Abstract
Long noncoding RNAs (lncRNAs), a class of ribonucleic molecules, participate in various cellular processes. They are highly expressed in several types of cancer and their expression was related to pathophysiological characteristics of tumor growth, therefore, they can be considered as a promising diagnostic tool and a convenient prognostic biomarker. SPRY4-IT1, belonging to a group of intron-retained lncRNAs, was reported to affect tumor development of many types of cancer. However, the expression and the role of SPRY4-IT1 in glioma are still unclear. Therefore, in this study, we examined for the first time the expression and role of SPRY4-IT1 in glioma cells. The results of our study showed that SPRY4-IT1 was up-regulated in human glioma tissues and cell lines. Knockdown of SPRY4-IT1 could inhibit glioma cell growth and migration. Moreover, knockdown of SPRY4-IT1 could inhibit epithelial-mesenchymal transition (EMT) phenotype in glioma cells. Based on these findings, SPRY4-IT1 may be used as a new target for diagnosis and treatment of glioma.
Keywords: SPRY4-IT1, glioma, proliferation, metastasis, epithelial-mesenchymal transition
Introduction
Glioma, accounting for 10% of tumors developing in the central nervous system, is the most common malignant brain tumor [1]. It is categorized into ependymoma, astrocytoma, oligodendroglioma and choroid plexus papilloma [2]. With the infiltrative and invasive features, glioma exhibits high recurrence and low cure rate [1,3]. Despite significant progress made in glioma treatment with surgery, chemotherapy and radiotherapy, the clinical treatment of gliomas is still unsatisfactory due to the relapse of residual nidus [4]. Therefore, a better understanding of the molecular mechanisms involved in glioma development will help identify new diagnostic and therapeutic targets.
Long noncoding RNAs (lncRNAs) are a class of ribonucleic molecules that are longer than 200 nucleotides but lack protein coding potential [5]. They share similar features with mRNAs, for example, some of them are transcribed by RNA polymerase II and undergo polyadenylation and splicing [6,7]. As important regulatory molecules, lncRNAs participate in various cellular processes. LncRNAs can be considered as a promising diagnostic tool and a convenient prognostic biomarker, for some of them are highly expressed in several types of cancer and their expression was associated with pathophysiological characteristics of tumor growth [8-11]. For example, the long noncoding RNA SPRY4-IT1 was reported to have high expression in melanoma cells and its downregulation inhibited invasion and proliferation of melanoma cells [12]. Zhang et al. reported high expression of SPRY4-IT1 in renal cancer cell lines and reduced renal cancer cell proliferation, migration, and invasion by knockdown of SPRY4-IT1 [13]. Additionally, SPRY4-IT1 was found significantly expressed in breast cancer cells and its suppression could inhibit proliferation and induce apoptosis of breast cancer cells [14]. However, the expression and the role of SPRY4-IT1 in glioma are still unclear.
In this study, we investigated the effects of SPRY4-IT1 expression on glioma cells. The results of our study indicated that knockdown of SPRY4-IT1 inhibited proliferation, migration and EMT of glioma cells.
Materials and methods
Patients and clinical sample collection
The primary glioma tissues and the adjacent normal brain tissues were obtained from 18 glioma patients at the Department of Neurosurgery or Oncology, the Second Hospital of Hebei Medical University, during 2006 to 2010. These patients included 10 women and 8 men, with median age of 61. All the tissues were collected and frozen in guanidinium thiocyanate solution at -80°C for future experiments. The informed consent was provided by all the patients and all the experiments were approved by the Institute Research Ethics Committee according to the Helsinki Declaration.
Cell culture
The human glioma cell lines (U251 and SF295) and the normal human astrocytes (NHA) were purchased from the American Type Culture Collection (ATCC, USA). All the cells were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% FBS (Gibco, USA), 50 U/mL penicillin and 50 μg/mL streptomycin (Sigma, St. Louis, MO, USA) in a humidified incubator with 5% CO2 at 37°C.
Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from the tissues or cells using Trizol reagent (Life Technologies). The synthesis of cDNA was performed with a reverse transcription kit (Takara). The primers for qRT-PCR were as follows: SPRY4-IT1, 5’-AGCCACATAAATTCAGCAGA-3’ (forward) and 5’-CGATGTAGTAGGATTCCTTTCA-3’ (reverse); E-cadherin, 5’-GGTTATTCCTCCCATCAGCT-3’ (forward) and 5’-CTTGGCTGAGGATGGTGTA-3’ (reverse); Fibronectin, 5’-GGACATGCATTGCCTACTCG-3’ (forward) and 5’-GAATCCTGGCATTGGTCGAC-3’ (reverse); Vimentin, 5’-GAGTCCACTGAGTACCGGAG-3’ (forward) and 5’-ACGAGCCATTTCCTCCTTCA-3’ (reverse); GAPDH, 5’-GACTCATGACCACAGTCCATGC-3’ (forward) and 5’-AGAGGCAGGGATGATGTTCTG-3’ (reverse). GAPDH was used for normalization of the qRT-PCR results. The qRT-PCR was performed for 40 cycles with the following conditions: 94°C for 10 min, 55°C for 30 s, and 72°C for 20 s. Data collection and analysis were performed with SDS2.3 Soft-ware (Applied Biosystems). The results were expressed by the comparative CT method (2-ΔΔCT) as previously described [15].
Western blot
Cells at the logarithmic phase were lysed in lysis buffer. The total proteins were extracted by 12% SDS-PAGE, transferred onto PVDF membranes (Pierce, Rockford, IL, USA) and then incubated overnight with specific antibodies (Abcam) against E-cadherin, Fibronectin and Vimentin followed by incubation with HRP-conjugated secondary antibodies (Abcam). β-actin (Santa Cruz) was used as control. Protein expression was detected by a chemiluminescence kit (Amersham Biosciences).
siRNA to knockdown SPRY4-IT1 in glioma cells and transfection
SPRY4-IT1 siRNA (si-SPRY4-IT1) and scrambled negative control siRNA (si-NC) were purchased from Life Technologies. The sequences of si-SPRY4-IT1 were 5’-CCCAGAATGTTGACAGCTGCCTCTT-3’. Human glioma U251 and SF295 cells were transfected with si-SPRY4-IT1 or si-NC, using Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instruction. After 48 h transfection with siRNA, the cells were harvested for qRT-PCR to detect the transfection efficiency.
Cell proliferation assay
The proliferative capacity of the glioma cells was examined by MTT assay. After transfection with si-SPRY4-IT1 or si-NC, the U251 and SF295 cells were seeded in the 96-well plate and cultured for 24 h, at a density of 1×103 cells/well. Then the cells were treated with 100 μg MTT solution (Sigma) after 24, 48, 72 and 96 h incubation. The absorbance was measured at 490 nm with a microplate reader (Thermo Scientific, Hudson, NH). Three individual experiments were performed.
Cell migration assay
Migration assays were performed in a 24-well transwell chamber (BD Biosciences, San Jose, CA, USA) according to the manufacturer’s instruction. After transfection, 1×103 cells were collected and suspended in serum-free medium. Then, the upper chamber is filled with cell suspensions and the bottom chamber with culture medium containing 10% FBS. After 48 h incubation at 37°C, the cells migrating to the lower surface of the insert were fixed and stained with 0.1% crystal violet and counted under a microscope (200×, Olympus, Tokyo, Japan).
Statistical analysis
SPSS software (SPSS, Inc., Chicago, IL, USA) was applied for statistical analysis. The results were expressed as mean ± standard deviation (SD). All experiments were conducted for at least three times and P < 0.05 was considered statistically significant.
Results
Expression of SPRY4-IT1 in glioma tissues and cell lines
The expression levels of SPRY4-IT1 in 18 patients with glioma were measured by qRT-PCR. In comparison with the adjacent normal brain tissues, the glioma tissues had a significantly increased SPRY4-IT1 mRNA expression (Figure 1A). The expression levels of SPRY4-IT1 in glioma cell lines (U251 and SF295) and NHA were also detected. The results indicated that the mRNA expression of SPRY4-IT1 was higher in glioma cell lines than in NHA (Figure 1B).
Figure 1.
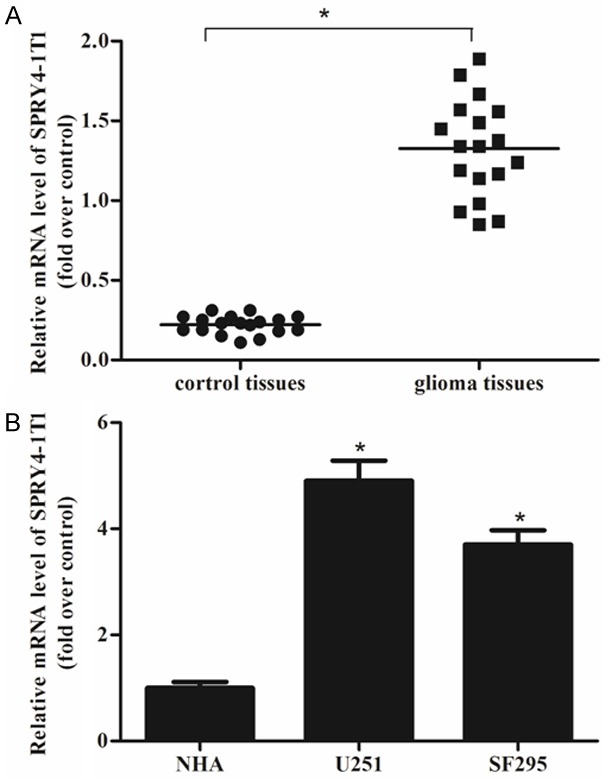
Expression of SPRY4-IT1 in glioma tissues and cell lines. The expression levels of SPRY4-IT1 were measured by qRT-PCR and demonstrated by the comparative CT method (2-ΔΔCT). A. Higher expression levels of SPRY4-IT1 were observed in glioma tissues than in the adjacent normal brain tissues. B. Higher expression levels of SPRY4-IT1 were observed in two glioma cell lines (U251 and SF295) than in NHA. The data was expressed as mean ± SD of three independent experiments. *P < 0.05.
Effects of SPRY4-IT1 expression on glioma cell proliferation
To investigate the potential role of SPRY4-IT1 in proliferation of glioma cells, we constructed SPRY4-IT1 knockdown glioma cell lines. As shown in Figure 2A, knockdown of SPRY4-IT1 significantly decreased SPRY4-IT1 mRNA expression in U251 cells, as compared with the si-NC group. Similar results were found in SF2925 cells (Figure 2B). Then, we performed MTT assays to detect the effects of SPRY4-IT1 on glioma cell proliferation. As shown in Figure 2C, si-SPRY4-IT1 exhibited a lower proliferative rate in U251 cells, in comparison with the si-NC group. And, si-SPRY4-IT1 also remarkably inhibited the proliferation of SF2925 cells (Figure 2D). The results indicated that downregulation of SPRY4-IT1 could reduce proliferation of glioma cells.
Figure 2.
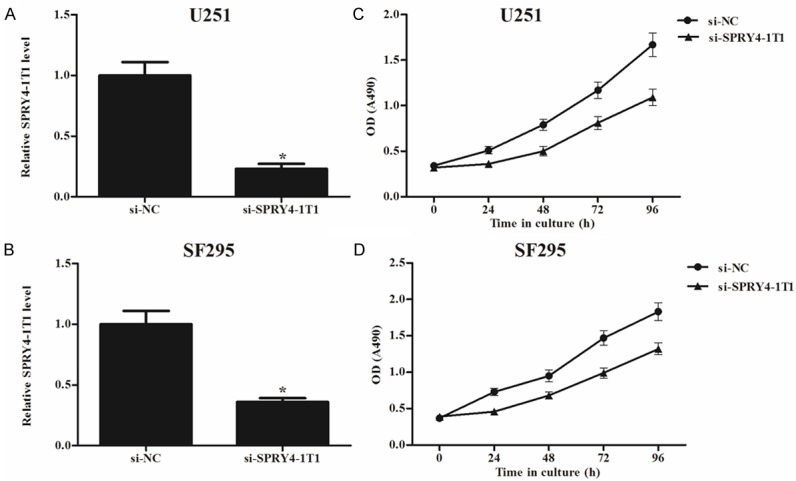
Effects of SPRY4-IT1 expression on glioma cell proliferation. After transfection with si-SPRY4-IT1, qRT-PCR analysis was performed to detect the expression levels of SPRY4-IT1 in glioma cells, and MTT assay was performed to determine glioma cell proliferation. For U251 cells, a remarkable decrease in SPRY4-IT1 expression (A) and cell proliferation (C) was found, compared to the si-NC group. For SF295 cells, a similar decrease in SPRY4-IT1 expression (B) and cell proliferation (D) was observed. The results were expressed as mean ± SD of three individual experiments. *P < 0.05.
Effects of SPRY4-IT1 expression on glioma cell migration
Then we performed migration assays to detect the effect of SPRY4-IT1 on cell migration in glioma cells. The migration assays showed that the migratory rate of U251 and SF295 cells transfected with si-SPRY4-IT1 was significantly reduced in comparison with si-NC groups (Figure 3). The results suggested that knockdown of SPRY4-IT1 could decrease migration of glioma cells.
Figure 3.
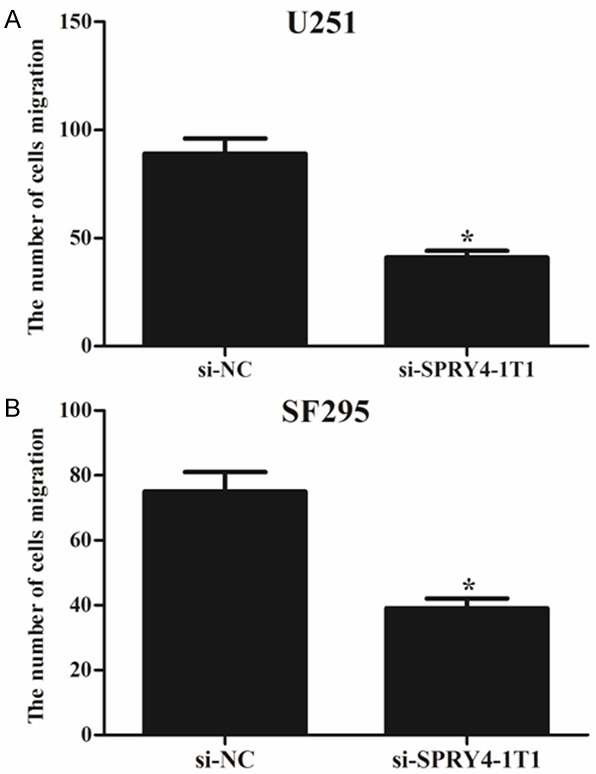
Effects of SPRY4-IT1 expression on glioma cell migration. Transwell assays were performed to investigate the migratory capacity of glioma cells. A. U251 cells transfected with si-SPRY4-IT1 exhibited a lower migratory capacity in comparison with those transfected with si-NC. B. SF295 cells transfected with si-SPRY4-IT1 exhibited a lower migratory capacity in comparison with those transfected with si-NC. The number of migrating cells was counted under a microscope and the results were shown as means ± SD of three replicate experiments. *P < 0.05.
Effects of SPRY4-IT1 expression on glioma cell EMT
To measure the effect of SPRY4-IT1 on EMT of glioma cells, both qRT-PCR and western blot were performed to examine the expression of EMT-related markers in U251 and SF295 cells after transfection with si-SPRY4-IT1. As shown in Figure 4, downregulation of SPRY4-IT1 in U251 and SF295 cells remarkably increased the expression of E-cadherin (marker of epithelial cells) and meanwhile greatly decreased the expression of fibronectin and vimentin (markers of mesenchymal cells), in comparison with control groups. The results displayed that downregulation of SPRY4-IT1 obviously blocked the EMT process.
Figure 4.
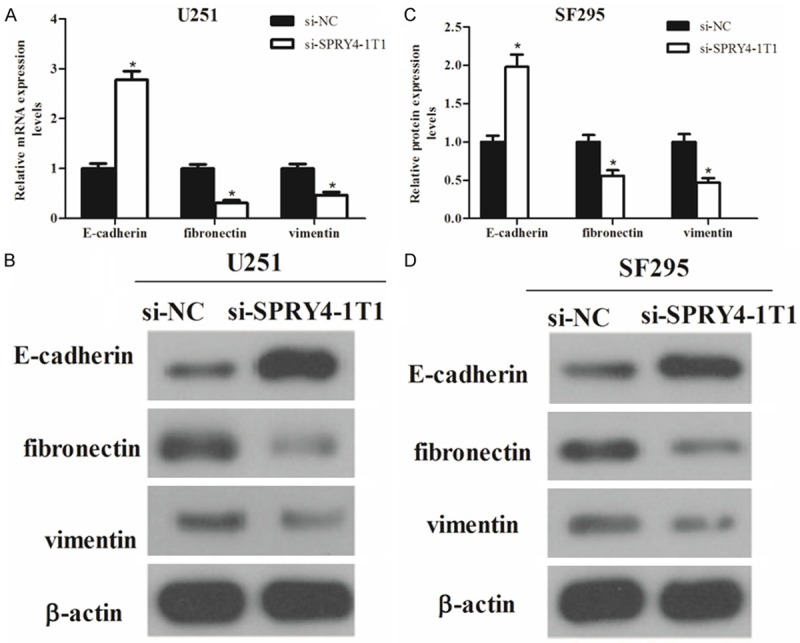
Effects of SPRY4-IT1 expression on glioma cell EMT. The expression of EMT-related molecular markers in U251 cells was analyzed by qRT-PCR (A) and western blot (B) respectively. The expression of EMT-related molecular markers in SF295 cells was analyzed by qRT-PCR (C) and western blot (D) respectively. The data was expressed as mean ± SD of three individual experiments. *P < 0.05.
Discussion
From genomic microarray analysis and cDNA cloning studies, we know that over 90% of the human genome transcriptional products are non-protein coding RNAs (ncRNAs) [16-18]. Among ncRNAs, microRNAs (miRNA) are proved to play a significant role in the post-transcriptional regulation of the target genes. Among these biological RNAs, the newly discovered lncRNAs are important ones. There is accumulating evidence showing the contribution of lncRNAs to tumorigenesis. Besides, a considerable progress has been made in identification of the structural and regulatory roles of lncRNAs in some biological processes such as cell proliferation and migration as well as epithelial-mesenchymal transition in many types of cancer [19-22].
SPRY4-IT1, belonging to a group of intron-retained lncRNAs, was processed by cleavage of a 244 nt 5’ fragment prior to transport to the cytoplasm [23]. Its dysregulation was reported to affect tumor development of many types of cancer. In this study, we examined for the first time the clinical significance of SPRY4-IT1 in glioma cells. We observed a remarkably higher expression of SPRY4-IT1 in glioma tissues and cell lines than in the normal brain tissues and NHA. These results showed that SPRY4-IT1 expression may be applied as a prognostic factor for patients with glioma.
Khaitan et al. reported that downregulation of SPRY4-IT1 reduced cell growth, invasion and migration and induced apoptosis in melanoma cells [24]. Besides, it was reported that SPRY4-IT1 affected the cell proliferation, invasion and migration of human esophageal squamous cell carcinoma and its knockdown could lead to a decrease in cell proliferation, invasion and migration [25]. Based on the experiment results of this study, we found the important role of SPRY4-IT1 in cell growth, migration and EMT of glioma cells and thus identify the biological function of SPRY4-IT1 in glioma.
To further understand the mechanism of SPRY4-IT1 in glioma cell process, we performed related experiments and found that knockdown of SPRY4-IT1 could greatly decrease proliferation, migration and EMT of glioma cells compared with the control group, indicating that downregulation of SPRY4-IT1 could inhibit the development of glioma.
Despite biological effects of SPRY4-IT1 on glioma cells, a cell function may be regulated by multiple lncRNAs. Therefore, other target lncRNAs may also be involved in the modulation of glioma cells and more studies will be needed to prove this point. Moreover, the time-consuming study is still being performed on molecular mechanisms of lncRNAs [26]. To date, there are limited hypothetical mechanisms proposed, via which lncRNAs exerted their effects: guiding chromatin-modifying complexes [27,28]; binding to miRNAs to block their binding to the target mRNAs [29]; binding to the promoters to modulate transcriptional factors [30,31]. However, which mechanism was applied by SPRY4- IT1 to regulate the glioma cells remains to be further examined.
In conclusion, we demonstrated that SPRY4-IT1 could promote proliferation, metastasis and EMT of glioma cells and its knockdown could inhibit glioma cell growth, migration and EMT. Therefore, SPRY4-IT1 may be used as a new target for diagnosis and treatment of glioma.
Disclosure of conflict of interest
None.
References
- 1.Wei J, Gabrusiewicz K, Heimberger A. The controversial role of microglia in malignant gliomas. Clin Dev Immunol. 2013;2013:285246. doi: 10.1155/2013/285246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gurney JG, Kadan-Lottick N. Brain and other central nervous system tumors: rates, trends, and epidemiology. Curr Opin Oncol. 2001;13:160–166. doi: 10.1097/00001622-200105000-00005. [DOI] [PubMed] [Google Scholar]
- 3.Jovčevska I, Kočevar N, Komel R. Glioma and glioblastoma-how much do we (not) know? Mol Clin Oncol. 2013;1:935–941. doi: 10.3892/mco.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting New Advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60:166–193. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng SY, Lin L, Soh BS, Stanton LW. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 2013;29:461–468. doi: 10.1016/j.tig.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Dinger ME, Pang KC, Mercer TR, Crowe ML, Grimmond SM, Mattick JS. NRED: a database of long noncoding RNA expression. Nucleic Acids Res. 2009;37:D122–D126. doi: 10.1093/nar/gkn617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinger ME, Pang KC, Mercer TR, Mattick JS. Differentiating Protein-Coding and Noncoding RNA: Challenges and Ambiguities. PLoS Comput Biol. 2008;4:e1000176. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder R, Trauner M, Zatloukal K. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 9.Wu Y, Zhang L, Wang Y, Li H, Ren X, Wei F, Yu W, Wang X, Zhang L, Yu J, Hao X. Long noncoding RNA HOTAIR involvement in cancer. Tumour Biol. 2014;35:9531–9538. doi: 10.1007/s13277-014-2523-7. [DOI] [PubMed] [Google Scholar]
- 10.Kondo M, Suzuki H, Ueda R, Osada H, Takagi K, Takahashi T. Frequent loss of imprinting of the H19 gene is often associated with its overexpression in human lung cancers. Oncogene. 1995;10:1193–1198. [PubMed] [Google Scholar]
- 11.Hu Y, Chen HY, Yu CY, Xu J, Wang JL, Qian J, Zhang X, Fang JY. A long non-coding RNA signature to improve prognosis prediction of colorectal cancer. Oncotarget. 2014;5:2230–2242. doi: 10.18632/oncotarget.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazar J, Zhao W, Khalil AM, Lee B, Shelley J, Govindarajan SS, Yamamoto F, Ratnam M, Aftab MN, Collins S, Finck BN, Han X, Mattick JS, Dinger ME, Perera RJ. The functional characterization of long noncoding RNA SPRY4-IT1 in human melanoma cells. Oncotarget. 2014;5:8959–8969. doi: 10.18632/oncotarget.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang HM, Yang FQ, Yan Y, Che JP, Zheng JH. High expression of long non-coding RNA SPRY4-IT1 predicts poor prognosis of clear cell renal cell carcinoma. Int J Clin Exp Pathol. 2014;7:5801–5809. [PMC free article] [PubMed] [Google Scholar]
- 14.Shi Y, Li J, Liu Y, Ding J, Fan Y, Tian Y, Wang L, Lian Y, Wang K, Shu Y. The long noncoding RNA SPRY4-IT1 increases the proliferation of human breast cancer cells by upregulating ZNF703 expression. Mol Cancer. 2015;14:51–63. doi: 10.1186/s12943-015-0318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G. Transcriptional Maps of 10 Human Chromosomes at 5-Nucleotide Resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 17.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C. The Transcriptional Landscape of the Mammalian Genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 18.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of Long Non-coding RNA HOTAIR Predicts Tumor Recurrence in Hepatocellular Carcinoma Patients Following Liver Transplantation. Ann Surg Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 19.Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- 20.Shen L, Chen L, Wang Y, Jiang X, Xia H, Zhuang Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J Neurooncol. 2014;121:101–108. doi: 10.1007/s11060-014-1613-0. [DOI] [PubMed] [Google Scholar]
- 21.Guo Q, Zhao Y, Chen J, Hu J, Wang S, Zhang D, Sun Y. BRAF-activated long non-coding RNA contributes to colorectal cancer migration by inducing epithelial-mesenchymal transition. Oncol Lett. 2014;8:869–875. doi: 10.3892/ol.2014.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibb EA, Brown CJ, Wan LL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38–54. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazar J, Zhao W, Khalil AM, Lee B, Shelley J, Govindarajan SS, Yamamoto F, Ratnam M, Aftab MN, Collins S. The functional characterization of long noncoding RNA SPRY4-IT1 in human melanoma cells. Oncotarget. 2014;5:8959–8969. doi: 10.18632/oncotarget.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, Perera RJ. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852–3862. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- 25.Xie HW, Wu QQ, Zhu B, Chen FJ, Ji L, Li SQ, Wang CM, Tong YS, Tuo L, Wu M. Long noncoding RNA SPRY4-IT1 is upregulated in esophageal squamous cell carcinoma and associated with poor prognosis. Tumour Biol. 2014;35:7743–7754. doi: 10.1007/s13277-014-2013-y. [DOI] [PubMed] [Google Scholar]
- 26.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang H. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khalil AM, Rinn JL. RNA-protein interactions in human health and disease. Semin Cell Dev Biol. 2011;22:359–365. doi: 10.1016/j.semcdb.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. RNA regulation of epigenetic processes. Bioessays. 2009;31:51–59. doi: 10.1002/bies.080099. [DOI] [PubMed] [Google Scholar]
- 29.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea MD, Thomas K, Presser A, Bernstein BE, Van OA. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]


