Abstract
Purpose: The purpose of this study was to investigate the effect of transmembrane-4-l-six-family-1 (TM4SF1) on breast cancer cell line MDA-MB-231 invasion and apoptosis and its mechanism through PI3K/AKT/mTOR pathway. Methods: siRNA-TM4SF1 and pcDNA-TM4SF1 plasma were constructed and then transfected into MDA-MB-231 cells respectively. Real time (RT)-PCR was used to measure the mRNA expression of TM4SF1 in each group. Also, matrigel method and Annexin V-FITC were used to detect the effect of TM4SF1 expression on MDA-MB-231 cell migration and apoptosis respectively. Besides, western blotting analyze was used to assay the effects of TM4SF1 expression on PI3K/AKT/mTOR pathway associated proteins expressions. Results: The results showed that after being transfected with siRNA-TM4SF1, TM4SF1 expression was significantly declined, while it was significantly increased after cells were transfected with pcDNA-TM4SF1 (P<0.05). Compared with the controls, TM4SF1 overexpression significantly contributed MDA-MB-231 cell migration but decreased apoptotic cells (P<0.05), which were opposite to the results when TM4SF1 was sliced in cells. Moreover, TM4SF1 slicing significantly decreased the expressions of phosphorylated (p)-AKT, p-mTOR, and p-P70 (P<0.05). Conclusion: Our study suggested that TM4SF1 may be a therapeutic target for breast cancer treatment and may loan insight into the mechanisms behind the development and metastasis of advanced breast cancer.
Keywords: Breast cancer, transmembrane-4-l-six-family-1 (TM4SF1), cell apoptosis, cell migration, PI3K/AKT/mTOR pathway
Introduction
Breast cancer is a worldwide female malignancy, which arises about 18% among female malignancies. Statistics data refers that about 1.2 million women are diagnosed as breast cancer and the 5-year survival rate is approximately to 50%-60% [1]. Traditional methods on breast cancer treatment are surgery, chemotherapy, radiotherapy and medicine for different subtypes [2,3]. Papers report that invasion and metastasis are the main challenges for breast cancer treatment in clinical [4,5]. However, mechanism of breast cancer invasion and metastasis still remain incompletely described. Thus, it is necessary to explore the deep mechanism for breast cancer invasion and metastasis and several target genes for the disease treatment.
Transmembrane4 L six family protein (TM4SF) are widely distribute in human cells and tissues, and have homologous sequences which are composed by four highly conserved hydrophotic transmembrane domain (TM) [6]. The four TM lead to the cell membrane glycoprotein family of TM4FS proteins involving in cell growth, proliferation, adhesion, migration and invasion in many tumors such as hepatocytes, endothelial cell, colon cancer, and ovarian cancer [7-10]. Lee and his colleagues reported that TM4FS5 was associated with migration and invasion of hepatocytes metastasis and might be a therapeutic target during dealing with TM4FS5-mediated hepatocellular cancers [11]. TM4SF member 1 (TM4SF1) is a member of the transmembrane4 superfamily protein, which is firstly discovered as an antigen for the immunotherapy in lung cancer [12]. Previous studies refer that TM4SF1 is pivotal for endothelial cell function and tumor angiogenesis, including combined with colon epithelial cells adhesion and E-cadherin, ovarian cancer, and hepatoma carcinoma metastasis [13-15]. Recent evidences prove that TM4SF1 is involved in tumor cell migration, invasion and apoptosis. For instance, Gao and Ramachandran refer that TM4SF1 could stimulate prostate cancer cell migration and invasion during its metastasis [16]. Varma reports that TM4SF1 differently expressed in ovarian cancer cell line A2780/C10 and may be a potential target gene for ovarian cancer A2780/C10 [17,18]. However, role of TM4SF1 in female breast cancer has not been reported.
In this study, we investigated the effect of TM4SF1 expression on breast cancer cell migration and invasion based on MDA-MB-231 cell line. Comprehensive experimental methods were used to analyze the effects of TM4SF1 expression on MDA-MB-231 cell proliferation, apoptosis and invasion. The purpose of this study was to investigate the effect of TM4SF1 expression on breast cancer cell apoptosis and invasion, and to illustrate the mechanism of TM4SF1 in breast cancer metastasis.
Materials and methods
Cell culture and plasma transfection
The human breast cancer cell line MDA-MB-231 (purchased from Basic medical cell center, Peking Union medical college) were cultured in Dulbecco’s modified Eagle’s medium (Sigma, USA) supplemented with 10% fetal bovine serum (FBS), 1% streptomycin, and 1% glutaMAX (Sigma, USA) at 37°C in an atmosphere of 5% CO2.
The full length TM4SF1 coding sequence was transfected into the pcDNA3.1 plasma (Invitrogen, USA) to produce a TM4SF1 expression vector of pcDNA-TM4SF1 and then confirmed by sequencing. Also, the empty pcDNA3.1 was transfected into MDA-MB-231 cells as a control group. In addition, target sequence of TM4SF1 that synthesized by Sangon Biotech (Shanghai, China) was linked onto the siRNA plasma to produce siRNA-TM4SF1, as well as the control siRNA (no slicing sequence) was transfected into cells as control group. G418 (Sigma, USA) was used for the stable TM4SF1 transfectants selection according to previously described [19]. Cell transfection was conducted with Lipofectamine 2000 reagent (Invitrogen, USA) based on the manufacture’s protocol.
Real time (RT)-PCR
mRNA expression of TM4SF1 in each group was detected using the RT-PCR analysis as previously described [20]. The main steps were as follows: MDA-MB-231 cells in different group collected at 24 h were grinded in liquid nitrogen and washed with PBS buffer for 3 times. Total RNA from MDA-MB-231 cells was isolated using the Trizol extraction reagent (Invitrogen, USA) according to the manufacturer’s protocol [21], followed by the addition of RNase-free Dnase I (Promega, Biotech) to remove the DNA. Concentration and purity for isolated RNA were measured with SMA4000 UV-VIS (Merinton, Shanghai, China) at 260 nm. Then 0.5 μg/μL purified RNA was used for cDNA synthesis using the Primer Script 1st Strand cDNA Synthesis Kit (Invitrogen, USA). Expression of mRNA was measured with SYBR green-based quantitative RT-PCR (Sangon, Shanghai, China). The total reaction system of 20 μL volume containing 1 μL cDNA from the above PCR, 10 μL SYBR Premix EX Taq, 1 μL each of the primers (10 μM), and 7 μL ddH2O. PCR reaction was carried out at 50°C for 2 min, 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Melting curve analysis of amplification products was performed at the end of each PCR to confirm that only one product was amplified and detected. Phosphoglyceraldehyde dehydrogenase (GAPDH, Sigma, USA) was chosen as the internal control. Primers used for target gene amplification were shown as follows: sense primer: sense primer: 5’-ACCACTATGTCTTGATTCCCTC-3’, anti-sense primer: 5’-ATTGTGGCTCTGTCCTGGGT-3’. GAPDH sense primer: 5’-GGGTGGAGCCAAACGGGTC-3’, anti-sense primer: 5’-GGAGTTGCTGTTGAAGTCGCA-3’.
Cell migration assay
For effect of TM4SF1 expression on MDA-MB-231 cells migration ability, Matrigel method was used as previously described [22]. Cells in each group cultured at 48 h were incubated with serum-free DMEM medium containing 0.01% serum albumin (BSA, Sigma, USA) for 24 h. The upper Transwell was enveloped with serum-free DMEM medium containing 50 mg/L Matrigel and then air-dried at 4°C. After being sucked out the medium, 50 μL fresh serum-free medium containing 10 g/L BSA was added and then cultured for at 37°C 30 min. Then Transwell was put into the 24-well plates and cultured with DMEM medium supplemented with 10% FBS. Cells in Transwell were suspended with serum-free DMEM medium. Transwell in control group was treated without Matrigel. After being cultured for 12 h, Transwell in each group was washed with PBS buffer to remove the upper cells on microporous membrane, followed with fixed in ice-cold alcohol. Finally, Transwell from each group was stained with 0.1% crystal violet for 30 min, and then decolorated with 33% acetic acid. The absorbance of eluents was observed at OD 570 nm using a microplate reader (Biotech, USA). Total experiments were conducted independently for three times.
Apoptosis assay
Influence of TM4SF1 expression on MDA-MB-231 cells apoptosis was assessed with the flow cytometry Annexin V-FITC cell apoptosis kit (Roche, USA) according to the manufacturer’s instructions [23]. Briefly, MDA-MB-231 cells were treated with siRNA-TM4SF1 and pcDNA-TM4SF1 plasma for 24 h, followed by the replacement of serum-free cell culture medium. Total cells were harvested and then washed with PBS buffer with 3 times for 5 min. Then cells were resuspended in the staining buffer. Consequently, 5 μL of annexin-V-FITC and 5 μL of propidium iodide (PI) were added with the cells. After being cultivated at room temperature for 10 min, cells in each group were analyzed using the FACScan flow cytometry. Annexin V-positive and propidium iodide-negative cells were considered to be apoptotic cells. The experiments were conducted three times independently.
Western blotting analysis
MDA-MB-231 breast cancer cells transfected with siRNA-TM4SF1 or pcDNA-TM4SF1 plasma cultured at 48 h in each group were lapped with radioimmunoprecipitation (RIPA) assay (Sangon Biotech, China) lysate containing phenylmethanesufonyl fluoride (PMSF, Sigma, USA), and then were centrifuged at 12,000 rpm for 10 min at 4°C. Supertanant was collected for the measurement of protein concentrations using bicinchoninic acid (BCA) protein assay kit (Pierce, Rochford, IL) [24]. For western blotting, 20 μg protein per cell lysate was subjected to a 12% sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed with transferred onto a polyvinylidencefluoride (PVDF) membrane (Mippore). Then the PVDF membrane was blocked in Tris buffered saline Tween (TBST) containing 5% non-fat milk for 1 h. After that, the membrane was incubated with rabbit anti-human antibodies (mTOR, p-mTOR, P-70, p-P70, AKT, and p-AKT 1:100 dilution, Invitrogen, USA) and overnight at 4°C. Then membrane was incubated with hoseradish peroxidase labeled goat anti-rat secondary antibody (1:1000 dilution) at room temperature for 1 h. Finally, PVDF membrane was washed with 1 × TBST buffer with 3 times for 10 min. Detection was conducted using the development of X-ray after chromogenic substrate with an enhanced chemiluminescence (CEL) method. Additionally, GAPDH served as the internal control.
Statistical analysis
All the data were presented as the mean ± standard deviation (SD). Statistical analyses were performed using SPSS 19.0 statistical software (Armonk, NY, USA). The P-values were calculated using a one-way analysis of variance (ANOVA). The P<0.05 was considered to be statistically significant.
Results
Expression of TM4SF1 in MDA-MB-231 breast cancer cells
After being transfected with pcDNA-TM4SF1 plasma, TM4SF1 level in MDA-MB-231 cells was significantly increased compared with that in normal cells (P<0.05, Figure 1A). Besides, when cells were transfected with siRNA-TM4SF1 plasma, TM4SF1 expression in MDA-MB-231 cells was significantly declined compared with its control (P<0.05, Figure 1B). The results were coincidence with the western blotting analysis (Figure 1).
Figure 1.
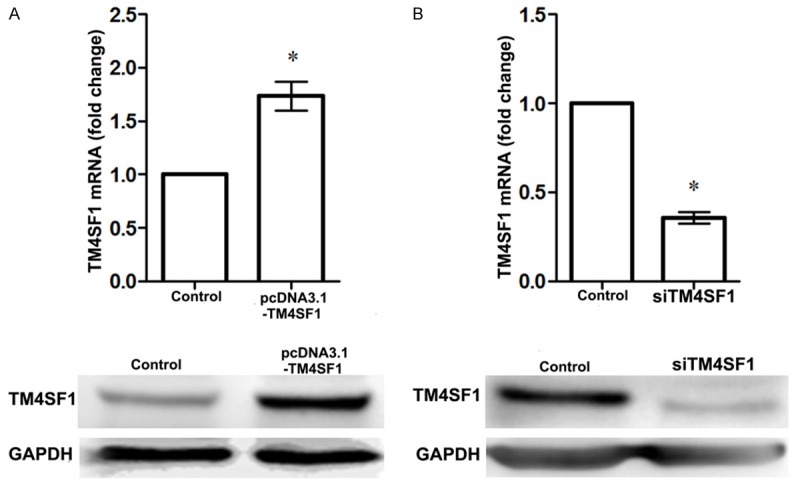
Expression of TM4SF1 in MDA-MB-231 breast cancer cells. A. TM4SF1 was up-regulated in MDA-MB-231 cells after being transfected with pcDNA-TM4SF1 plasma; B. TM4SF1 was down-regulated in MDA-MB-231 cells after being transfected with siRNA-TM4SF1 plasma. *P<0.05, compared with the control cells.
Cell migration assay
Effect of TM4SF1 expression on MDA-MB-231 breast cancer cells was detected using the Matrigel method (Figure 2). After being transfected with siRNA-TM4SF1 plasma, TM4SF1 expression in MDA-MB-231 breast cancer cells was declined but migrate cells were significantly decreased compared with the control (P<0.01, Figure 2B). Besides, when MDA-MB-231 breast cancer cells were transfected with pcDNA-TM4SF1, TM4SF1 expression was up-regulated and migrate breast cells were increased compared that in control group (P<0.01, Figure 2A), indicating that MDA-MB-231 breast cancer cells migrate ability may be positive correlated with TM4SF1 expression.
Figure 2.
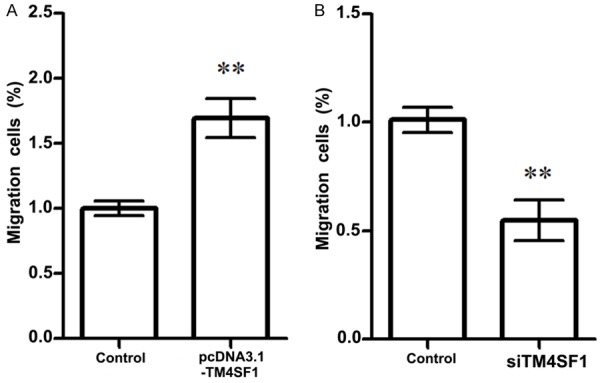
Effects of TM4SF1 expression on MDA-MB-231 breast cancer cell migration. A. Overexpression of TM4SF1 significantly increased MDA-MB-231 breast cancer cell migration; B. TM4SF1 slicing significantly decreased MDA-MB-231 breast cancer cell migration. **P<0.01, compared with the control cells.
Cell apoptosis assay
Flow cytometry was used to assess whether TM4SF1 expression was correlated to MDA-MB-231 breast cancer cell apoptosis or not (Figure 3). Our results showed that when MDA-MB-231 cells were transfected with siRNA-TM4SF1, apoptotic cells in the siRNA-TM4SF1 group were significantly increased compared with the control group (P<0.05, Figure 3B). Besides, when MDA-MB-231 cells were transfected with pcDNA-TM4SF1, apoptotic cells were significantly decreased compared with its control (P<0.05), indicating that overexpression of TM4SF1 was negatively correlated with MDA-MB-231 cell apoptosis (Figure 3A).
Figure 3.
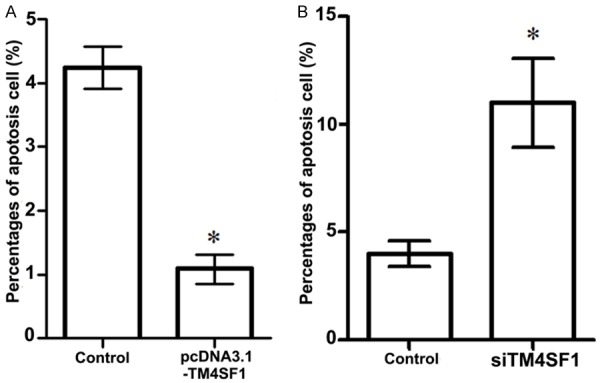
Influences of TM4SF1 expression on MDA-MB-231 breast cancer cell apoptosis. A. Overexpression of TM4SF1 decreased MDA-MB-231 breast cancer cell apoptosis; B. TM4SF1 slicing significantly increased MDA-MB-231 breast cancer cell apoptosis. *P<0.05, compared with the control cells.
Analysis of TM4SF1 expression and PI3K/AKT/mTOR pathway
Western blotting analysis was used to assess the effect of TM4SF1 expression on PI3K/AKT/mTOR pathway associated proteins expression (Figure 4). The results showed that when TM4SF1 was up-regulated in MDA-MB-231 cells, expressions of p-mTOR (mammalian target of rapamycin), P-P70, and p-phosphoinositide-3-kinase (PI3K) were all increased compared with the control group (Figure 4A). Our data showed that when MDA-MB-231 cells were transfected with siRNA-TM4SF1 plasma, expressions of p-mTOR, P-P70, and p-AKT were all decreased compared with the controls (Figure 4B). In addition, the results also performed that migrate cells in TM4SF1 slicing group was significantly decreased compared with the control group (P<0.05), which was the same tendency as migrate cells in inhibitor group (P<0.05). However, when MDA-MB-231 cells were transfected with siRNA-TM4SF1 plasma and with addition of inhibitor, the pathway associated proteins expression such as p-mTOR, P-P70, and p-AKT were all significantly decreased compared with the inhibitor group or TM4SF1 group, suggesting that TM4SF1 functioned as a cell migration inhibitor in breast cells (Figure 4C and 4D).
Figure 4.
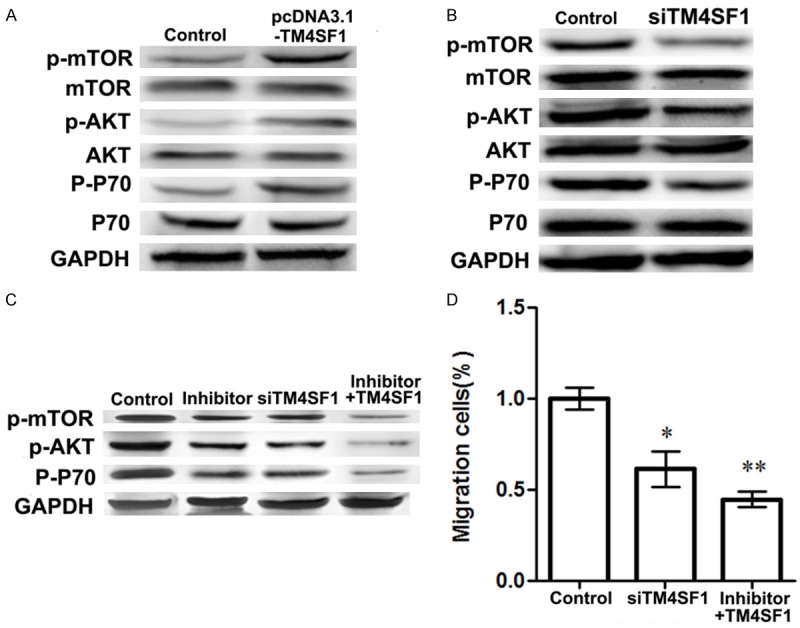
Influence of TM4SF1 expression on PI3K/AKT/mTOR pathway associated proteins expressions. A. Effects of TM4SF1 overexpression on expressions of p-mTOR, p-AKT, and p-P70 in MDA-MB-231 breast cancer cells; B. Effects of TM4SF1 slicing on expressions of p-mTOR, p-AKT, and p-P70 in MDA-MB-231 breast cancer cells; C. Effects of siRNA-TM4SF1 inhibitor on expressions of p-mTOR, p-AKT, and p-P70; D. Migrate MDA-MB-231 breast cancer cells in different group. *P<0.05, **P<0.01, compared with the control cells.
Discussion
Breast cancer is a worldwide female malignancy, which has increasing morbidity [1,4,5]. TM4SF1 has been reported to be associated with many tumors invasion and metastasis, but has not been reported in breast cancer. In this study, we used human breast cancer cell line MDA-MB-231 to analyze the effect of TM4SF1 expression on breast cancer cell invasion and apoptosis. The results presented in this study showed that TM4SF1 overexpression significantly enhanced MDA-MB-231 cells migration but decreased cell apoptosis (P<0.05), which are opposite to the results when TM4SF1 slicing. Moreover, TM4SF1 overexpression significantly increased PI3K/AKT/mTOR pathway associated proteins expressions including p-mTOR, p-AKT, and p-P70, indicating that TM4SF1 may be correlated with breast cancer invasion and migration through PI3K/AKT/mTOR pathway.
Previous papers have shown that cell migration, apoptosis, and invasion were associated with malignancies metastasis and progression [25,26]. Roles of TM4SF1 in breast cancer migration and invasion have not been fully illustrated except for in other tumors. Zukauskas and his colleagues proved that the endothelial cell selectively expressed TM4SF1 was necessary for endothelial cells migration and nanopodia formation [27], and TM4SF1 was overexpressed in prostate cancer and was involved in cell migration [28]. The same functions of TM4SF1 in cell migration and invasion have been reported in pancreatic cancer from Cao’s study [16]. Coincidence with former studies, our results showed that TM4SF1 overexpression could contribute breast cancer cell migration, suggesting that TM4SF1 may be a contributor in breast cancer metastasis. On the other hand, our data showed that TM4SF1 overexpression could inhibit breast cancer cell apoptosis. In Gordon’s study, TM4SF1 was a negative regulator of mesothelioma cell apoptosis, which indicated its potential of therapeutic target for tumor treatment [29]. Thus, we speculated that TM4SF1 may be a negative regulator for breast cancer cell apoptosis and overexpression of TM4SF1 may contribute breast cancer progression through inhibiting cell apoptosis.
Meanwhile, AKT family proteins are involved in many processes including metabolism, proliferation, cell survival, growth and angiogenesis [30], while PI3K family proteins regulate PIP3, which is pivotal in recruiting PH domain-containing proteins to membrane including AKT, and then activating signaling cascades involved in cell growth, migration and function [31]. PI3K signaling pathway is involved in cell growth, survival and movement, and p-AKT of the apoptotic-inducing protein Bad creates a binding site for proteins and prevents Bad from binding to Bcl-2 family, therefore resulting a cell survival response [32]. It has been proved that TM4SF1 could activating the Ras signal and then inducing the downstream activation of PI3K for epithelial mesenchmal transition (EMT) in colon cancers [33]. In Morgensztern’s paper, PI3K/AKT/mTOR pathway has been proved to be correlated to regulate several normal cellular functions that are critical for tumorigenesis, and components in this pathway are frequently abnormal in a variety of tumors, making them an attractive target for cancer therapy [34]. In this study, overexpression of TM4SF1 significantly enhanced p-PI3K, p-AKT, and p-mTOR expressions in MDA-MB-231 cells which were contrary to the results when TM4SF1 was sliced, indicating that TM4SF1 overexpression may be associated with breast cancer migration and metastasis through influencing PI3K/AKT/mTOR signal pathway.
In conclusion, the data presented in this study performed that TM4SF1 expression was involved in breast cancer MDA-MB-231 cell migration and apoptosis through down-regulating the PI3K/AKT/mTOR pathway. TM4SF1 slicing inhibits MDA-MB-231 cell migration, promotes cell apoptosis, and down-regulates PI3K/AKT/mTOR pathway associated proteins expressions including p-PI3K, p-AKT, and p-mTOR. Our study suggests that TM4SF1 may be a therapeutic target for breast cancer treatment and may loan insight into the mechanisms behind the development and metastasis of advanced breast cancer. However, further investigate studies are still needed to explore the deep mechanism for the clinical application of TM4SF1 in breast cancer.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Harder H, Langridge C, Solis-Trapala I, Zammit C, Grant M, Rees D, Burkinshaw L, Jenkins V. Post-operative exercises after breast cancer surgery: Results of a RCT evaluating standard care versus standard care plus additional yoga exercise. European Journal of Integrative Medicine. 2015 [Google Scholar]
- 3.Courneya KS, Segal RJ, Gelmon K, Mackey JR, Friedenreich CM, Yasui Y, Reid RD, Proulx C, Trinh L, Dolan LB. Predictors of adherence to different types and doses of supervised exercise during breast cancer chemotherapy. Int J Behav Nutr Phys Act. 2014;11:85. doi: 10.1186/s12966-014-0085-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, Yu M, Pely A, Engstrom A, Zhu H. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou F, Drabsch Y, Dekker TJ, de Vinuesa AG, Li Y, Hawinkels LJ, Sheppard KA, Goumans MJ, Luwor RB, de Vries CJ. Nuclear receptor NR4A1 promotes breast cancer invasion and metastasis by activating TGF-β signalling. Nature Commun. 2014;5:3388. doi: 10.1038/ncomms4388. [DOI] [PubMed] [Google Scholar]
- 6.Berditchevski F, Tolias KF, Wong K, Carpenter CL, Hemler ME. A novel link between integrins, transmembrane-4 superfamily proteins (CD63 and CD81), and phosphatidylinositol 4-kinase. J Biol Chem. 1997;272:2595–2598. doi: 10.1074/jbc.272.5.2595. [DOI] [PubMed] [Google Scholar]
- 7.Lee S, Kim TY, Kwak TK, Kim H, Kim S, Lee HJ, Kim SH, Park KH, Kim HJ, Cho M. Transmembrane 4 L six family member 5 (TM4SF5) enhances migration and invasion of hepatocytes for effective metastasis. J Cell Biochem. 2010;111:59–66. doi: 10.1002/jcb.22662. [DOI] [PubMed] [Google Scholar]
- 8.Shih SC, Zukauskas A, Li D, Liu G, Ang LH, Nagy JA, Brown LF, Dvorak HF. The L6 protein TM4SF1 is critical for endothelial cell function and tumor angiogenesis. Cancer Res. 2009;69:3272–3277. doi: 10.1158/0008-5472.CAN-08-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhn S, Koch M, Nübel T, Ladwein M, Antolovic D, Klingbeil P, Hildebrand D, Moldenhauer G, Langbein L, Franke WW. A complex of EpCAM, claudin-7, CD44 variant isoforms, and tetraspanins promotes colorectal cancer progression. Mol Cancer Res. 2007;5:553–567. doi: 10.1158/1541-7786.MCR-06-0384. [DOI] [PubMed] [Google Scholar]
- 10.Giordano TJ, Shedden KA, Schwartz DR, Kuick R, Taylor JM, Lee N, Misek DE, Greenson JK, Kardia SL, Beer DG. Organ-specific molecular classification of primary lung, colon, and ovarian adenocarcinomas using gene expression profiles. Am J Pathol. 2001;159:1231–1238. doi: 10.1016/S0002-9440(10)62509-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SA, Kim TY, Kwak TK, Kim H, Kim S, Lee HJ, Kim SH, Park KH, Kim HJ, Cho M, Lee JW. Transmembrane 4 L six family member 5 (TM4SF5) enhances migration and invasion of hepatocytes for effective metastasis. J Cell Biochem. 2010;111:59–66. doi: 10.1002/jcb.22662. [DOI] [PubMed] [Google Scholar]
- 12.Seo DC, Sung JM, Cho HJ, Yi H, Seo KH, Choi IS, Kim DK, Kim JS, El-Aty A, Shin HC. Gene expression profiling of cancer stem cell in human lung adenocarcinoma A549 cells. Mol Cancer. 2007;6:75. doi: 10.1186/1476-4598-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang MY, Wang HM, Chang HJ, Hsiao CP, Wang JY, Lin SR. Overexpression of S100B, TM4SF4, and OLFM4 genes is correlated with liver metastasis in Taiwanese colorectal cancer patients. DNA Cell Biol. 2012;31:43–49. doi: 10.1089/dna.2011.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greco C, Bralet MP, Ailane N, Dubart-Kupperschmitt A, Rubinstein E, Le Naour F, Boucheix C. E-cadherin/p120-catenin and tetraspanin Co-029 cooperate for cell motility control in human colon carcinoma. Cancer Res. 2010;70:7674–7683. doi: 10.1158/0008-5472.CAN-09-4482. [DOI] [PubMed] [Google Scholar]
- 15.Macintyre G, Bailey J, Gustafsson D, Haviv I, Kowalczyk A. Using Gene Ontology annotations in exploratory microarray clustering to understand cancer etiology. Pattern Recognition Letters. 2010;31:2138–2146. [Google Scholar]
- 16.Cao J, Ramachandran V, Arumugam T, Nast F, Logsdon C. TM4SF1 stimulates pancreatic cancer cell migration and invasion. Journal. 2009;38:986–986. [Google Scholar]
- 17.Cody NA, Zietarska M, Filali-Mouhim A, Provencher DM, Mes-Masson AM, Tonin PN. Influence of monolayer, spheroid, and tumor growth conditions on chromosome 3 gene expression in tumorigenic epithelial ovarian cancer cell lines. BMC Med Genom. 2008;1:34. doi: 10.1186/1755-8794-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varma RR, Hector SM, Clark K, Greco WR, Hawthorn L, Pendyala L. Gene expression profiling of a clonal isolate of oxaliplatin-resistant ovarian carcinoma cell line A2780/C10. Oncol Rep. 2005;14:925–932. [PubMed] [Google Scholar]
- 19.Zhang HZ, Wang Y, Gao P, Lin F, Liu L, Yu B, Ren JH, Zhao H, Wang R. Silencing stathmin gene expression by survivin promoter-driven siRNA vector to reverse malignant phenotype of tumor cells. Cancer Biol Ther. 2006;5:1457–1461. doi: 10.4161/cbt.5.11.3272. [DOI] [PubMed] [Google Scholar]
- 20.Huang GL, Zhang XH, Guo GL, Huang KT, Yang KY, Shen X, You J, Hu XQ. Clinical significance of miR-21 expression in breast cancer: SYBR-Green I-based real-time RT-PCR study of invasive ductal carcinoma. Oncol Rep. 2009;21:673–679. [PubMed] [Google Scholar]
- 21.Taylor DD, Zacharias W, Gercel-Taylor C. Serum/plasma proteomics. Springer; 2011. Exosome isolation for proteomic analyses and RNA profiling; pp. 235–246. [DOI] [PubMed] [Google Scholar]
- 22.Poincloux R, Collin O, Lizárraga F, Romao M, Debray M, Piel M, Chavrier P. Contractility of the cell rear drives invasion of breast tumor cells in 3D Matrigel. Proc Natl Acad Sci U S A. 2011;108:1943–1948. doi: 10.1073/pnas.1010396108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faber AC, Corcoran RB, Ebi H, Sequist LV, Waltman BA, Chung E, Incio J, Digumarthy SR, Pollack SF, Song Y. BIM expression in treatmentnaive cancers predicts responsiveness to kinase inhibitors. Cancer Discov. 2011;1:352–365. doi: 10.1158/2159-8290.CD-11-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker JM. The Protein Protocols Handbook. Springer; 2009. The bicinchoninic acid (BCA) assay for protein quantitation; pp. 11–15. [DOI] [PubMed] [Google Scholar]
- 25.Zetter P, Bruce R. Angiogenesis and tumor metastasis. Ann Rev Med. 1998;49:407–424. doi: 10.1146/annurev.med.49.1.407. [DOI] [PubMed] [Google Scholar]
- 26.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 27.Zukauskas A, Merley A, Li D, Ang LH, Sciuto TE, Salman S, Dvorak AM, Dvorak HF, Jaminet SC. TM4SF1: a tetraspanin-like protein necessary for nanopodia formation and endothelial cell migration. Angiogenesis. 2011;14:345–354. doi: 10.1007/s10456-011-9218-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allioli N, Vincent S, Vlaeminck-Guillem V, Decaussin-Petrucci M, Ragage F, Ruffion A, Samarut J. TM4SF1, a novel primary androgen receptor target gene over-expressed in human prostate cancer and involved in cell migration. Prostate. 2011;71:1239–1250. doi: 10.1002/pros.21340. [DOI] [PubMed] [Google Scholar]
- 29.Gordon GJ, Bueno R, Sugarbaker DJ. Genes associated with prognosis after surgery for malignant pleural mesothelioma promote tumor cell survival in vitro. BMC Cancer. 2011;11:169. doi: 10.1186/1471-2407-11-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho C, Wang C, Mattu S, Destefanis G, Ladu S, Delogu S, Armbruster J, Fan L, Lee SA, Jiang L. AKT (v-akt murine thymoma viral oncogene homolog 1) and N-Ras (neuroblastoma ras viral oncogene homolog) coactivation in the mouse liver promotes rapid carcinogenesis by way of mTOR (mammalian target of rapamycin complex 1), FOXM1 (forkhead box M1)/SKP2, and c-Myc pathways. Hepatology. 2012;55:833–845. doi: 10.1002/hep.24736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brader S, Eccles SA. Phosphoinositide 3-kinase signalling pathways in tumor progression, invasion and angiogenesis. Tumori. 2004;90:2–8. doi: 10.1177/030089160409000102. [DOI] [PubMed] [Google Scholar]
- 32.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 33.Golubkov VS, Strongin AY. Downstream signaling and genome-wide regulatory effects of PTK7 pseudokinase and its proteolytic fragments in cancer cells. Cell Commun Signal. 2014;12:15. doi: 10.1186/1478-811X-12-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morgensztern D, McLeod HL. PI3K/Akt/mTOR pathway as a target for cancer therapy. Anticancer Drugs. 2005;16:797–803. doi: 10.1097/01.cad.0000173476.67239.3b. [DOI] [PubMed] [Google Scholar]


