Abstract
Aims: The present study was designed to evaluate the different expression of ubiquitin-like with PHD and ring finger domains 1 (UHRF1) in hepatocellular carcinoma (HCC) tissues and the adjacent normal tissues, further explore the correlation between UHRF1 expression and the prognosis of HCC patients. Methods: The UHRF1 expression at protein level in HCC tissues and the adjacent normal tissues were measured by high performance liquid chromatography (HPLC). Chi-square test was used to estimate the relationship between UHRF1 expression and clinicopathologic characteristics of HCC patients. The overall survival of HCC patients with diverse expression of UHRF1 was measured by Kaplan-Meier analysis. Cox regression analysis was conducted to judge the prognostic value of UHRF1 in HCC patients. Results: The UHRF1 was over-expressed in HCC tissues compared with the adjacent normal tissues according to the outcome of HPLC (P<0.001). Besides, the UHRF1 expression was tightly related to distant metastasis, cancer area, and HBV (P<0.05), but shared no correlation with gender, cirrhosis, and bilirubin (P>0.05). Patients with high UHRF1 expression had a shorter overall survival time than those with low UHRF1 expression (P<0.001). Cox regression analysis showed that UHRF1 was significantly linked with the prognosis of HCC patients (P=0.002, HR=5.807, 95% CI=1.901-17.742). Conclusion: UHRF1 was over-expressed in HCC tissues compared to the adjacent normal tissues and UHRF1 expression shared significant relevance with distant metastasis, cancer area and HBV. It could be an important and independent prognostic biomarker for HCC patients.
Keywords: Hepatocellular carcinoma, HPLC, UHRF1, prognosis
Introduction
Hepatocellular carcinoma (HCC) is a malignant cancer that accounts for the majority of primary liver cancers [1]. HCC is one of the most common cancers and one of the leading causes of cancer-related deaths all over the world [2]. Every year, more than 600,000 patients are diagnosed as HCC and the 5-year survival rate of HCC patients is poor [3]. Virus infection including HBV and HCV has been regarded as the crucial factor for HCC in china [4,5]. However, the pathogenesis of HCC is still unclear and concealed, the process of HCC is aggressive, invasive and rapid, and the prognosis of HCC is poor with a high mortality because of the diagnosis at an advanced stage [6,7]. A diagnostic or prognostic marker is useful for the diagnosis or improving the overall survival of patients with HCC. However, the accurate indicators for the diagnosis and prognosis were scarce. Consequently, it is urgent to develop a novel biomarker for therapy and prognosis of HCC.
Ubiquitinlike with PHD and ring finger domain 1 (UHRF1), which is also known as ICBP90/Np95, encodes multi-functional domain containing proteins with a length of 793 amino acids [8,9]. UHRF1 could not only regulate the cell proliferation but also maintain DNA methylation and play an important role on the high order of chromatin via the SRA domain and TTD domain [10-15]. It is also considered as an apoptotic-gene and abnormal expressed in various cancers including breast cancer, colorectal cancer, bladder cancer and pancreatic cancer [16-19]. All these studies have suggested that UHRF1 may play an important role in the progression of human cancers. Besides, Zhuo et al. have demonstrated that MEG3 regulates HCC cell proliferation and apoptosis and its over-expression was thought to be a mechanism underlying DNA hypomethylation in HCC. Meanwhile, UHRF1 had been proven to be a controlling gene for MEG3 which might be a prognostic marker for predicting the survival of HCC patients. [20,21]. However, according to the information we have, the prognostic role of UHRF1 in HCC has never been reported.
In this study, we aimed at detecting the expression of UHRF1 in HCC and evaluating whether it could be an innovated prognostic marker for HCC patients.
Materials and methods
Patients and specimens
A total of sixty eight HCC patients, including 29 cases infected with HBV, were recruited from Hepatobiliary Department of Internal Medicine in Henan Provincial People’s Hospital from 2008 to 2009 in this study. All patients had not received any radiotherapy or chemotherapy before surgery. The study was performed upon the approval of Ethics Committee of Henan Provincial People’s Hospital and all patients were asked to sign the informed consents in advance.
The HCC tissues and adjacent tissues were collected from HCC patients and frozen by liquid nitrogen immediately. Then the tissues were stored at -80°C for use. In addition, a 5-years follow-up was conducted with a telephone or questionnaire. The overall survival time of patients was defined from the day of surgery to the day of death. The clinicopathologic characteristics including gender, liver cirrhosis, bilirubin, distant metastasis, cancer area and HBV were recorded. Patients died from unexpected events or other disease were eliminated from our study.
The detection of UHRF1 protein via high performance liquid chromatography
10 g HCC tissues and the adjacent normal tissues were selected from each specimen, respectively. The homogenate was prepared with the addition of 20 mL methyl cyanides to the tissues and then centrifugated at 4000 r/min for 10 min. The supernatant was collected and the precipitation was dropped. The extracting solution was affiliated and dried with rotary evaporator, followed by dissolution with 1 mL mobile phase for use. 100 RP C18 column (12×4 mm, 5 µm) was applied and the column temperature was maintained at 35°C. The mobile phase included 20% methyl cyanides, 30% methanol and 50% water with a flow speed of 1 mL/min, and the detection wavelength was at 365 nm. 0.5 g UHRF1 protein was dissolved with mobile phase in a 10 mL volumetric flask and used as standard solution for the HPLC.
Statistical analysis
All data were analyzed by SPSS 18.0 software. The difference of UHRF1 protein expression in HCC tissues and adjacent tissues was estimated by students t test. While the relationship between UHRF1 protein expression and clinicopathologic characteristics was estimated by Chi-square test. Kaplan-Meier analysis was adopted to delineate the overall survival time of HCC patients with different UHRF1 protein expression. The prognostic value of UHRF1 was evaluated by Cox regression analysis. It was considered to be significant different when P value was less than 0.05.
Results
Up-regulation of UHRF1 protein in HCC tissues
The UHRF1 protein expression in HCC tissues and adjacent normal tissues was determined by HPLC method. As shown in Figure 1, the concentration of UHRF1 protein in HCC tissues was 66.98±13.46 (mean ± SD), which was significantly higher than that in the adjacent normal tissues (36.26±9.72), indicating that UHRF1 was increased in HCC tissues (P<0.001). This might intimate that UHRF1 was an oncogene in HCC.
Figure 1.
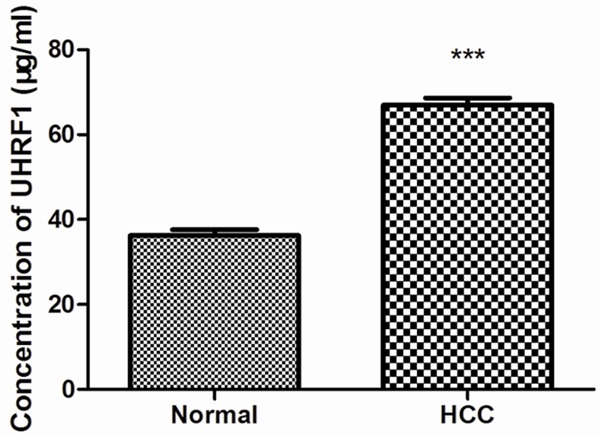
The expression of UHRF1 in HCC tissues and the adjacent normal tissues were detected by HPLC. The result presented that UHRF1 expression was strongly increased in HCC tissues compared with the adjacent normal tissues (P<0.001).
Association between UHRF1 protein expression and clinicopathologic characteristics of HCC patients
Further investigations were performed to evaluate the association between UHRF1 expression and the clinicopathologic characteristics of HCC patients. All the enrolled patients were divided into two groups: the high group with UHRF1 protein expression of more than 42.3, and the low group with UHRF1 protein expression of less than or equal to 42.3. It was confirmed that UHRF1 expression was significantly related to distant metastasis (P=0.032), cancer area (P=0.041), and HBV (P=0.036) (Table 1). However, there was no statistical difference between UHRF1 protein expression and gender, cirrhosis as well as the concentration of bilirubin (P>0.05).
Table 1.
Correlation between UHRF1 expression and clinicopathologic characteristics
| Characteristics | Case (n) | UHRF1 protein Expression | χ2 | P value | |
|---|---|---|---|---|---|
|
| |||||
| High (n) | Low (n) | ||||
| Gender | 0.180 | 0.671 | |||
| Male | 35 | 25 | 10 | ||
| Female | 33 | 22 | 11 | ||
| Liver cirrhosis | 1.240 | 0.265 | |||
| Yes | 36 | 27 | 9 | ||
| No | 32 | 20 | 12 | ||
| Bilirubin (µmol/L) | 1.722 | 0.189 | |||
| ≤17.1 | 34 | 21 | 13 | ||
| >17.1 | 34 | 26 | 8 | ||
| Distant metastasis | 4.607 | 0.032 | |||
| Yes | 39 | 31 | 8 | ||
| No | 29 | 16 | 13 | ||
| Cancer area (cm2) | 4.168 | 0.041 | |||
| ≤10 | 36 | 21 | 15 | ||
| >10 | 32 | 26 | 6 | ||
| HBV | 4.408 | 0.036 | |||
| Yes | 29 | 24 | 5 | ||
| No | 39 | 23 | 16 | ||
Elevated protein expression of UHRF1 was related to poor prognosis of HCC patients
A follow-up ranged from 0 to 60 months was adapted to deeply research the prognostic value of UHRF1 in HCC patients. During the follow-up, 33 (70.2%) patients in high UHRF1 protein expression group died, while only 4 (19.0%) died among those with low UHRF1 protein expression. Kaplan-Meier analysis showed that the overall survival time of patients with high UHRF1 protein expression was significantly shorter than those with low UHRF1 protein expression (Figure 2, log rank test, P=0.000). Cox regression analysis manifested that there was significant correlation between UHRF1 protein expression and the prognosis of HCC patients (Table 2, P=0.002, HR=5.807, 95% CI=1.901-17.742). The results indicated that UHRF1 could act as an independent prognostic biomarker for HCC patients.
Figure 2.
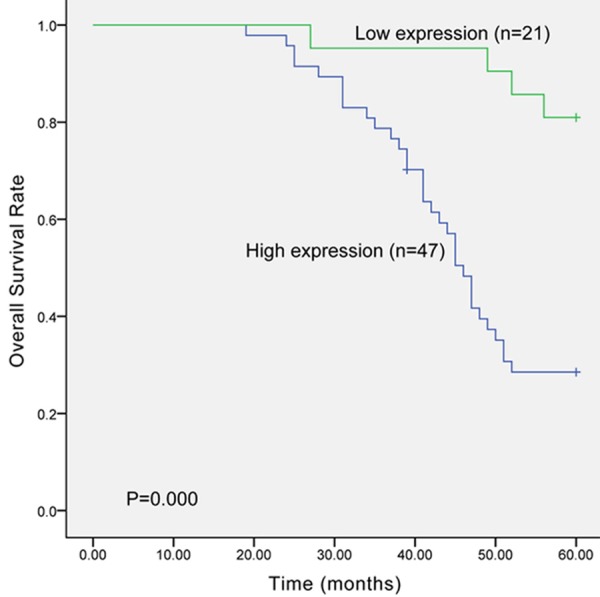
The overall survival of HCC patients was evaluated by Kaplan-Meier analysis. The result indicated that the survival time of patients with high UHRF1 protein expression was shorter than those with low UHRF1 protein expression (P=0.000).
Table 2.
Multivariate analysis of UHRF1 protein expression and clinicopathologic characteristics in the prognostic of HCC
| Clinical characteristics | P value | HR | 95% CI |
|---|---|---|---|
| Bilirubin | 0.346 | 0.713 | 0.353-1.441 |
| Cancer area | 0.250 | 1.598 | 0.719-3.551 |
| HBV | 0.157 | 1.725 | 0.811-3.668 |
| High UHRF1 expression | 0.002 | 5.807 | 1.901-17.742 |
| Low UHRF1 expression | - | - | - |
Discussion
HCC is of high malignancy and rapid development with a short average survival time [22]. Early HCC has no obvious symptoms thus patients are often diagnosed with advanced stage. The 5-year survival rate of HCC is only 14% [23]. Therapies for HCC in clinical are predominantly surgery, radiotherapy and chemotherapy. However, these therapies have weak effects on advanced stage patients and the prognosis of HCC patients was poor [24]. Therefore, early discovery, early diagnosis and early treatment are of urgent needs for HCC patients. Many researchers have engaged in finding some molecules that can be candidate biomarkers for therapy and prognosis of HCC and various biomarkers had already been found such as AFP, miRNA and lncRNA [25-28].
UHRF1 encodes a series of a subfamily of ring-finger type E3 ubiquitin ligases. The protein can bind to specific DNA sequences to regulate chromatin structure and gene expression. Recently, much attention from investigators has been focused on the relationship between UHRF1 expression and various cancers. A quantity of reports manifested that over-expression of UHRF1 was observed in many kinds of cancer. For example, Cui et al. found that the up-regulated UHRF1 in pancreatic cancer cells not only be an oncogene, but also could promote the cell growth, migration and metastasis [29]. In bladder cancer although UHRF1 expression was also increased, but it played a role in inducing cell invasion [30]. The over-expression of UHRF1 was also discovered in lung cancer, and it played an important role in the diagnosis of this cancer [31]. In addition, UHRF1 over-expression was observed in HCC in previous study, but the relationship between UHRF1 expression and clinicopathological characteristics as well as the prognostic value of UHRF1 in HCC patients was still unclear.
In the present study, we validated the expression level of UHRF1 in HCC by HPLC. The over-expression of UHRF1 was observed in HCC cases, especially in HBV infected cases, which was accordant with the previous description [21]. So we inferred that UHRF1 might be an oncogene of HCC. Moreover, previous studies have found that UHRF1 expression level was linked to the presence of lymph node metastasis, distal metastasis, and T stage [17,31]. Yang et al. [18] have shown that in bladder cancer, UHRF1 expression was positively related with tumor grade. Thus in our study, we analyzed the relationship between UHRF1 expression and clinicopathologic characteristics in HCC. Distant metastasis, cancer area and HBV infection were proved to be influential factors for the protein expression of UHRF1. Based on the result, UHRF1 was suggested to be a participator in the development of HCC.
The prognostic value of UHRF1 was studied in several cancers including bladder cancer [18], lung cancer [31], and prostate cancer [32]. Furthermore, in bladder cancer [18] and lung cancer [31] patients, UHRF1 could be used as an independent prognostic biomarker. However, the potential role of UHRF1 in the prognosis of HCC is still unclear, and it is necessary to explore an effective prognostic marker for HCC patients. Thus we estimated the prognostic value of UHRF1 in HCC. The overall survival time of patients with high UHRF1 expression was shorter than those with low expression according to Kaplan-Meier analysis which manifested that UHRF1 was related to the prognosis of HCC. Hence, the multivariate analysis was performed and the result exhibited that high UHRF1 expression increased the risk of HCC. The UHRF1 was a vital factor for the prognosis of HCC and would be an independent prognostic biomarker for HCC patients.
In conclusion, combing with the previous findings, we have verified that the expression of UHRF1 was increased in HCC and it could act as a potential prognostic indicator for HCC patients. However, the mechanism of UHRF1 on HCC remains unclear which needs to be further investigated.
Disclosure of conflict of interest
None.
References
- 1.Amaddeo G, Guichard C, Imbeaud S, Zucman-Rossi J. Next-generation sequencing identified new oncogenes and tumor suppressor genes in human hepatic tumors. Oncoimmunology. 2012;1:1612–1613. doi: 10.4161/onci.21480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Jhunjhunwala S, Jiang Z, Stawiski EW, Gnad F, Liu J, Mayba O, Du P, Diao J, Johnson S, Wong KF, Gao Z, Li Y, Wu TD, Kapadia SB, Modrusan Z, French DM, Luk JM, Seshagiri S, Zhang Z. Diverse modes of genomic alteration in hepatocellular carcinoma. Genome Biol. 2014;15:436. doi: 10.1186/s13059-014-0436-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang P, Li QJ, Feng Y, Zhang Y, Markowitz GJ, Ning S, Deng Y, Zhao J, Jiang S, Yuan Y, Wang HY, Cheng SQ, Xie D, Wang XF. TGF-beta-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell. 2012;22:291–303. doi: 10.1016/j.ccr.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willimsky G, Schmidt K, Loddenkemper C, Gellermann J, Blankenstein T. Virus-induced hepatocellular carcinomas cause antigen-specific local tolerance. J Clin Invest. 2013;123:1032–1043. doi: 10.1172/JCI64742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sherman M. Recurrence of hepatocellular carcinoma. N Engl J Med. 2008;359:2045–2047. doi: 10.1056/NEJMe0807581. [DOI] [PubMed] [Google Scholar]
- 7.Yamashita T, Wang XW. Cancer stem cells in the development of liver cancer. J Clin Invest. 2013;123:1911–1918. doi: 10.1172/JCI66024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo T, Cui S, Bian C, Yu X. Uhrf2 is important for DNA damage response in vascular smooth muscle cells. Biochem Biophys Res Commun. 2013;441:65–70. doi: 10.1016/j.bbrc.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H, Zhang C, Sheng Y, Yao S, Liu Z, Zhang T. Frequent SOCS3 and 3OST2 promoter methylation and their epigenetic regulation in endometrial carcinoma. Am J Cancer Res. 2015;5:180–190. [PMC free article] [PubMed] [Google Scholar]
- 10.Fang L, Shanqu L, Ping G, Ting H, Xi W, Ke D, Min L, Junxia W, Huizhong Z. Gene therapy with RNAi targeting UHRF1 driven by tumor-specific promoter inhibits tumor growth and enhances the sensitivity of chemotherapeutic drug in breast cancer in vitro and in vivo. Cancer Chemother Pharmacol. 2012;69:1079–1087. doi: 10.1007/s00280-011-1801-y. [DOI] [PubMed] [Google Scholar]
- 11.Karagianni P, Amazit L, Qin J, Wong J. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol Cell Biol. 2008;28:705–717. doi: 10.1128/MCB.01598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothbart SB, Dickson BM, Ong MS, Krajewski K, Houliston S, Kireev DB, Arrowsmith CH, Strahl BD. Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation. Genes Dev. 2013;27:1288–1298. doi: 10.1101/gad.220467.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 14.Rottach A, Frauer C, Pichler G, Bonapace IM, Spada F, Leonhardt H. The multi-domain protein Np95 connects DNA methylation and histone modification. Nucleic Acids Res. 2010;38:1796–1804. doi: 10.1093/nar/gkp1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Unoki M, Nishidate T, Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23:7601–7610. doi: 10.1038/sj.onc.1208053. [DOI] [PubMed] [Google Scholar]
- 16.Geng Y, Gao Y, Ju H, Yan F. Diagnostic and prognostic value of plasma and tissue ubiquitin-like, containing PHD and RING finger domains 1 in breast cancer patients. Cancer Sci. 2013;104:194–199. doi: 10.1111/cas.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang F, Yang YZ, Shi CZ, Zhang P, Moyer MP, Zhang HZ, Zou Y, Qin HL. UHRF1 promotes cell growth and metastasis through repression of p16 (ink(4)a) in colorectal cancer. Ann Surg Oncol. 2012;19:2753–2762. doi: 10.1245/s10434-011-2194-1. [DOI] [PubMed] [Google Scholar]
- 18.Yang GL, Zhang LH, Bo JJ, Chen HG, Cao M, Liu DM, Huang YR. UHRF1 is associated with tumor recurrence in non-muscle-invasive bladder cancer. Med Oncol. 2012;29:842–847. doi: 10.1007/s12032-011-9983-z. [DOI] [PubMed] [Google Scholar]
- 19.Crnogorac-Jurcevic T, Gangeswaran R, Bhakta V, Capurso G, Lattimore S, Akada M, Sunamura M, Prime W, Campbell F, Brentnall TA, Costello E, Neoptolemos J, Lemoine NR. Proteomic analysis of chronic pancreatitis and pancreatic adenocarcinoma. Gastroenterology. 2005;129:1454–1463. doi: 10.1053/j.gastro.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 20.Zhuo H, Tang J, Lin Z, Jiang R, Zhang X, Ji J, Wang P, Sun B. The aberrant expression of MEG3 regulated by UHRF1 predicts the prognosis of hepatocellular carcinoma. Mol Carcinog. 2015 doi: 10.1002/mc.22270. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 21.Mudbhary R, Hoshida Y, Chernyavskaya Y, Jacob V, Villanueva A, Fiel MI, Chen X, Kojima K, Thung S, Bronson RT, Lachenmayer A, Revill K, Alsinet C, Sachidanandam R, Desai A, SenBanerjee S, Ukomadu C, Llovet JM, Sadler KC. UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell. 2014;25:196–209. doi: 10.1016/j.ccr.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bosch FX, Ribes J, Diaz M, Cleries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127:S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 23.Cao K, Lu C, Han S, Zou Q, Li J, Xie D, He S, Yu L, Zhou J, Peng X, Cao P. Expression of Girdin in primary hepatocellular carcinoma and its effect on cell proliferation and invasion. Int J Clin Exp Pathol. 2015;8:551–559. [PMC free article] [PubMed] [Google Scholar]
- 24.Hao K, Luk JM, Lee NP, Mao M, Zhang C, Ferguson MD, Lamb J, Dai H, Ng IO, Sham PC, Poon RT. Predicting prognosis in hepatocellular carcinoma after curative surgery with common clinicopathologic parameters. BMC Cancer. 2009;9:389. doi: 10.1186/1471-2407-9-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shang S, Plymoth A, Ge S, Feng Z, Rosen HR, Sangrajrang S, Hainaut P, Marrero JA, Beretta L. Identification of osteopontin as a novel marker for early hepatocellular carcinoma. Hepatology. 2012;55:483–490. doi: 10.1002/hep.24703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, Wang JF, Zhang Z, Lu S, Huang X, Wang Z, Qiu S, Wang X, Yang G, Sun H, Tang Z, Wu Y, Zhu H, Fan J. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J. Clin. Oncol. 2011;29:4781–4788. doi: 10.1200/JCO.2011.38.2697. [DOI] [PubMed] [Google Scholar]
- 27.Yamashita T, Kitao A, Matsui O, Hayashi T, Nio K, Kondo M, Ohno N, Miyati T, Okada H, Mizukoshi E, Honda M, Nakanuma Y, Takamura H, Ohta T, Nakamoto Y, Yamamoto M, Takayama T, Arii S, Wang X, Kaneko S. Gd-EOB-DTPA-enhanced magnetic resonance imaging and alpha-fetoprotein predict prognosis of early-stage hepatocellular carcinoma. Hepatology. 2014;60:1674–1685. doi: 10.1002/hep.27093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z, Boldanova T, Andersen JB, Hammerle M, Tornillo L, Heim MH, Diederichs S, Cillo C, Terracciano LM. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911–923. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui L, Chen J, Zhang Q, Wang X, Qu J, Zhang J, Dang S. Up-regulation of UHRF1 by oncogenic Ras promoted the growth, migration, and metastasis of pancreatic cancer cells. Mol Cell Biochem. 2015;400:223–232. doi: 10.1007/s11010-014-2279-9. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Huang Z, Zhu Z, Zheng X, Liu J, Han Z, Ma X. Upregulated UHRF1 promotes bladder cancer cell invasion by epigenetic silencing of KiSS1. PLoS One. 2014;9:e104252. doi: 10.1371/journal.pone.0104252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Unoki M, Daigo Y, Koinuma J, Tsuchiya E, Hamamoto R, Nakamura Y. UHRF1 is a novel diagnostic marker of lung cancer. Br J Cancer. 2010;103:217–222. doi: 10.1038/sj.bjc.6605717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Babbio F, Pistore C, Curti L, Castiglioni I, Kunderfranco P, Brino L, Oudet P, Seiler R, Thalman GN, Roggero E, Sarti M, Pinton S, Mello-Grand M, Chiorino G, Catapano CV, Carbone GM, Bonapace IM. The SRA protein UHRF1 promotes epigenetic crosstalks and is involved in prostate cancer progression. Oncogene. 2012;31:4878–4887. doi: 10.1038/onc.2011.641. [DOI] [PubMed] [Google Scholar]


