Abstract
Paxillin (PXN) gene has been reported to act as an oncogene in many malignancies and play important roles in the development of human carcinomas. However, the relationship between the expression of PXN and clinicopathological characteristics in human laryngeal carcinoma remains unclear. This study aimed to examine the expression of PXN, and to evaluate the clinical significance of its expression in human laryngeal squamous cell carcinoma (LSCC). Real-time quantitative PCR (qRT-PCR), Western blotting and immunohistochemistry were performed to analyze the expression of PXN in LSCC tissues and corresponding paracancerous normal tissues. Kaplan-Meier survival and Cox regression analyses were performed to evaluate the prognosis of patients with LSCC. The expression of PXN was significantly higher in LSCC than in matched paracancerous normal tissues. Immunohistochemical analysis was performed in human LSCC samples and the data were correlated with clinicopathologic features. Levels of PXN in LSCC were related to histopathological grade (P = 0.016), Lymph node metastasis (P = 0.029) and TNM stage (P < 0.001). Kaplan-Meier analysis revealed that survival curves of the overall survival of patients with high PXN expression was significantly worse than that of low PXN expression (P = 0.035). Cox regression analysis revealed that PXN expression level was an independent prognostic factor of the overall survival rate of patients with LSCC (P = 0.002). These findings suggest that PXN expression has potential use as a novel biomarker of LSCC patients and may serve as an independent predictive factor for prognosis of LSCC patients.
Keywords: Laryngeal squamous cell carcinoma, paxillin, prognosis
Introduction
Laryngeal cancer, one of the most common malignancies in the head and neck region, is the 11th most common cancer worldwide and has a high mortality rate and a poor prognosis [1]. Laryngeal squamous cell carcinomas (LSCC) represent approximately 85 to 90% of all the malignant tumors of the larynx [2]. Over the recent years, the incidence rate of LSCC has gradually increased; the invasion and metastasis of LSCC are the main factors that severely affect patients’ life quality and overall survival [3]. Although great progress has been achieved in the studies on LSCC, there are no ideal markers for the decision of prognosis and the guidance of treatment in patients of LSCC. Therefore, it is urgent to develop novel and valuable markers to distinguish patients with poor prognosis or at high risk of early recurrence and guide chemotherapy and radiotherapy.
Paxillin (PXN) is a focal adhesion-associated, phosphotyrosine-containing 68-kDa adaptor protein discovered in 1990 [4]. As with focal adhesion signaling, paxillin phosphorylation appears to play a critical role, which might control cell movement through the regulation of adhesion dynamics [5]. And not only that, the focal adhesion points also play an important role in the regulation of cell motility because they involve cycles of formation of cell adhesion and cell spreading by disassembling the components of cell adhesion [6]. In addition to a role for focal adhesion, paxillin is essential for adhesion-mediated activation of ERK [7], and ERK phosphorylation of paxillin then increases the association of focal adhesion kinase (FAK) with paxillin to potentiate motility and cell spreading. Increased cell spreading contributes to an increased cell migration and invasiveness [8]. Interestingly, paxillin have all been linked to malignant progression through both tissue microarray and histologic analyses. As yet paxillin mutations and changes in protein expression are also associated with alterations in a variety of invasive or metastatic cancers [9] ,including breast [10,11], lung [12-14], prostate [15], melanoma [16], ovarian cancer [17] and colorectal cancer [18]. However, the expression and function of PXN and its correlations with clinical parameters and influence on prognosis of LSCC have still remained poorly understood.
In this study, we examined the expression levels of PXN in LSCC tissues and matched paracancerous normal tissues by qRT-PCR, western blotting and immunohistochemistry. In addition, to explore the exact role of PXN in LSCC, it is important to further study the role of PXN in the pathogenesis of LSCC and understand the clinical significances of PXN in LSCC.
Material and methods
Patients and clinical tissue specimens
A total of 84 paraffin-embedded LSCC tissue samples and 18 corresponding adjacent non-tumorous tissue samples were obtained from 84 patients and collected from the archives of the People’s Hospital of Liaocheng. These patients who had not been performed radiotherapy, chemotherapy or immunotherapy before the surgery were all retrospectively analyzed from January 2006 to December 2008. In addition, fresh cancer specimens and corresponding paracancerous normal tissues between 2012 and 2014 were also kept in liquid nitrogen and sectioned for protein and RNA extraction. According to the International Union against Cancer Tumor-Node-Metastasis staging system, all tumors were also clinically classified as supraglottic, glottic, and subglottic, and were histologically graded as well, moderately, and poorly differentiated. The relevant clinical reports of patients during 5-year follow-up were obtained with prior patient consent and the approval by the Ethics Committee of the People’s Hospital of Liaocheng.
Real-time quantitative PCR
Total RNA was extracted from tissues lysate using a Trizol kit (Invitrogen, Carlsbad, CA). Omniscript RT kit 50 (QIAGEN) was used for the complementary DNA (cDNA) synthesis following the manufacturer’s instruction. The amplification of cDNA by PCR was carried out in a total volume of 25 μL containing 1 μL of cDNA, 12.5 μL of 2 × Fast Eva GreenTM qPCR Master Mix (Biotium Inc, Hayward, CA), 1 μL primers (10 mM), and 10.5 μL of RNase/DNase-free water. The reaction was heated in a thermocycle for 2 min at 96°C and then submitted to 40 cycles of amplification. The conditions of amplification were 15 s at 96°C, and 1 min at 60°C. The Ct value comparison method was used in this study to evaluate the mRNA expression level. The Ct values were defined as referring to the number of cycles when the fluorescence signal reached the set threshold in each reaction tube. The β-actin was used as the internal reference gene. The relative expression levels of PXN were calculated by the Ct values using a 2ΔΔCt quantification method. The primers as follows: PXN forward: 5’-ACGTCTACAGCTTCCCCAACAA-3’; PXN reverse: 5’-AGCAGGCGGTCGAGTTCA-3’.
Western blot
Matched tumor tissues and the adjacent non-tumor tissues were lysed in RIPA lysis buffer (Sigma Vetec, USA). After centrifugation at 12,000 rpm for 20 min, the supernatant was collected for determination of total protein concentration by DC-protein assay method to maintain the same loads. Protein samples were electrophoretically separated on a 12% SDS-PAGE, and transferred to a PVDF membrane. The membrane was blocked with 5% non-fat dry milk in TBST buffer and incubated with a rabbit anti-human Paxillin polyclonal antibody (1:500, Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 4°C overnight. The membranes were washed three times with TBST buffer for 5 minutes, and further incubated with HRP-conjugated secondary antibody (1:5000) at room temperature for 2 h. The proteins were detected by ECL and exposed to X-ray film. The densities of protein bands were analyzed using NIH image software.
Immunohistochemistry
For immunohistochemical staining, paraffin sections were cut serially at 4 μm, dried for 50 min at 60°C and incubated overnight at 37°C. Sections were dewaxed with xylene and rehydrated through a graded alcohol series, and then dipped into 0.3% H2O2 for 10 min in methanol to inactivate endogenous peroxidase. Heat-induced epitope reparation was performed in citrate buffer (pH 6.0) for 5 min in an autoclave at 96°C. After cooling to room temperature, the sections were rinsed with phosphate-buffered saline (PBS) three times for 10 min each time and then incubated with rabbit anti-human Paxillin polyclonal antibody (1:100, Santa Cruz Biotechnology, Inc, USA) for 24 h at 4°C. The sections were rinsed with phosphate-buffered saline (PBS) three times for 10 min, and then incubated overnight at 4°C with HRP-conjugated secondary antibody. After washing in PBS, the visualization signal was developed with 3, 3’-diaminobenzidine (DAB) solution, and all sections were counterstained with hematoxylin.
The PXN immunopositivity was semiquantitatively classified into five score groups: score 0, 0-5%; score 1, > 5-25%; score 2, > 26-50%; score 3, > 51-75%; and score 4, > 75-100% positive tumor cells. Intensity was graded as 0 (no staining), 1 (weak staining), 2 (moderate staining), or 3 (strong staining). The final score was calculated as a sum total score of central parts and invasive marginal front. Based on our standard, the final score was: 0, negative (–); 1-4, weakly positive (+); 5-8, moderately positive (++) or 9-12, strongly positive (+++).
Statistical analysis
All statistical analyses were carried out by using a statistical software package (SPSS17.0, Chicago, IL). The chi-square test was used to analyze the relationship between PXN expression and clinicopathological characteristics. Overall survival curves were evaluated using the Kaplan-Meier method and compared by the log-rank test. The significance of various variables for survival was analyzed by multivariate survival analysis using Cox proportional hazards regression model. P-value less than 5 percent were considered to be statistically significant difference.
Results
Up-regulated expression of PXN gene in LSCC tissues
Real-time RT-PCR was performed to reveal the potential changes in the mRNA levels of PXN expression in LSCC as well as its corresponding paracancerous normal tissues, which showed significant differences in gene expression between the cancerous and paired adjacent normal tissues (Figure 1D, P = 0.0155). The expression of PXN in the cancerous tissue significantly increased in 13 of 18 compared with those in corresponding paracancerous normal tissues (Figure 1B). The above results were further confirmed by means of Western blot. We examined protein expression of PXN in 24 LSCC tissues and corresponding paracancerous normal tissues. As shown in Figure 1A, the PXN protein expression was significantly upregulated in LSCC samples compared with that in paracancerous normal tissues in 18 of 24 LSCC patients. Also the average PXN protein level in 24 LSCC tissues was significantly higher than that in paracancerous normal tissues (Figure 1C, P = 0.00018).
Figure 1.
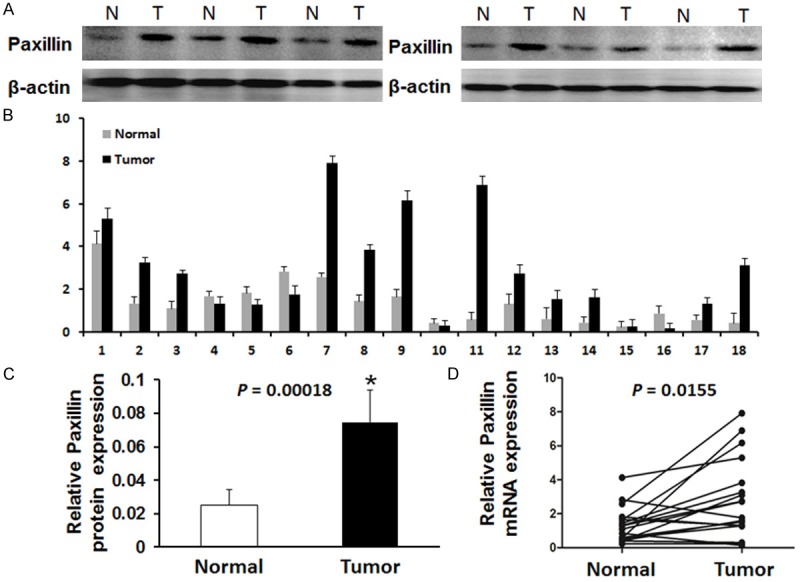
Expression PXN in LSCC tissues and adjacent non-tumor tissues. A. Western blot analysis of PXN proteins expressed in six paired representative LSCC tissues and their matched adjacent non-tumor tissues. β-actin was used as a control for equal loading (T: tumor tissues, N: non-tumor tissues). B. The relative mRNA level of PXN expression in LSCC tissues compared to paired adjacent non-tumor tissues (n = 18) assessed by real time quantitative RT-PCR after normalizing to β-actin. C. Relative PXN protein expression levels was significantly increased in 18 of 24 LSCC tissues compared with the corresponding adjacent non-tumor tissues (P = 0.00018). D. The mean relative expression of mRNA level of PXN in LSCC tissues compared to paired adjacent normal tissues (P = 0.0155).
Immunohistochemical analysis of PXN expression in LSCC tissue samples
We investigated the expression of PXN in LSCC by using immunohistochemical analysis. Upregulated PXN expression was detected in 48 of 84 (57.14%) LSCC tissues, however, only 6 cases of 18 corresponding adjacent non-tumorous tissue samples (33.3%) showed PXN expression. We found that positive staining was mainly localized in the cytoplasm of cancer tissues while strong staining was hardly ever observed in the adjacent non-cancerous tissue areas (Figure 2).
Figure 2.
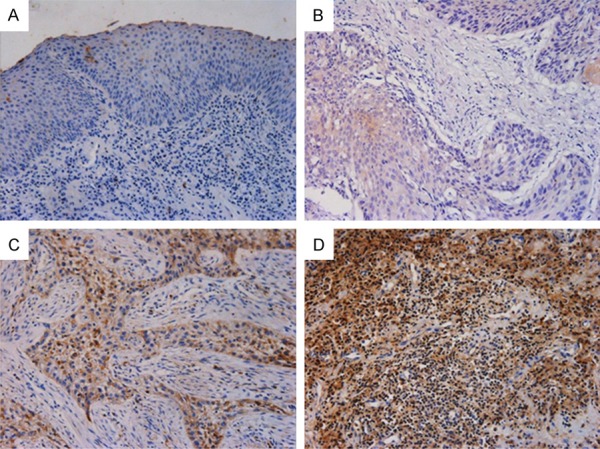
Representative images of the PXN protein expression in LSCC tissues and their corresponding adjacent non-tumor tissues were detected via immunohistochemical staining. A. Negative staining of PXN in adjacent non-tumor tissues, scored as PXN (-); B. Weak staining of PXN in well-differentiated LSCC tissues, scored as PXN (+); C. Strong staining of PXN in moderate-differentiated LSCC tissues, scored as PXN (++); D. Strong staining of PXN in poor-differentiated LSCC tissues, scored as PXN (+++). Original magnification: 200×.
Correlations between the expression of PXN and various clinicopathological characteristics
We analyzed the relationships between PXN expression levels in LSCC tissues and the clinical data from 84 patients by the Chi square analysis (Table 1). There were no differences between gender, age, alcohol consumption, smoking consumption, tumor Location and tumor size regarding PXN expression, but PXN expression in LSCC was positively correlated with histological differentiation, lymph node metastasis, and TNM stage. Patients with higher PXN expression had poor differentiation, lymph node metastases, or more advanced TNM stage, strongly supporting that PXN can be considered as a new marker of poor prognosis in human LSCC.
Table 1.
Relationship between PXN expression level and clinicopathologic parameters of LSCC
| Characteristics | All cases | PXN expression | P-value | |
|---|---|---|---|---|
|
|
||||
| Low (n = 36) | High (n = 48) | |||
| Gender | 0.350 | |||
| Male | 56 | 26 | 30 | |
| Female | 28 | 10 | 18 | |
| Age (years) | 0.312 | |||
| < 55 | 39 | 19 | 20 | |
| ≥ 55 | 45 | 17 | 28 | |
| Alcohol consumption | 0.067 | |||
| Yes | 44 | 23 | 21 | |
| No | 40 | 13 | 27 | |
| Smoking consumption | 0.847 | |||
| Yes | 50 | 21 | 29 | |
| No | 34 | 15 | 19 | |
| Tumor Location | 0.159 | |||
| Superaglottic | 34 | 11 | 23 | |
| Glottic | 46 | 24 | 22 | |
| Subglottic | 4 | 1 | 3 | |
| Tumor size (cm) | 0.180 | |||
| < 2.5 | 35 | 18 | 17 | |
| ≥ 2.5 | 49 | 18 | 31 | |
| Histological differentiation | 0.016* | |||
| Well/moderate | 53 | 28 | 25 | |
| Poor | 31 | 8 | 23 | |
| Lymph node metastasis | 0.029* | |||
| Negative | 71 | 34 | 37 | |
| Positive | 13 | 2 | 11 | |
| Invasive depth | 0.529 | |||
| T1/T2 | 43 | 17 | 26 | |
| T3/T4 | 41 | 19 | 22 | |
| TNM stage | < 0.001 | |||
| I-II | 37 | 25 | 12 | |
| III-IV | 47 | 11 | 36 | |
P-value < 0.05 was considered statistically significant.
Association of PXN expression with clinical outcome
The prognostic effect of PXN on the overall survival rate of LSCC patients was investigated by comparing the 5-year survival rate of patients with high or low levels of PXN expression in tumors using Kaplan-Meier survival curves and the log-rank test. According to the immunohistochemical results of PXN staining in tumor cells, LSCC patients were divided into two groups including PXN low level expression (−~ +) group and PXN high level expression (++~ +++) group. The overall survival of patients with high PXN expression was significantly worse than that of low PXN expression (P = 0.035, Figure 3). Furthermore, a multivariate analysis confirmed the PXN expression, Lymph node metastasis and TNM stage as independent predictors of the overall survival of LSCC patients by a Cox proportional-hazard model (Table 2).
Figure 3.
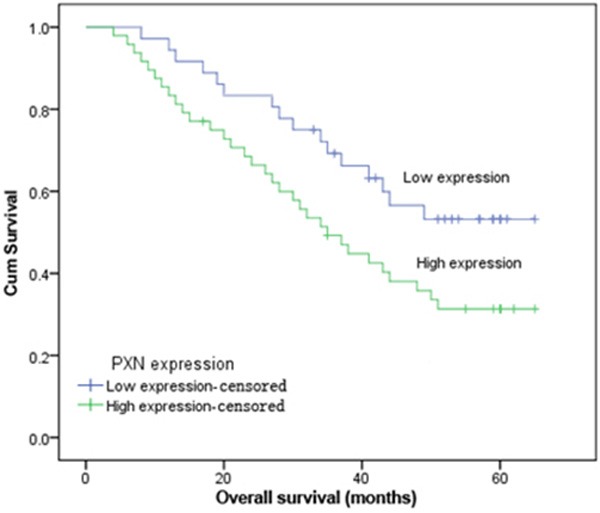
Kaplan-Meier curves for overall survival in LSCC patients grouped by immunohistochemical levels of PXN. Patients with higher PXN expression exhibit a significantly poorer prognosis (χ2 = 4.433, P = 0.035) than those with lower PXN expression.
Table 2.
Univariate and multivariate analysis of prognostic factors in LSCC for overall survival
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| PXN expression | 2.748 | 1.224-4.061 | 0.006* | 2.572 | 1.056-3.861 | 0.002* |
| Gender (Male vs. Female) | 1.969 | 1.024-2.316 | 0.582 | |||
| Age (< 55 vs. ≥ 55) | 1.546 | 0.986-2.176 | 0.065 | |||
| Alcohol consumption (Yes vs. No) | 1.228 | 0.593-2.038 | 0.486 | |||
| Smoking consumption (Yes vs. No) | 0.964 | 0.495-1.698 | 0.078 | |||
| Tumor Location (Superaglottic vs. Glottic vs. Subglottic) | 1.508 | 1.045-3.254 | 0.118 | |||
| Tumor size (< 2.5 vs. ≥ 2.5) | 0.896 | 0.526-1.640 | 0.086 | |||
| Histological differentiation (Well/moderate vs. Poor) | 1.956 | 1.023-3.243 | 0.046* | 1.450 | 0.516-2.356 | 0.479 |
| Lymph node metastasis (Negative vs. Positive) | 1.893 | 1.289-3.905 | 0.018* | 1.345 | 0.912-4.129 | 0.013* |
| Invasive depth (T1/T2 vs. T3/T4) | 1.378 | 0.855-1.897 | 0.022* | 1.495 | 0.925-2.120 | 0.207 |
| TNM stage (I-II vs. III-IV) | 1.192 | 0.846-1.806 | 0.023* | 1.163 | 0.875-2.062 | 0.006* |
HR, hazard ratio; CI, confidence interval;
P-value < 0.05 was considered statistically significant.
Discussion
Although several cutting edge treatment strategies have been developed for LSCC, no treatment could achieve a satisfactory therapeutic outcome and the mortality rate of LSCC is still high [19]. Better understanding of molecular mechanisms underlying proliferation, differentiation, and overall survival of LSCC is crucial to the development of optimal therapeutic strategies. Nowadays, extensive researches are focused on the identification of useful biologic and molecular markers in the diagnosis and therapy of LSCC.
We previously showed that paxillin, a focal adhesion associated protein, has been shown to play an important role in controlling cell migration [5,20], proliferation [12] and apoptosis [21]. Our previous studies have demonstrated that paxillin expression varies among different cancer types. Given its roles in cell adhesion and migration, paxillin is thought to play an important role in tumor migration, invasion, and metastasis [22]. Previously, it was identified that overexpression of wild-type paxillin promoted cell proliferation and enhanced migration, invasive capacity and metastasis of the colorectal cancer cells [23]. In human cervical cancer tissues, PXN overexpression may be associated with cervical tumor metastasis [24]. Moreover, a recent report indicated that increased expression of paxillin directly correlates with HER2/3 receptor expression in both aggressive breast cancer lines and grade III human breast tumors [25]. In another study, we found that paxillin expression was closely correlated with the prognosis of non-small cell lung carcinoma [26]. Similarly, paxillin upregulation has been reported in metastatic renal carcinoma [27]. Furthermore, we also demonstrate that PXN is a critical regulator of both androgen- and growth factor-mediated proliferation in prostate cancer cell lines by modulating ERK signaling and subsequent downstream effects [28]. In this study, this result is consistent with previous studies, the results of real-time RT-PCR and western blot analysis indicated that PXN mRNA and protein expression was higher in LSCC tissues than in paracancerous normal tissues. Subsequently, we also evaluated strong cytoplasmic expression of PXN was detected in 48 of 84 LSCC specimens by immunohistochemistry analysis. This analysis revealed higher PXN expression in the cytoplasm of LSCC compared with matched tumor adjacent normal tissues.
In the current research, we also demonstrate that high cytoplasmic expression of PXN in LSCC is correlated with histopathological grade. Moreover, using immunohistochemistry analysis, we evaluated whether the level of PXN expression correlates with clinicopathological parameters and the prognosis of LSCC patients, which indicated PXN expression correlated with lymph node metastasis and TNM stage. Multivariate analysis further demonstrated that PXN expression, lymph node metastasis and TNM stage were also independent factors of poor prognosis in patients with LSCC. These data are in keeping with recent studies showing that high PXN expression is independently associated with poor survival in patients with colorectal carcinoma [29]. Furthermore, the relationship between PXN expression and clinical survival was also investigated using a log-rank test which compares the overall survival time of patients as a function of PXN expression. Survival curve analysis showed patients of PXN high expression group was significantly shorter than those of PXN low expression group. These results strongly support the hypothesis that PXN can be considered as a new marker of poor prognosis in human LSCC. Additionally, paxillin is able to promote cell survival signaling through its direct interaction with the pro-survival, proto-oncogene bcl-2 [30], which is known to function to prevent apoptosis in the absence of cell adhesion in both normal and cancerous cells, in part through maintaining adhesion related signaling through FAK [31,32]. Thus, the involvement of paxillin in regulating cell survival and apoptosis suggests that it plays an essential role during normal developmental processes and could also provide a novel cellular target for cancer therapeutics [33].
In conclusion, this study showed the first evidences of the expression and clinical significances of PXN in LSCC, suggesting that PXN gene might involve in the development and progression of LSCC. Moreover, we have showed that PXN served as both a potential negative prognostic biomarker for clinical prognosis and a promising molecular target for more selective therapeutic treatment in LSCC. However, further larger studies are still necessary for validation of our findings and an investigation of the molecular mechanisms underlying the observed correlations.
Conclusions
These findings suggests that PXN expression have potential use as novel biomarkers of LSCC patients and may serve as an independent predictive factor for prognosis of LSCC patients.
Disclosure of conflict of interest
None.
References
- 1.Marioni G, Marchese-Ragona R, Cartei G, Marchese F, Staffieri A. Current opinion in diagnosis and treatment of laryngeal carcinoma. Cancer Treat Rev. 2006;32:504–515. doi: 10.1016/j.ctrv.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Genden EM, Ferlito A, Silver CE, Jacobson AS, Werner JA, Suarez C, Leemans CR, Bradley PJ, Rinaldo A. Evolution of the management of laryngeal cancer. Oral Oncol. 2007;43:431–439. doi: 10.1016/j.oraloncology.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y, Su Z, Li G, Yu C, Ren S, Huang D, Fan S, Tian Y, Zhang X, Qiu Y. Increased expression of metadherin protein predicts worse disease-free and overall survival in laryngeal squamous cell carcinoma. Int J Cancer. 2013;133:671–679. doi: 10.1002/ijc.28071. [DOI] [PubMed] [Google Scholar]
- 4.Turner CE, Glenney JR Jr, Burridge K. Paxillin: a new vinculin-binding protein present in focal adhesions. J Cell Biol. 1990;111:1059–1068. doi: 10.1083/jcb.111.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C, Rajfur Z, Borchers C, Schaller MD, Jacobson K. JNK phosphorylates paxillin and regulates cell migration. Nature. 2003;424:219–223. doi: 10.1038/nature01745. [DOI] [PubMed] [Google Scholar]
- 6.Burridge K, Chrzanowska-Wodnicka M, Zhong C. Focal adhesion assembly. Trends Cell Biol. 1997;7:342–347. doi: 10.1016/S0962-8924(97)01127-6. [DOI] [PubMed] [Google Scholar]
- 7.Hagel M, George EL, Kim A, Tamimi R, Opitz SL, Turner CE, Imamoto A, Thomas SM. The adaptor protein paxillin is essential for normal development in the mouse and is a critical transducer of fibronectin signaling. Mol Cell Biol. 2002;22:901–915. doi: 10.1128/MCB.22.3.901-915.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu ZX, Yu CF, Nickel C, Thomas S, Cantley LG. Hepatocyte growth factor induces ERK-dependent paxillin phosphorylation and regulates paxillin-focal adhesion kinase association. J Biol Chem. 2002;277:10452–10458. doi: 10.1074/jbc.M107551200. [DOI] [PubMed] [Google Scholar]
- 9.Deakin NO, Pignatelli J, Turner CE. Diverse roles for the paxillin family of proteins in cancer. Genes Cancer. 2012;3:362–370. doi: 10.1177/1947601912458582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madan R, Smolkin MB, Cocker R, Fayyad R, Oktay MH. Focal adhesion proteins as markers of malignant transformation and prognostic indicators in breast carcinoma. Hum Pathol. 2006;37:9–15. doi: 10.1016/j.humpath.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 11.Short SM, Yoder BJ, Tarr SM, Prescott NL, Laniauskas S, Coleman KA, Downs-Kelly E, Pettay JD, Choueiri TK, Crowe JP, Tubbs RR, Budd TG, Hicks DG. The expression of the cytoskeletal focal adhesion protein paxillin in breast cancer correlates with HER2 overexpression and may help predict response to chemotherapy: a retrospective immunohistochemical study. Breast J. 2007;13:130–139. doi: 10.1111/j.1524-4741.2007.00389.x. [DOI] [PubMed] [Google Scholar]
- 12.Jagadeeswaran R, Surawska H, Krishnaswamy S, Janamanchi V, Mackinnon AC, Seiwert TY, Loganathan S, Kanteti R, Reichman T, Nallasura V, Schwartz S, Faoro L, Wang YC, Girard L, Tretiakova MS, Ahmed S, Zumba O, Soulii L, Bindokas VP, Szeto LL, Gordon GJ, Bueno R, Sugarbaker D, Lingen MW, Sattler M, Krausz T, Vigneswaran W, Natarajan V, Minna J, Vokes EE, Ferguson MK, Husain AN, Salgia R. Paxillin is a target for somatic mutations in lung cancer: implications for cell growth and invasion. Cancer Res. 2008;68:132–142. doi: 10.1158/0008-5472.CAN-07-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mackinnon AC, Tretiakova M, Henderson L, Mehta RG, Yan BC, Joseph L, Krausz T, Husain AN, Reid ME, Salgia R. Paxillin expression and amplification in early lung lesions of high-risk patients, lung adenocarcinoma and metastatic disease. J Clin Pathol. 2011;64:16–24. doi: 10.1136/jcp.2010.075853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salgia R, Li JL, Ewaniuk DS, Wang YB, Sattler M, Chen WC, Richards W, Pisick E, Shapiro GI, Rollins BJ, Chen LB, Griffin JD, Sugarbaker DJ. Expression of the focal adhesion protein paxillin in lung cancer and its relation to cell motility. Oncogene. 1999;18:67–77. doi: 10.1038/sj.onc.1202273. [DOI] [PubMed] [Google Scholar]
- 15.Sen A, De Castro I, DeFranco DB, Deng FM, Melamed J, Kapur P, Raj GV, Rossi R, Hammes SR. Paxillin mediates extranuclear and intranuclear signaling in prostate cancer proliferation. J Clin Invest. 2012;122:2469. doi: 10.1172/JCI62044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Velasco-Velázquez MA, Salinas-Jazmín N, Mendoza-Patiño N, Mandoki JJ. Reduced paxillin expression contributes to the antimetastatic effect of 4-hydroxycoumarin on B16-F10 melanoma cells. Cancer Cell Int. 2008;8:8. doi: 10.1186/1475-2867-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim G, Davidson B, Henning R, Wang J, Yu M, Annunziata C, Hetland T, Kohn EC. Adhesion molecule protein signature in ovarian cancer effusions is prognostic of patient outcome. Cancer. 2012;118:1543–1553. doi: 10.1002/cncr.26449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y, Zhang X, Guda K, Lawrence E, Sun Q, Watanabe T, Iwakura Y, Asano M, Wei L, Yang Z, Zheng W, Dawson D, Willis J, Markowitz SD, Satake M, Wang Z. Identification and functional characterization of paxillin as a target of protein tyrosine phosphatase receptor T. Proc Natl Acad Sci U S A. 2010;107:2592–2597. doi: 10.1073/pnas.0914884107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyle P, Ferlay J. Cancer incidence and mortality in Europe, 2004. Ann Oncol. 2005;16:481–488. doi: 10.1093/annonc/mdi098. [DOI] [PubMed] [Google Scholar]
- 20.Chen HY, Shen CH, Tsai YT, Lin FC, Huang YP, Chen RH. Brk activates rac1 and promotes cell migration and invasion by phosphorylating paxillin. Mol Cell Biol. 2004;24:10558–10572. doi: 10.1128/MCB.24.24.10558-10572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sorenson CM. Interaction of bcl-2 with Paxillin through its BH4 domain is important during ureteric bud branching. J Biol Chem. 2004;279:11368–11374. doi: 10.1074/jbc.M310079200. [DOI] [PubMed] [Google Scholar]
- 22.Brown MC, Turner CE. Paxillin: adapting to change. Physiol Rev. 2004;84:1315–1339. doi: 10.1152/physrev.00002.2004. [DOI] [PubMed] [Google Scholar]
- 23.Jun Q, Zhiwei W, Lilin M, Jing K, Qichao N. Effects of paxillin on HCT-8 human colorectal cancer cells. Hepatogastroenterology. 2010;58:1951–1955. doi: 10.5754/hge11352. [DOI] [PubMed] [Google Scholar]
- 24.McCormack SJ, Brazinski SE, Moore JL Jr, Werness BA, Goldstein DJ. Activation of the focal adhesion kinase signal transduction pathway in cervical carcinoma cell lines and human genital epithelial cells immortalized with human papillomavirus type 18. Oncogene. 1997;15:265–274. doi: 10.1038/sj.onc.1201186. [DOI] [PubMed] [Google Scholar]
- 25.Vadlamudi R, Adam L, Tseng B, Costa L, Kumar R. Transcriptional up-regulation of paxillin expression by heregulin in human breast cancer cells. Cancer Res. 1999;59:2843–2846. [PubMed] [Google Scholar]
- 26.Zuo W, Li H. [Relationship of the expression of CD44v6 and paxillin to the prognosis of non-small cell lung carcinoma] . Sichuan Da Xue Xue Bao Yi Xue Ban. 2003;34:484–485. [PubMed] [Google Scholar]
- 27.Jenq W, Cooper DR, Ramirez G. Integrin expression on cell adhesion function and up-regulation of P125FAK and paxillin in metastatic renal carcinoma cells. Connect Tissue Res. 1996;34:161–174. doi: 10.3109/03008209609000696. [DOI] [PubMed] [Google Scholar]
- 28.Sen A, De Castro I, Defranco DB, Deng FM, Melamed J, Kapur P, Raj GV, Rossi R, Hammes SR. Paxillin mediates extranuclear and intranuclear signaling in prostate cancer proliferation. J Clin Invest. 2012;122:2469–2481. doi: 10.1172/JCI62044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin H, Zhang Q, Wang X, Li T, Wan Y, Liu Y, Zhu J. Role of paxillin in colorectal carcinoma and its relationship to clinicopathological features. Chin Med J (Engl) 2014;127:423–429. [PubMed] [Google Scholar]
- 30.Sheibani N, Tang Y, Sorenson CM. Paxillin’s LD4 motif interacts with bcl-2. J Cell Physiol. 2008;214:655–661. doi: 10.1002/jcp.21256. [DOI] [PubMed] [Google Scholar]
- 31.Frisch SM, Vuori K, Ruoslahti E, Chan-Hui PY. Control of adhesion-dependent cell survival by focal adhesion kinase. J Cell Biol. 1996;134:793–799. doi: 10.1083/jcb.134.3.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin HM, Lee YJ, Li G, Pestell RG, Kim HR. Bcl-2 induces cyclin D1 promoter activity in human breast epithelial cells independent of cell anchorage. Cell Death Differ. 2001;8:44–50. doi: 10.1038/sj.cdd.4400770. [DOI] [PubMed] [Google Scholar]
- 33.Deakin NO, Turner CE. Paxillin comes of age. J Cell Sci. 2008;121:2435–2444. doi: 10.1242/jcs.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]


