expression is associated with diverse human cancers [7]. The abnormally expressed miRNAs in human cancers target transcripts of essential protein-coding genes involved in tumorigenesis, including oncogenes and tumor-suppressor genes [8]. MiR-34a is a gene which can not only suppress tumor, but also take part in the proliferation, migration and metastasis of tumor cells. In human hepatocellular carcinoma, it inhibited the scattering, migration and invasion of hepatocellular carcinoma cells by down-regulation of c-Met expression [9]. In prostate cancer, it inhibited the metastasis of prostate cancer stem cells by directly repressing CD44, which establishes a strong rationale for developing miR-34a as a novel therapeutic agent against prostate CSCs [10]. In p53-deficient human gastric cancer cells, restoration of functional miR-34 inhibits cell growth and induces chemosensitization and apoptosis, indicating that miR-34 may restore p53 function [11].
Tgif2 was found related with many cancers. The over-expression of TGIF2 played an important role in the development and/or progression of some ovarian tumors [12]. Its expression could be regulated by miR-34a. Study showed miR-34a could block osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2 [13]. In gastric cancer cells, the relationship between miR-34a and Tgif2 still remained unclear.
In this paper, we aimed to study the mechanism that miR-34a inhibited the invasion and metastasis of gastric cancer cells. MiR-34a and Tgif2 expression in gastric cancer tissues and the adjacent non-tumor tissues were analyzed by real-time PCR and western blot analysis. Then, gastric cancer cells were transfected with miR-34a mimic/Tgif2 siRNA to study the influence of miR-34a and Tgif2 on the biological behavior of gastric cancer cells. After transfection, real-time PCR and western blot analysis were used to analyze Tgif2 expression. Cell proliferation and apoptosis were also analyzed.
Materials and methods
Cell culture
The human gastric cancer cells were purchased from Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. The immortalized normal gastric mucosal epithelial cell line GES-1 was a gift from Professor Feng Bin (Sichuan University, Chengdu, China). The cells were routinely cultured in RPMI-1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin in a humidified cell incubator with an atmosphere of 5% CO2 at 37°C. Cells growing at an exponential rate were used for the experiments.
Tissue collection
Fresh frozen human tumor samples and human non-neoplastic gastric tissues were obtained from 30 patients with gastric cancer undergoing radical gastrectomy at the department of Surgery, Ruijin Hospital Shanghai Jiao Tong University School of Medicine. None of the patients had undergone preoperative treatment, such as radiation therapy or chemotherapy.
RNA extraction and real-time PCR
Total RNA was extracted and isolated from tissue samples or cell lines using either the mirVana miRNA isolation kit (Ambion Inc, Austin, TX, USA) or the TRIzol method. Standard electrophoresis and spectrophotometric methods were used to assess the quality and quantity of the RNA samples. The expression levels of mature miR-34a and Tgif2 were measured by real-time PCR according to the TaqMan® MicroRNA Assay protocol (PE Applied Biosystems, Foster City, CA, USA) and normalized using U6 small nuclear RNA by the 2-ΔCt method. The relative expressions of Tgif2 in normal gastric mucosal epithelial cells, gastric cancer cells and gastric cancer cells transfected with miR-34a mimic and gastric cancer cells transfected with Tgif2 siRNA were calculated using the 2-ΔΔCt method.
Western blot analysis
Cultured cells were lysed using RIPA buffer in the presence of Protease Inhibitor Cocktail. Tissue samples were lysed using the T-PER Tissue Protein Extraction Reagent (Pierce Chemical Co., LTD. Rockford, IL, USA) in the presence of Protease Inhibitor Cocktail. BCA Protein Assay Kit (Pierce Chemical Co., LTD. Rockford, IL, USA) was used to measure the protein concentration of the lysates. Equivalent amounts of protein were resolved and mixed with 5X Lane Marker Reducing Sample Buffer, electrophoresed in a 12.5% SDS-acrylamide gel, and transferred to Immobilon-P Transfer Membrane (Millipore Corp., Bedford, MA, USA). The membranes were blocked with 5% non-fat milk in Tris-buffered saline and then incubated with mouse anti-human PTEN monoclonal antibody (Santa Cruz Biotech, Santa Cruz, CA, USA) followed by horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotech, Santa Cruz, CA, USA). Signals were detected with Immobilon Western chemiluminescent HRP Substrate (Millipore Corp., Bedford, MA, USA).
Transfection with miR-34a mimic
MiR-34a mimic was purchased from Shanghai GenePharma Co., Ltd (Shanghai, China). Gastric cancer cells were trypsinised, counted, and seeded onto 6-well plates the day prior to transfection to ensure 50% cell confluence on the day of transfection. MiR-34a mimic was transfected into gastric cancer cells using Lipofectamine 2000 (Invitrogen Corp., Carlsbad, CA, USA) according to the manufacturers’ advised procedure. The final concentration of miR-34a mimic was 100 nM. At 48 h post-transfection, real-time PCR and western blot analysis were performed. Transfection efficiency was monitored by the transfection of Cy3-labeled pre-miR™ negative control (Ambion Inc, Austin, TX, USA).
RNA interference targeting Tgif2
RNA interference (RNAi) is an evolutionarily conserved surveillance mechanism that responds to double-stranded RNA by sequence-specific silencing of homologous genes [14]. To investigate the role of Tgif2 in cell proliferation and apoptosis, we used RNAi-based strategy to specifically silence Tgif2 expression. SiRNA common to Tgif2 mRNA was designed. Tgif2 siRNA transfection (50 nmol/L) was performed in cultured gastric cancer cells. Then, RT-PCR and western blot analysis were used to detect relative mRNA level. Colorimetric water-soluble tetrazolium salt (CCK8) assay using a Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan) was used for cell proliferation detection and Annexin-V-FITC Apoptosis Detection Kit I (BD PharMingen, Heidelberg, Germany) was used for apoptosis analysis.
Cell proliferation assay
Cell proliferation was analyzed by the colorimetric water-soluble tetrazolium salt (CCK8) assay using a Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan) in accordance with the manufacturer’s instructions. At 12 h post-transfection with miR-34a mimic or control oligotides, Gastric cancer cells were seeded onto 96-well plates (2×103 cells/well), and cell proliferation was recorded every 12 h for 50 h. The number of viable cells was obtained by measuring the absorbance at 450 nm with a Safire2 microplate reader (Tecan Austria GmbH, Austria).
Apoptosis analysis
At 48 h post-transfection with the miR-34a mimic, Gastric cancer cells were collected by trypsinisation and washed with phosphate-buffered saline (PBS). In apoptosis analysis, an Annexin-V-FITC Apoptosis Detection Kit I (BD PharMingen, Heidelberg, Germany) was used according to the manufacturer’s instructions. After washed with PBS, cells were resuspended in 1X binding buffer at a concentration of 1×106 cells/ml. Then, 5 μl of FITC Annexin-V and 5 μl PI were added to 100 μl of the cell suspension and the samples were incubated for 15 min in the dark. After incubation, 400 μl 1X binding buffer was added. Apoptosis was analyzed by flow cytometry (FACSCalibur, Becton-Dickinson) using the Cell-Quest software (Becton Dickinson, Mountain View, CA, USA). The cells undergoing apoptosis were Annexin-V-FITC-positive and PI-negative.
Statistical analysis
The relationships between the miR-34a expression level and clinicopathological parameters were analyzed using the Pearson χ2 test. For comparisons between two different groups, statistical significance was determined using the Student’s t-test. All statistical analyses were performed using the SAS 6.12 software package. A two-tailed value of P<0.05 was considered statistically significant.
Results
Expression of miR-34a and Tgif2 in tissue samples
To explore the role of miRNAs in gastric cancer, the expression level of miRNAs was a primary consideration. The relative expression levels of miR-34a in gastric cancer tissues and the adjacent non-tumor tissues were shown in Figure 1. As shown, miR-34a was significantly downregulated in gastric cancer tissues compared with its matched non-tumor tissues. The relative expression levels of Tgif2 in gastric cancer tissues and the adjacent non-tumor tissues were shown in Figure 2, which were obtained by real-time PCR and western blot analysis, respectively. As shown, Tgif2 expression value in gastric cancer tissues was higher than that in the adjacent non-tumor tissues. Collectively, these results provided strong evidence that miR-34a and Tgif2 is prominently upregulated and downregulated in gastric cancer, respectively.
Figure 1.
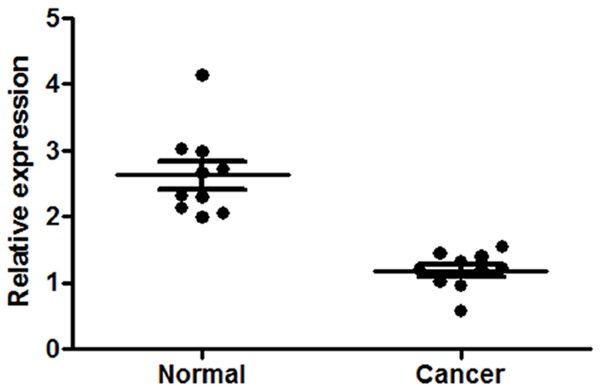
Real-time PCR for the relative expression level of miR-34a in gastric cancer tissues and the adjacent non-tumor tissues.
Figure 2.
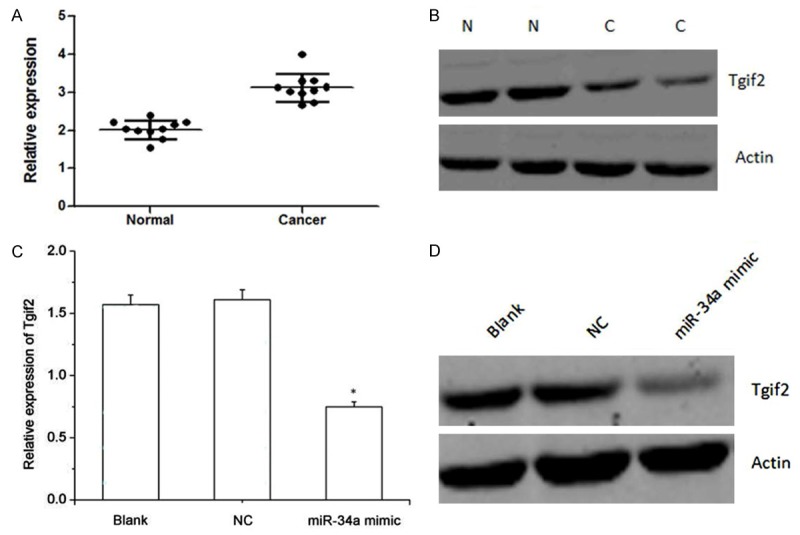
A, B. The relative expression level of Tgif2 in gastric cancer tissues and the adjacent non-tumor tissues. A: Real-time PCR; B: Western blot analysis. N: Normal; C: Cancer. C, D. The relative expression of Tgif2 in gastric cancer transfected with miR-34a mimic and the controls after transfection with miR-34a mimic. C: Real-time PCR; D: Western blot analysis. NC: Normal cancer.
Tgif2 expression level after transfection with miR-34a mimic
The expression level of Tgif2 after gastric cancer cells transfected with miR-34a mimic was shown in Figure 2C, 2D, which was obtained by real-time PCR and western blot. As shown, Tgif2 expression in gastric cancer cells transfected with miR-3a mimic was significantly downregulated compared with the blank and normal gastric cancer cells.
Cell proliferation assay after transfection with miR-34a mimic
Because miR-34a is markedly downregulated in gastric cancer, it may thus function as a tumor suppressor. Therefore, we tested whether over-expression of miR-34a in gastric cancer cells affects cell growth and the result obtain by CCK8 assay was shown in Figure 3. As shown, cells transfected with miR-34a mimic grew more slowly than the control group. The dramatic contrast in proliferative activity indicated that over-expression of miR-34a inhibited the gastric cancer cell growth activity. These results suggested that over-expression of miR-34a inhibited cell growth in vitro. The transfection efficiency was monitored using a Cy3-labeled pre-miR™ negative control.
Figure 3.
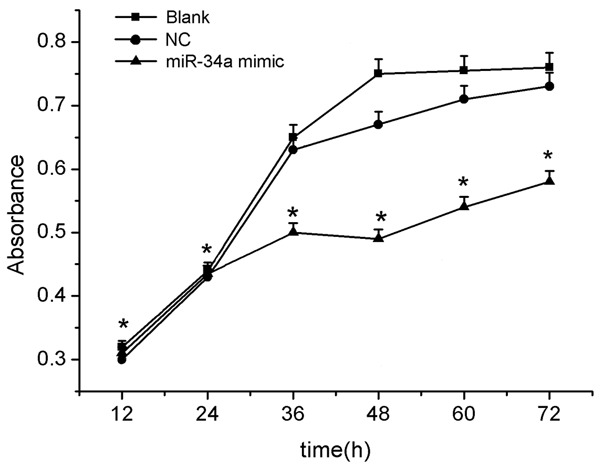
The effect of miR-34a on the proliferation of gastric cancer cells after transfection with miR-34a mimic. Cell proliferation was measured by the CCK8 assay.
Overexpression of miR-34a affects gastric cancer cell apoptosis
To study the mechanism of miR-34a-mediated cell growth reduction in gastric cancer cell, flow cytometry was performed. The effect of miR-34a on the apoptosis of gastric cancer cells was shown in Figure 4A. As shown, the percentage of apoptosis in gastric cancer cells transfected with miR-34a was much higher compared to the controls.
Figure 4.
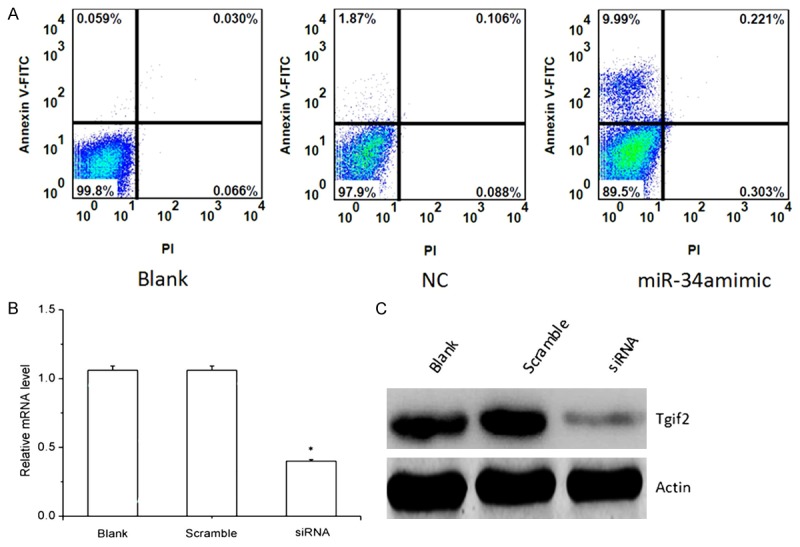
A. The effect of miR-34a on the apoptosis of gastric cancer cells after transfection with miR-34a mimic. Cells were stained with PI and analyzed by flow cytometry at 48 h post-transfection. Cells staining positive for Annexin-V-FITC and negative for PI at 48 h post-transfection were considered to have undergone apoptosis. B, C. The relative expression of Tgif2 in gastric cancer transfected with Tgif2 siRNA and the controls after transfection with Tgif2 siRNA. B: real-time PCR; C: western blot analysis.
Interference of RNA onTgif2
The relative mRNA level of Tgif2 obtained by real-time PCR and western-blot analysis was shown in Figure 4B, 4C. As shown, Tgif2 mRNA level in Tgif2 siRNA transfection cells was lower compared to the blank and scrambled siRNA-transfected cells. Results of gastric cancer cell proliferation and apoptosis after RNAi was shown in Figures 5 and 6, respectively. As shown, cell proliferation was significantly lower in gastric cancer cells transfected with Tgif2 siRNA than in those transfected with scrambled siRNA. The percentage of apoptosis in gastric cancer cells transfected with Tgif2 siRNA was much higher compared to the controls.
Figure 5.
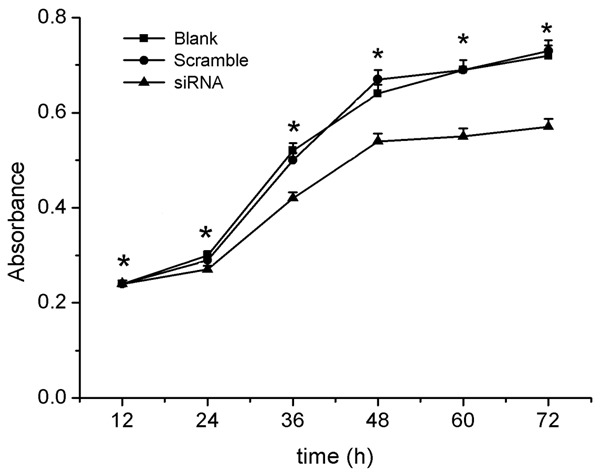
The effect of Tgif2 on the proliferation of gastric cancer cells after transfection with Tgif2 siRNA. Cell proliferation was measured by the CCK8 assay.
Figure 6.
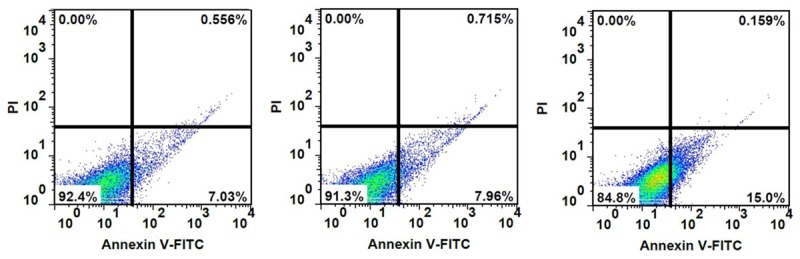
The effect of Tgif2 on the apoptosis of gastric cancer cells after transfection with Tgif2 siRNA. Cells were stained with PI and analyzed by flow cytometry at 48 h post-transfection. Cells staining positive for Annexin-V-FITC and negative for PI at 48 h post-transfection were considered to have undergone apoptosis.
Discussion
In China, gastric cancer is common with high rate of mortality. However, the prognosis for gastric cancer is quite poor. Despite progress in the development of new management strategies, gastric cancer remains difficult to diagnose at an early stage [15]. Thus, the discovery of new biomarkers is of critical importance to allow for early diagnosis of gastric cancer. MicroRNAs are a type of non-coding RNA and important regulators. Recent studies have linked deregulation of miRNAs to various diseases including cancer [16]. Reports have indicated that dysregulation of miRNAs is associated with the formation and progression of gastric cancer [17]. They have been implicated as oncogenes and tumor suppressor genes in several human neoplasms [18]. Therefore, miRNAs are potentially useful biomarkers for clinical diagnosis. MiR-34a, a pivotal member of the p53 network, was found to be down-regulated in multiple types of tumors and further reported as a tumor suppressor microRNA [19]. Reduced expression of miR-34a is a very frequent feature of pancreatic cancer cells, and it causes dramatic reprogramming of gene expression and promotes apoptosis [20]. MiR-34a is generally expressed at lower levels in unfavorable primary neuroblastoma tumors and cell lines relative to normal adrenal tissue [21]. It is downregulated in human glioma tumors as compared to normal brain, and its expression in glioma and medulloblastoma cells inhibits cell proliferation [22].
They are associated with tumorigenesis and tumor progression. MicroRNAs are involved in numerous bioprocesses of an organism, and play as oncogenes or tumor suppressor genes. Many previous reports indicated miR-34a can act as tumor suppressors in diverse cancers. In this study, we investigate whether miR-34a acts as a tumor suppressor to inhibit invasion and metastasis in gastric cancers, and study the expression of Tgif2 in gastric cancer cells. In this study, miR-34a was significantly downregulated in gastric cancer tissues compared with its matched non-tumour tissues. Although miR-34a may be a suppressor for gastric cancer, the mechanism remains unclear.
In order to investigate how miR-34a exerts its role in gastric cancer, we confirmed other experiments and found Tgif2 was up-regulated in gastric cancer tissues compared with its matched non-tumour tissues. Hence, we speculated Tgif2 may be the authentic target gene of miR-34a in gastric cancer. Lujambio et al. reported the reintroduction of miR-148a and miR-34b/c in cancer cells could reduce tumor growth, and inhibit metastasis formation in xenograft models, with an associated down-regulation of the miRNA oncogenic target genes, such as C-MYC, E2F3, CDK6, and TGIF2 [23]. TGIF2 was identified as a modifier of early endoderm, and required for pancreas development [24]. Tgif2 was up-regulated in diffuse-type gastric cancers [25].
To experimentally validate whether Tgif2 was a target gene of miR-34a in gastric cancer, first, gastric cancer was transfected with miR-34a mimic, and study the expression level of Tgif2, gastric cancer cells growth and apoptosis. Then, a RNAi-based strategy was used to specifically silence Tgif2 expression. Gastric cancer cell proliferation and apoptosis were analyzed after transfection with Tgif2 siRNA. In gastric cancer cells transfected with miR-34a mimic experiment, RT-PCR and western blot analysis showed Tgif2 was down-regulated. Cell proliferation assay showed cells transfected with miR-34a mimic grew more slowly than the control group. Apoptosis analysis showed the percentage of apoptotic cells increased after transfection with miR-34a mimic. The results above indicated the over-expression of miR-34a could down-regulated Tgif2, inhibit the gastric cancer cell growth activity and increased the percentage of apoptotic cells. Therefore, we obtained miR-34a could inhibit tumor invasion and metastasis in gastric cancer. In gastric cancer cells transfected with Tgif2 siRNA experiment, gastric cancer cells transfected with Tgif2 siRNA grew more slowly than the control group and the percentage of apoptotic cells increased, which were in consistent with the results in transfection with miR-34a mimic experiment. So, we concluded miR-34a inhibited tumor invasion and metastasis in gastric cancer by targeting Tgif2.
Disclosure of conflict of interest
None.
References
- 1.Murray CJ, Lopez AD. Alternative projections of mortality and disability by cause 1990-2020: Global Burden of Disease Study. Lancet. 1997;349:1498–1504. doi: 10.1016/S0140-6736(96)07492-2. [DOI] [PubMed] [Google Scholar]
- 2.Fidler IJ. The pathogenesis of cancer metastasis: the seed and soil hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 3.Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Krützfeldt J, Poy MN, Stoffel M. Strategies to determine the biological function of microRNAs. Nat Gene. 2006;38:S14–S19. doi: 10.1038/ng1799. [DOI] [PubMed] [Google Scholar]
- 5.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 6.Hwang H, Mendell J. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y, Zheng X. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009;275:44–53. doi: 10.1016/j.canlet.2008.09.035. [DOI] [PubMed] [Google Scholar]
- 10.Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, Wiggins JF, Bader AG, Fagin R, Brown D, Tang DG. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji Q, Hao X, Meng Y, Zhang M, Desano J, Fan D, Xu L. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer. 2008;8:266. doi: 10.1186/1471-2407-8-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imoto I, Pimkhaokham A, Watanabe T, Saito-Ohara F, Soeda E, Inazawa J. Amplification and overexpression of TGIF2, a novel homeobox gene of the TALE superclass, in ovarian cancer cell lines. Biochem Biophys Res Commun. 2000;276:264–270. doi: 10.1006/bbrc.2000.3449. [DOI] [PubMed] [Google Scholar]
- 13.Krzeszinski JY, Wei W, Huynh H, Jin Z, Wang X, Chang TC, Xie XJ, He L, Mangala LS, Lopez-Berestein G, Sood AK, Mendell JT, Wan Y. miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature. 2014;512:431–435. doi: 10.1038/nature13375. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. Gene expression: RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- 15.Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol. 2006;12:354. doi: 10.3748/wjg.v12.i3.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang P, Zou F, Zhang X, Li H, Dulak A, Tomko RJ Jr, Lazo JS, Wang Z, Zhang L, Yu J. microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res. 2009;69:8157–8165. doi: 10.1158/0008-5472.CAN-09-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheung H, Davis A, Lee TL, Nagrani S, Rennert OM, Chan WY. Methylation of an intronic region regulates miR-199a in testicular tumor malignancy. Oncogene. 2011;30:3404–3415. doi: 10.1038/onc.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole KA, Attiyeh EF, Mosse YP, Laquaglia MJ, Diskin SJ, Brodeur GM, Maris JM. A functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol Cancer Res. 2008;6:735–742. doi: 10.1158/1541-7786.MCR-07-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, Yuan L, Luo J, Gao J, Guo J, Xie X. MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin Exp Med. 2013;13:109–117. doi: 10.1007/s10238-012-0186-5. [DOI] [PubMed] [Google Scholar]
- 20.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welch C, Chen Y, Stallings R. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 22.Guessous F, Zhang Y, Kofman A, Catania A, Li Y, Schiff D, Purow B, Abounader R. microRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle. 2010;9:1031–6. doi: 10.4161/cc.9.6.10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lujambio A, Calin GA, Villanueva A, Ropero S, Sánchez-Céspedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, Gallagher WM, Eccles SA, Croce CM, Esteller M. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spagnoli FM, Brivanlou AH. The Gata5 target, TGIF2, defines the pancreatic region by modulating BMP signals within the endoderm. Development. 2008;135:451–461. doi: 10.1242/dev.008458. [DOI] [PubMed] [Google Scholar]
- 25.Jinawath N, Furukawa Y, Hasegawa S, Li M, Tsunoda T, Satoh S, Yamaguchi T, Imamura H, Inoue M, Shiozaki H, Nakamura Y. Comparison of gene-expression profiles between diffuse-and intestinal-type gastric cancers using a genome-wide cDNA microarray. Oncogene. 2004;23:6830–6844. doi: 10.1038/sj.onc.1207886. [DOI] [PubMed] [Google Scholar]


