Abstract
Objective: To explore the methylation status of DNA-binding inhibitor 4 (ID4) in tamoxifen-refractory (TR) breast cancer. Methods: From January 2012 to December 2014, breast cancer patients managed by radical mastectomy previously and receiving tamoxifen treatment for at least 12 months were enrolled. According to the response to tamoxifen, patients were divided into TR group and tamoxifen-sensitive (TS) group. Genomic DNA was isolated from fasting venous blood, and methylight technique was applied to determine the methylation status of ID4. Results: 43 patients with TS breast cancer and 31 patients with TR breast cancer were enrolled. No significant difference between groups was observed in term of patients’ characteristics, such as age (P=0.693), progesterone receptor (P=0.970), menopausal status (P=0.784) and histological type (P=0.537), while the stage of cancer in TR group was significantly higher than TS group (P<0.001). Compared to TS group, PMR of ID4 was significantly higher in TR group (P=0.002). ROC curve analysis indicated that ID4 yielded an AUC of 0.716 with 77.4% sensitivity and 62.79% specificity in distinguishing TR breast cancer at the cut point of 3.8%. The PMR cut point of ID4 was set at 6.8% in survival analysis, log-rank test indicated the risk of disease progression was comparable between patients with ID4 hypermethylation or hypomethylation (P=0.287). Conclusion: ID4 hypomethylation is present in TR breast cancer, and it may serve as a potential biomarker in distinguishing TR breast cancer. However, the results need further validation in larger studies.
Keywords: DNA-binding inhibitor 4, tamoxifen, breast cancer, mastectomy, methylight, survival analysis
Introduction
Estrogen plays a key role in female physiology, reproduction and behavior, as well as the development of breast cancer [1]. It is well-known that the block of estrogen signaling by antiestrogen therapy could result in tumor suppression [2]. Tamoxifen, as a selective estrogen receptor (ER) modulator, has been widely used for the management of ER-positive breast cancer. It is reported that tamoxifen contributes a 40%-50% reduction in the odds of recurrence and reduced mortality [3], and provides temporary remission in 30% to 50% of patients with metastasis disease [4]. Unfortunately, one-third of females receiving tamoxifen treatment for 5 years will experience recurrence or metastasis within 15 years [5]. Therefore, developing specific biomarkers distinguishing acquired tamoxifen-refractory (TR) breast cancer will benefit these patients by shifting endocrine therapy strategy, especially before the detection of recurrence or metastasis.
DNA-binding inhibitor 4 (ID4), as a member of ID family, negatively regulates basic helix-loop-helix transcription factors, therapy plays an important role in the regulation of complex transcriptional network [6]. ID4 acts as a tumor suppressor or tumor promoter according different histologic types, and clear picture of ID4 expression pattern has not emerged in breast cancer. It is reported that ID4 expression is present in ERα negative mammary epithelial cells, basal cell-like breast cancer, triple negative breast cancer, but absent in ERα positive atypical ductal hyperplasia, ductal carcinoma in situ and invasive carcinomas [7-9]. Fan M et al. found that hypomethylation and overexpression of ID4 was observed in TR MCF7 cells [10].
In the present study, we would like to investigate the methylation status of ID4 in acquired TR breast cancer and temoxifen-sensitive (TS) breast cancer, therapy explore whether it could serve as a biomarker in distinguish acquired TR breast cancer.
Patients and methods
The research was conducted in accordance with the declaration of Helsinki, and got the approval of the ethic committee of our institution. Additionally, all participants signed informed consents.
Patients
From January 2012 to December 2014, breast cancer patients treated with tamoxifen were screened in our center. The inclusion criteria were as follows: previously treated with radical mastectomy and receiving tamoxifen treatment more than 12 months. Patients meeting the following items were excluded: receiving combination therapy of tamoixfen and other antiestrogen agents, taking immunosuppressive therapy, and suffering other kind of malignant tumor at the same time. TR was defined as breast cancer recurrence or metastasis while receiving tamoxifen therapy, while TS was defined as no recurrence or metastasis was observed during tamoxifen therapy. Patients in the TS group were followed up by magnetic resonance imaging, contrast-enhanced computerized tomography, ultrasound, X-ray or bone scans every 3 months, with the purpose of detecting recurrence or metastasis timely.
Samples and DNA isolation
Fasting venous blood was collected at the time of recruitment and detection of recurrence or metastasis for patients in TS group, and centrifuged at 2000 g at 4°C for 10 minutes within 30 minutes. The supernatant serum was transferred into Eppendorf tubes, and stored at -80°C until further analysis.
Genomic DNA was extracted from 200 μl serum by using High Pure Viral Nucleic Acid Kit (Roche Applied Science, Mannheim, Germany), which has been verified previously [11,12]. The isolation procedures were performed according to manufacturer’s instruction, and genomic DNA was eluted in 50 μl of Elution Buffer.
Methylight analysis
Sodium bisulfite conversion of genomic DNA was conducted using EZ DNA Methylation-Gold KitTM (Zymo Research, Orange, CA, USA) to convert unmethylated cytosines to uracils and leave methylated cytosines unmodified. Then, the genomic DNA was analyzed by Methylight technique. The primer and probe of ID4 has been verified in acute leukemia [13], which are as follows: forward primer, 5’-TCGGAGTTTTCGTTTTCGTT-3’; reverse primer, 5’-CGATACTACTCACAACCGCG-3’; probe, 5’-ATAAATATAGTTGCGCGGCG-3’. PCRs were performed in 20 μl volumes containing 4 μl gemonic DNA, 250 μmol/l deoxynucleotide triphosphate mixture, 1 × PCR buffer (Qiagen, Hilden, Germany), 0.05 units/μl Taq DNA polymerase (HotStar Taq, Qiagen), 4 mmol/l MgCl2, 0.6 μmol primers and 0.2 μmol probes, using the following PCR program: 95°C for 15 min, then 45 cycles of 94°C for 50 s, 56°C for 40 s, and 72°C for 60 s.
AluC4 was used to control for DNA amplification and normalize for input DNA, and completely methylated reference DNA with methylation of all the CpG islands was purchased from Millipore (#S7821) as a reference DNA. The methylation status of target gene was presented as percentage of methylated reference (PMR) values. PMR=100% × 2 exp-(CT, Target-CT, AluC4)sample-(CT, Target-CT, AluC4)fully methylated. Each methylation reaction was performed in triple, and the final PMR values represent the mean.
Statistical analysis
SPSS 13.0 (SPSS Inc., Chicago, IL, USA) was applied for statistical analysis. Comparison between groups was calculated by t test for measurement data and Mann-Whitney U test for enumeration data. Samples containing >4% fully methylated molecules were taken as hypermethylation, while samples containing ≤4% were designated as hypomethylation [14]. Receiver-operating characteristic (ROC) curves analysis and the area under the curve (AUC) was calculated to determine the efficacy of ID4 in distinguishing TR breast cancer. PMR of ID4 in patients suffering recurrence or metastasis in TS group was measured twice using the blood samples collected at enrollment and detection of recurrence or metastasis, PMR was compared by paired t test. In TS group, the mean of PMR at enrollment was taken as the cut point to distinguish hypomethylation and hypermethylation in the survival analysis. The association between progression-free survival and PMR of ID4 was computed by Log-rank test. Two-tailed P<0.05 was considered statistically significant.
Results
43 patients with TS breast cancer and 31 patients with TR breast cancer were enrolled. Estrogen receptor was positive in all participants, and the stage of cancer in TR group was significantly higher than TS group (P<0.001). No significant difference between groups was observed in term of patients’ characteristics, such as age (P=0.693), progesterone receptor (P=0.970), menopausal status (P=0.784) and histological type (P=0.537, Table 1).
Table 1.
Comparison of patients’ parameters between groups
| Characteristic | TS (n=43) | TR (n=31) | P |
|---|---|---|---|
| Age (years) | 47.9±12.5 | 49.0±11.1 | 0.693 |
| PR status | 0.970 | ||
| Negative | 22 | 16 | |
| Positive | 21 | 15 | |
| Menopausal status | 0.784 | ||
| Premenopausal | 23 | 14 | |
| Perimenopausal | 2 | 5 | |
| Postmenopausal | 18 | 12 | |
| AJCC stage | <0.001 | ||
| I | 15 | ||
| II | 17 | ||
| III | 11 | 5 | |
| IV | 26 | ||
| Histological type | 0.537 | ||
| Invasive ductal | 37 | 25 | |
| Invasive lobular | 6 | 6 | |
| Brain metastasis | 10 | ||
| PMR (%) | 6.8±5.3 | 3.4±2.3 | 0.002 |
| Methylation status | 0.001 | ||
| Hypermethylation | 27 | 7 | |
| Hypomethylation | 16 | 24 |
TS: temoxifen-sensitive breast cancer; TR: tamoxifen-refractory breast cancer; PR: progestogen receptor; AJCC: American Joint Committee on Cancer; PMR: percentage of methylated reference.
Compared to TS group, PMR of ID4 was significantly higher in TR group (6.8±5.3% vs. 3.4±2.3%, P=0.002, Figure 1A). At the cut point of 4%, the prevalence of hypomethylation of ID4 was 37.2% and 77.4% in TS and TR group (P=0.001). Further, ROC curve analysis was performed to evaluate the efficacy of ID methylation status in distinguishing TR breast cancer, which demonstrated 77.4% (95% CI: 58.9%-90.4%) sensitivity, 62.79% (95% CI: 46.7%-77.0%) specificity and AUC of 0.716 (95% CI: 0.599-0.815) at the cut point of 3.8% (Figure 1B).
Figure 1.
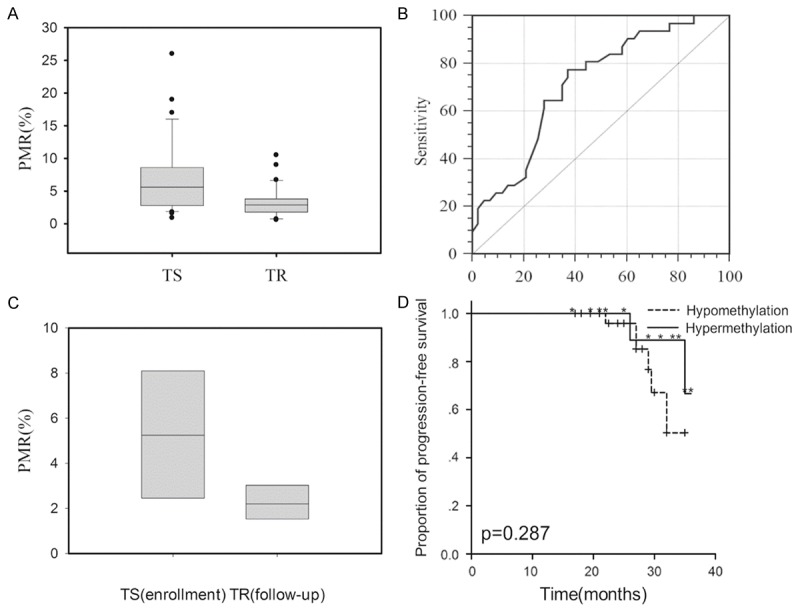
Comparison of parameters between different groups. A. Comparison of ID4 PMR between TS and TR group; B. ROC curve analysis of ID4 in distinguishing TR breast cancer; C. Comparison of ID4 PMR before and after the recurrence or metastasis in TS group; D. Kaplan-Meier cure of progression-free survival for patients in TS group according to ID4 methylation status.
During the follow-up, 8 (18.6%) patients in TS group suffered recurrence or metastasis of breast cancer. Compared to the blood sample collected at enrollment, PMR of ID4 significantly decreased after the recurrence or metastasis (5.4±2.8% vs. 2.2±0.9%, P=0.005, Figure 1C). The PMR cut point of ID4 was set at 6.8% in survival analysis, log-rank test indicated the risk of disease progression was comparable between patients with ID4 hypermethylation and hypomethylation (P=0.287, Figure 1D).
Discussion
The present study indicated that the methylation status of ID4 decreased along with TS breast cancer became TR, and ROC curve showed that hypomethylation of ID4 had moderate efficacy in distinguishing TR breast cancer. Unfortunately, the above perspectives didn’t get the support of survival analysis.
Even though the antiestrogen agents are diverse, tamoxifen is recommended for ER+ patients as the first line treatment by most doctors and the American Joint Committee of Cancer [15]. Therefore, de novo and acquired resistance to tamoxifen become a severe challenge. The mechanism of intrinsic and acquired resistance to tamoxifen is complicated and hasn’t been well elaborated, which refers to estrogen receptor and its co-regulators, receptor tyrosine kinase signaling, cell cycle regulators, cell survival signaling and apoptosis and so on [1]. More than 40 candidate tamoxifen resistance genes have been identified by gain-of-function screens in breast cancer, such as genes encoding estrogen receptor co-repressor NCOR2, intracellular signaling molecules (BCAR1, BCAR3, AKT1, AKT2, SRC and GRB7), cell surface receptors (EGFR, ERBB2, platelet-derived growth factor receptor-β) and colony-stimulating factor 1 receptor) and genes of poorly understood function (for example, BCAR4 and TLE3) [1].
Previous studies indicated that hypermethylation of ID4 was observed in 69% primary breast cancer sample, and the hypermethylation of ID4 was associated with decreased recurrence-free survival and increased risk of lymph node metastasis [16,17]. Fan M et al. established tamoxifen-refractory sublines from a single colony of hormone-dependent breast cancer MCF7 cells and conducted differential methylation hybridization, then found hypomethylation and overexpression of ID4 in tamoxifen-refractory sublines compared to MCF7 cell [10]. Our results also found that ID4 was hypermethylated in most TS cases, and then become hypomethylated along with the loss of response to tamoxifen treatment. Early study indicated that marked decreases of estrogen receptor was observed in patients receiving tamoxifen treatment [18,19], and it has been reported that ID4 expression was inversely correlated with estrogen receptor expression [7,8]. A possible explanation may be that ID4 expression is suppressed by estrogen signal, while the suppressive efficacy gradually decreased along with the progressive loss of estrogen receptor during the period of tamoxifen treatment. Of course, it is still unclear that it is ID4 regulates the expression of estrogen receptor or be regulated by estrogen signal. ID4 is frequently inactivated through promoter hypermethylation and serves as a tumor suppressor in ER+ breast cancer, while it acts like an oncogene in ERα-breast cancer cells MDA-MB-237 and SKBr3 that express mutant p53 [20]. Further study indicates that a protein complex including mutant p53 R273H-E2F1-p65 positively controls ID4 expression by assembling on specific regions of the ID4 promoter [21]. Moreover, adriamycin and cisplatin treatments could result in the increase of ID4 expression in mutant p53/ID4 expressing cells, further to achieve ‘mutant p53 associated’ chemo-resistance [20]. On contrary to the results from previous study in primary breast cancer [16,17]. Kaplan-Meier curve indicates that it seems as if ID4 serves as an oncogene and promotes the progression of breast cancer in tamoxifen-treated cases. Even though statistical difference wasn’t calculated between hypermethylation group and hypomethylation group in the survival analysis, a general trend could be seen that patients with hypomethylation of ID4 suffer shorter progression-free survival. We believe that the limited cohort and follow-up duration reduce the efficacy of log-rank test, larger studies with longer follow-up duration will provide more persuasive evidence.
There are several limitations needing to be considered. First, all participants are Asians and from a single center, whether they could represent the general population is still in doubt. Second, TS breast cancer was defined no recurrence or metastasis was observed in patients receiving tamoxifen treatment for at least 12 months. However, it couldn’t exclude patients with minor metastasis which couldn’t be detected by present techniques and intrinsic tamoxifen-refractory breast cancer which didn’t relapse at enrollment, it will bring bias to the results. Third, we propose a hypothesis to explain the results we observed, but the expressions of ER and mutant p53 weren’t evaluated in the present research due to economic and institutional reasons, regrettably. Fourth, the cohort of the study and duration of follow-up are limited. It is certain that larger studies with sufficient follow-up will provide more persuasive evidence.
In conclusion, ID4 hypomethylation was observed in TR breast cancer, it may serve as a potential biomarker in distinguishing TR breast cancer. However, the results need further validation in larger studies.
Acknowledgements
This study was supported by the Tianjin municipal natural science foundation (No. 13JCYBJC21800).
Disclosure of conflict of interest
None.
References
- 1.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9:631–43. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 2.Jensen EV, Jordan VC. The estrogen receptor: a model for molecular medicine. Clin Cancer Res. 2003;9:1980–9. [PubMed] [Google Scholar]
- 3.Osborne CK. Tamoxifen in the treatment of breast cancer. N Engl J Med. 1998;339:1609–18. doi: 10.1056/NEJM199811263392207. [DOI] [PubMed] [Google Scholar]
- 4.Fisher B, Costantino JP, Wickerham DL, Redmond CK, Kavanah M, Cronin WM, Vogel V, Robidoux A, Dimitrov N, Atkins J, Daly M, Wieand S, Tan-Chiu E, Ford L, Wolmark N. Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Inst. 1998;90:1371–88. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 5.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 6.Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13:410–8. doi: 10.1016/s0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- 7.Roldan G, Delgado L, Muse IM. Tumoral expression of BRCA1, estrogen receptor alpha and ID4 protein in patients with sporadic breast cancer. Cancer Biol Ther. 2006;5:505–10. doi: 10.4161/cbt.5.5.2597. [DOI] [PubMed] [Google Scholar]
- 8.de Candia P, Akram M, Benezra R, Brogi E. Id4 messenger RNA and estrogen receptor expression: inverse correlation in human normal breast epithelium and carcinoma. Hum Pathol. 2006;37:1032–41. doi: 10.1016/j.humpath.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Turner NC, Reis-Filho JS, Russell AM, Springall RJ, Ryder K, Steele D, Savage K, Gillett CE, Schmitt FC, Ashworth A, Tutt AN. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene. 2007;26:2126–32. doi: 10.1038/sj.onc.1210014. [DOI] [PubMed] [Google Scholar]
- 10.Fan M, Yan PS, Hartman-Frey C, Chen L, Paik H, Oyer SL, Salisbury JD, Cheng AS, Li L, Abbosh PH, Huang TH, Nephew KP. Diverse gene expression and DNA methylation profiles correlate with differential adaptation of breast cancer cells to the antiestrogens tamoxifen and fulvestrant. Cancer Res. 2006;66:11954–66. doi: 10.1158/0008-5472.CAN-06-1666. [DOI] [PubMed] [Google Scholar]
- 11.Philipp AB, Nagel D, Stieber P, Lamerz R, Thalhammer I, Herbst A, Kolligs FT. Circulating cell-free methylated DNA and lactate dehydrogenase release in colorectal cancer. BMC Cancer. 2014;14:245. doi: 10.1186/1471-2407-14-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gobel G, Auer D, Gaugg I, Schneitter A, Lesche R, Muller-Holzner E, Marth C, Daxenbichler G. Prognostic significance of methylated RASSF1A and PITX2 genes in blood- and bone marrow plasma of breast cancer patients. Breast Cancer Res Treat. 2011;130:109–17. doi: 10.1007/s10549-010-1335-8. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Kang HY, Wang LL, Lu XC, Zhu HL, Yu L. [Establishment of methylation-specific quantitative PCR system for ID4 gene in acute leukemia cells and its specificity and sensitivity] . Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2014;22:269–74. doi: 10.7534/j.issn.1009-2137.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Xu X, Gammon MD, Zhang Y, Cho YH, Wetmur JG, Bradshaw PT, Garbowski G, Hibshoosh H, Teitelbaum SL, Neugut AI, Santella RM, Chen J. Gene promoter methylation is associated with increased mortality among women with breast cancer. Breast Cancer Res Treat. 2010;121:685–92. doi: 10.1007/s10549-009-0628-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burstein HJ, Temin S, Anderson H, Buchholz TA, Davidson NE, Gelmon KE, Giordano SH, Hudis CA, Rowden D, Solky AJ, Stearns V, Winer EP, Griggs JJ. Adjuvant endocrine therapy for women with hormone receptor-positive breast cancer: american society of clinical oncology clinical practice guideline focused update. J. Clin. Oncol. 2014;32:2255–69. doi: 10.1200/JCO.2013.54.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noetzel E, Veeck J, Niederacher D, Galm O, Horn F, Hartmann A, Knüchel R, Dahl E. Promoter methylation-associated loss of ID4 expression is a marker of tumour recurrence in human breast cancer. BMC Cancer. 2008;8:154. doi: 10.1186/1471-2407-8-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Umetani N, Mori T, Koyanagi K, Shinozaki M, Kim J, Giuliano AE, Hoon DS. Aberrant hypermethylation of ID4 gene promoter region increases risk of lymph node metastasis in T1 breast cancer. Oncogene. 2005;24:4721–7. doi: 10.1038/sj.onc.1208538. [DOI] [PubMed] [Google Scholar]
- 18.Hull DF 3rd, Clark GM, Osborne CK, Chamness GC, Knight WA 3rd, McGuire WL. Multiple estrogen receptor assays in human breast cancer. Cancer Res. 1983;43:413–6. [PubMed] [Google Scholar]
- 19.Encarnacion CA, Ciocca DR, McGuire WL, Clark GM, Fuqua SA, Osborne CK. Measurement of steroid hormone receptors in breast cancer patients on tamoxifen. Breast Cancer Res Treat. 1993;26:237–46. doi: 10.1007/BF00665801. [DOI] [PubMed] [Google Scholar]
- 20.Dell’Orso S, Ganci F, Strano S, Blandino G, Fontemaggi G. ID4: a new player in the cancer arena. Oncotarget. 2010;1:48–58. doi: 10.18632/oncotarget.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fontemaggi G, Dell’Orso S, Trisciuoglio D, Shay T, Melucci E, Fazi F, Terrenato I, Mottolese M, Muti P, Domany E, Del Bufalo D, Strano S, Blandino G. The execution of the transcriptional axis mutant p53, E2F1 and ID4 promotes tumor neo-angiogenesis. Nat Struct Mol Biol. 2009;16:1086–93. doi: 10.1038/nsmb.1669. [DOI] [PubMed] [Google Scholar]


