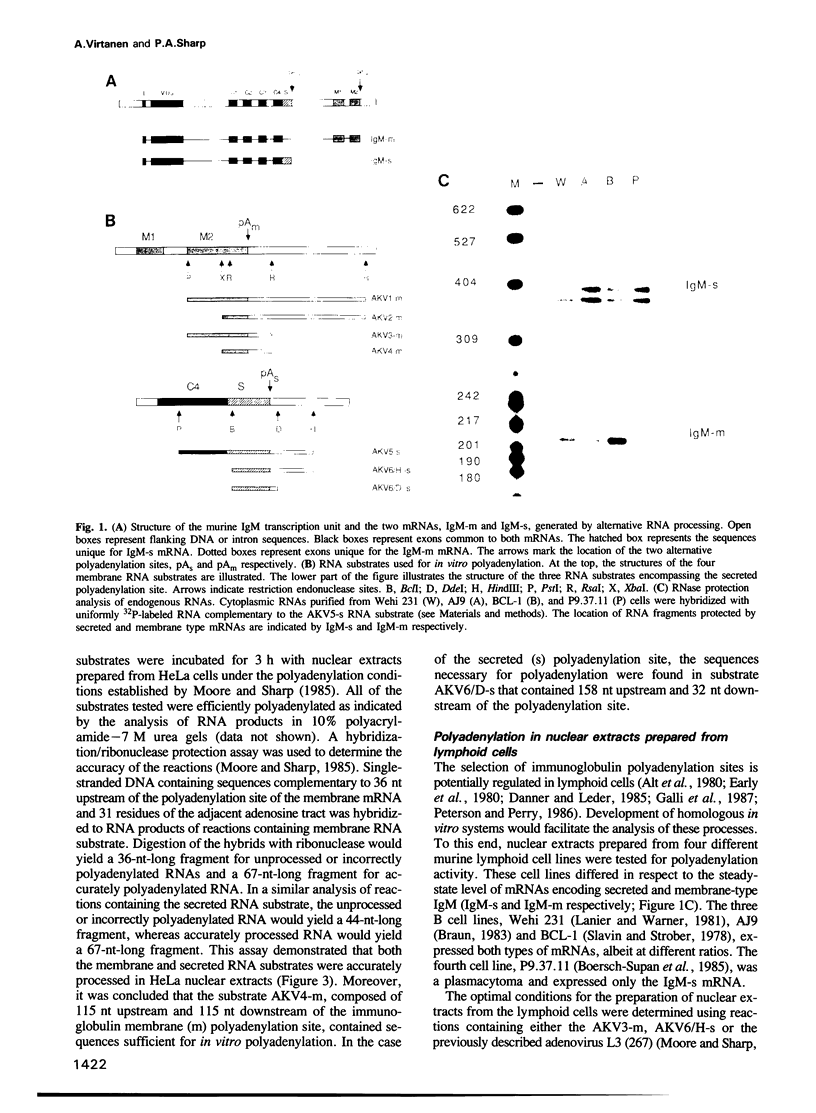
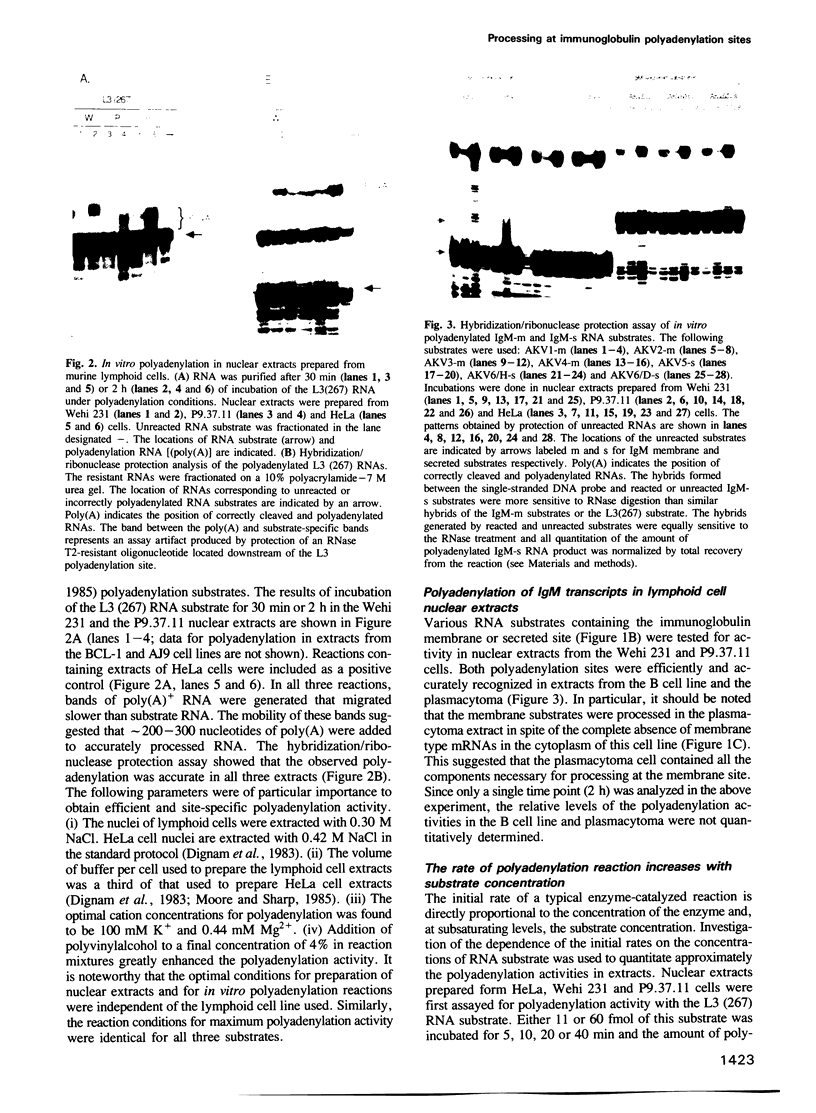
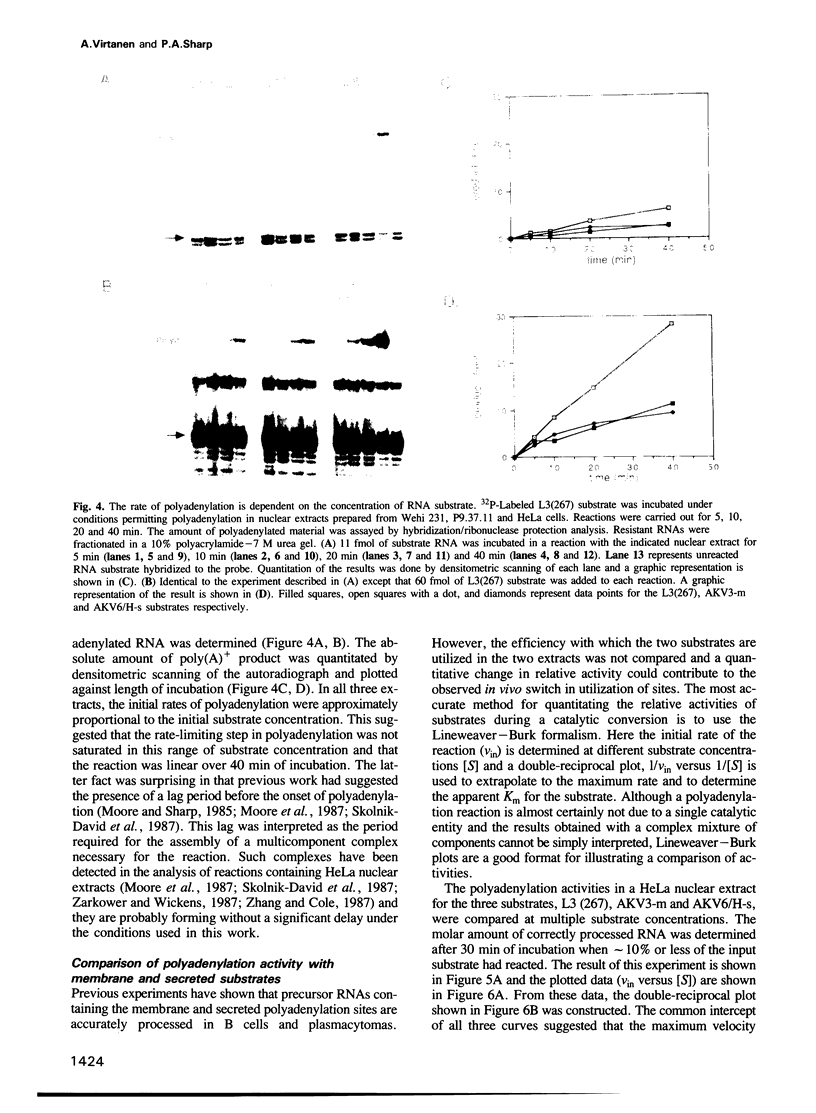
Abstract
We have developed an in vitro system for polyadenylation of RNA substrates in cell-free nuclear extracts prepared from murine cells of lymphoid origin. RNA substrates containing the adenovirus L3, murine immunoglobulin (IgM) secreted and membrane polyadenylation sites were accurately polyadenylated in these extracts. Kinetic analysis showed that the rate of polyadenylation in vitro responds proportionally to the substrate concentration. Quantitation of the initial rate of polyadenylation at the three sites permitted comparison of the activities of extracts prepared from HeLa cells, B cells (Wehi 231) and plasmacytoma cells (P9.37.11). From this analysis, we concluded that in all three extracts the polyadenylation activity at the L3 site was higher than that of either of the IgM sites. In contrast to the preferential utilization of the secreted site in vivo in plasmacytomas, this site was not selectively processed in plasmacytoma as compared to B cell extracts. The efficiency of polyadenylation at both IgM sites in the plasmacytoma extract was significantly lower than that in the B cell extract. The common low activity at the IgM sites in the plasmacytoma cell extract suggests that the rate-limiting step for polyadenylation at these two sites differs from that at the L3 site.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alt F. W., Bothwell A. L., Knapp M., Siden E., Mather E., Koshland M., Baltimore D. Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3' ends. Cell. 1980 Jun;20(2):293–301. doi: 10.1016/0092-8674(80)90615-7. [DOI] [PubMed] [Google Scholar]
- Berget S. M. Are U4 small nuclear ribonucleoproteins involved in polyadenylation? Nature. 1984 May 10;309(5964):179–182. doi: 10.1038/309179a0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Structure of the adenovirus 2 early mRNAs. Cell. 1978 Jul;14(3):695–711. doi: 10.1016/0092-8674(78)90252-0. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Boersch-Supan M. E., Agarwal S., White-Scharf M. E., Imanishi-Kari T. Heavy chain variable region. Multiple gene segments encode anti-4-(hydroxy-3-nitro-phenyl)acetyl idiotypic antibodies. J Exp Med. 1985 Jun 1;161(6):1272–1292. doi: 10.1084/jem.161.6.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J. Spontaneous in vitro occurrence and long-term culture of murine B lymphoblast cell lines. J Immunol. 1983 May;130(5):2113–2116. [PubMed] [Google Scholar]
- Brawerman G., Mendecki J., Lee S. Y. A procedure for the isolation of mammalian messenger ribonucleic acid. Biochemistry. 1972 Feb 15;11(4):637–641. doi: 10.1021/bi00754a027. [DOI] [PubMed] [Google Scholar]
- Conway L., Wickens M. A sequence downstream of A-A-U-A-A-A is required for formation of simian virus 40 late mRNA 3' termini in frog oocytes. Proc Natl Acad Sci U S A. 1985 Jun;82(12):3949–3953. doi: 10.1073/pnas.82.12.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danner D., Leder P. Role of an RNA cleavage/poly(A) addition site in the production of membrane-bound and secreted IgM mRNA. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8658–8662. doi: 10.1073/pnas.82.24.8658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early P., Rogers J., Davis M., Calame K., Bond M., Wall R., Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980 Jun;20(2):313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Galli G., Guise J. W., McDevitt M. A., Tucker P. W., Nevins J. R. Relative position and strengths of poly(A) sites as well as transcription termination are critical to membrane versus secreted mu-chain expression during B-cell development. Genes Dev. 1987 Jul;1(5):471–481. doi: 10.1101/gad.1.5.471. [DOI] [PubMed] [Google Scholar]
- Gil A., Proudfoot N. J. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit beta-globin mRNA 3' end formation. Cell. 1987 May 8;49(3):399–406. doi: 10.1016/0092-8674(87)90292-3. [DOI] [PubMed] [Google Scholar]
- Grabowski P. J., Padgett R. A., Sharp P. A. Messenger RNA splicing in vitro: an excised intervening sequence and a potential intermediate. Cell. 1984 Jun;37(2):415–427. doi: 10.1016/0092-8674(84)90372-6. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Baltimore D. Cell-type specificity of immunoglobulin gene expression is regulated by at least three DNA sequence elements. Cell. 1985 Jul;41(3):885–897. doi: 10.1016/s0092-8674(85)80069-6. [DOI] [PubMed] [Google Scholar]
- Hart R. P., McDevitt M. A., Nevins J. R. Poly(A) site cleavage in a HeLa nuclear extract is dependent on downstream sequences. Cell. 1985 Dec;43(3 Pt 2):677–683. doi: 10.1016/0092-8674(85)90240-5. [DOI] [PubMed] [Google Scholar]
- Hashimoto C., Steitz J. A. A small nuclear ribonucleoprotein associates with the AAUAAA polyadenylation signal in vitro. Cell. 1986 May 23;45(4):581–591. doi: 10.1016/0092-8674(86)90290-4. [DOI] [PubMed] [Google Scholar]
- Hernandez N., Keller W. Splicing of in vitro synthesized messenger RNA precursors in HeLa cell extracts. Cell. 1983 Nov;35(1):89–99. doi: 10.1016/0092-8674(83)90211-8. [DOI] [PubMed] [Google Scholar]
- Kemp D. J., Morahan G., Cowman A. F., Harris A. W. Production of RNA for secreted immunoglobulin mu chains does not require transcriptional termination 5' to the microM exons. Nature. 1983 Jan 6;301(5895):84–86. doi: 10.1038/301084a0. [DOI] [PubMed] [Google Scholar]
- Kobrin B. J., Milcarek C., Morrison S. L. Sequences near the 3' secretion-specific polyadenylation site influence levels of secretion-specific and membrane-specific IgG2b mRNA in myeloma cells. Mol Cell Biol. 1986 May;6(5):1687–1697. doi: 10.1128/mcb.6.5.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M. M., Padgett R. A., Sharp P. A. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984 Oct;38(3):731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- Lanier L. L., Warner N. L. Cell cycle related heterogeneity of Ia antigen expression on a murine B lymphoma cell line:analysis by flow cytometry. J Immunol. 1981 Feb;126(2):626–631. [PubMed] [Google Scholar]
- Leff S. E., Rosenfeld M. G., Evans R. M. Complex transcriptional units: diversity in gene expression by alternative RNA processing. Annu Rev Biochem. 1986;55:1091–1117. doi: 10.1146/annurev.bi.55.070186.005303. [DOI] [PubMed] [Google Scholar]
- Manley J. L., Yu H., Ryner L. RNA sequence containing hexanucleotide AAUAAA directs efficient mRNA polyadenylation in vitro. Mol Cell Biol. 1985 Feb;5(2):373–379. doi: 10.1128/mcb.5.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt M. A., Imperiale M. J., Ali H., Nevins J. R. Requirement of a downstream sequence for generation of a poly(A) addition site. Cell. 1984 Jul;37(3):993–999. doi: 10.1016/0092-8674(84)90433-1. [DOI] [PubMed] [Google Scholar]
- McLauchlan J., Gaffney D., Whitton J. L., Clements J. B. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3' termini. Nucleic Acids Res. 1985 Feb 25;13(4):1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Inhibition of RNA cleavage but not polyadenylation by a point mutation in mRNA 3' consensus sequence AAUAAA. Nature. 1983 Oct 13;305(5935):600–605. doi: 10.1038/305600a0. [DOI] [PubMed] [Google Scholar]
- Moore C. L., Sharp P. A. Accurate cleavage and polyadenylation of exogenous RNA substrate. Cell. 1985 Jul;41(3):845–855. doi: 10.1016/s0092-8674(85)80065-9. [DOI] [PubMed] [Google Scholar]
- Moore C. L., Sharp P. A. Site-specific polyadenylation in a cell-free reaction. Cell. 1984 Mar;36(3):581–591. doi: 10.1016/0092-8674(84)90337-4. [DOI] [PubMed] [Google Scholar]
- Moore C. L., Skolnik-David H., Sharp P. A. Analysis of RNA cleavage at the adenovirus-2 L3 polyadenylation site. EMBO J. 1986 Aug;5(8):1929–1938. doi: 10.1002/j.1460-2075.1986.tb04446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. L., Skolnik-David H., Sharp P. A. Sedimentation analysis of polyadenylation-specific complexes. Mol Cell Biol. 1988 Jan;8(1):226–233. doi: 10.1128/mcb.8.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R. A., Hardy S. F., Sharp P. A. Splicing of adenovirus RNA in a cell-free transcription system. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5230–5234. doi: 10.1073/pnas.80.17.5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M. L., Perry R. P. Regulated production of mu m and mu s mRNA requires linkage of the poly(A) addition sites and is dependent on the length of the mu s-mu m intron. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8883–8887. doi: 10.1073/pnas.83.23.8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Ruether J. E., Maderious A., Lavery D., Logan J., Fu S. M., Chen-Kiang S. Cell-type-specific synthesis of murine immunoglobulin mu RNA from an adenovirus vector. Mol Cell Biol. 1986 Jan;6(1):123–133. doi: 10.1128/mcb.6.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Ryner L. C., Manley J. L. Requirements for accurate and efficient mRNA 3' end cleavage and polyadenylation of a simian virus 40 early pre-RNA in vitro. Mol Cell Biol. 1987 Jan;7(1):495–503. doi: 10.1128/mcb.7.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets M. D., Stephenson P., Wickens M. P. Products of in vitro cleavage and polyadenylation of simian virus 40 late pre-mRNAs. Mol Cell Biol. 1987 Apr;7(4):1518–1529. doi: 10.1128/mcb.7.4.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skolnik-David H., Moore C. L., Sharp P. A. Electrophoretic separation of polyadenylation-specific complexes. Genes Dev. 1987 Sep;1(7):672–682. doi: 10.1101/gad.1.7.672. [DOI] [PubMed] [Google Scholar]
- Slavin S., Strober S. Spontaneous murine B-cell leukaemia. Nature. 1978 Apr 13;272(5654):624–626. doi: 10.1038/272624a0. [DOI] [PubMed] [Google Scholar]
- Wickens M., Stephenson P. Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3' end formation. Science. 1984 Nov 30;226(4678):1045–1051. doi: 10.1126/science.6208611. [DOI] [PubMed] [Google Scholar]
- Woychik R. P., Lyons R. H., Post L., Rottman F. M. Requirement for the 3' flanking region of the bovine growth hormone gene for accurate polyadenylylation. Proc Natl Acad Sci U S A. 1984 Jul;81(13):3944–3948. doi: 10.1073/pnas.81.13.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkower D., Stephenson P., Sheets M., Wickens M. The AAUAAA sequence is required both for cleavage and for polyadenylation of simian virus 40 pre-mRNA in vitro. Mol Cell Biol. 1986 Jul;6(7):2317–2323. doi: 10.1128/mcb.6.7.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkower D., Wickens M. Formation of mRNA 3' termini: stability and dissociation of a complex involving the AAUAAA sequence. EMBO J. 1987 Jan;6(1):177–186. doi: 10.1002/j.1460-2075.1987.tb04736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Cole C. N. Identification of a complex associated with processing and polyadenylation in vitro of herpes simplex virus type 1 thymidine kinase precursor RNA. Mol Cell Biol. 1987 Sep;7(9):3277–3286. doi: 10.1128/mcb.7.9.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]