Abstract
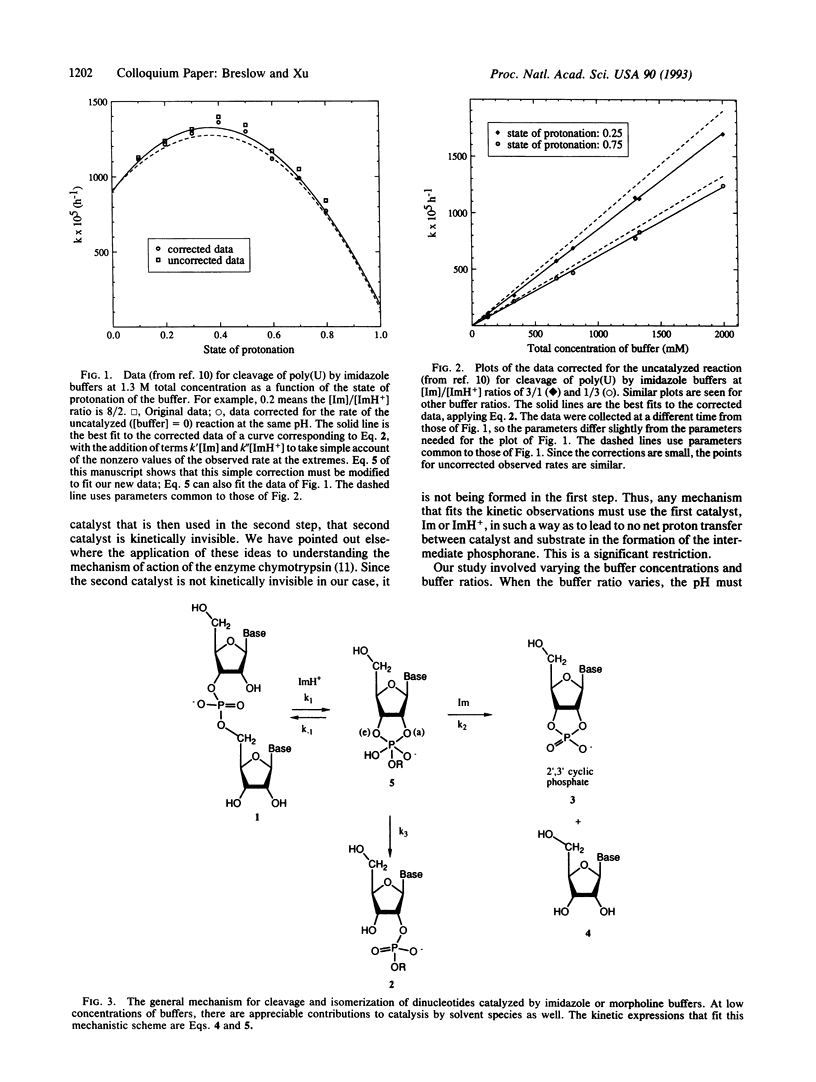
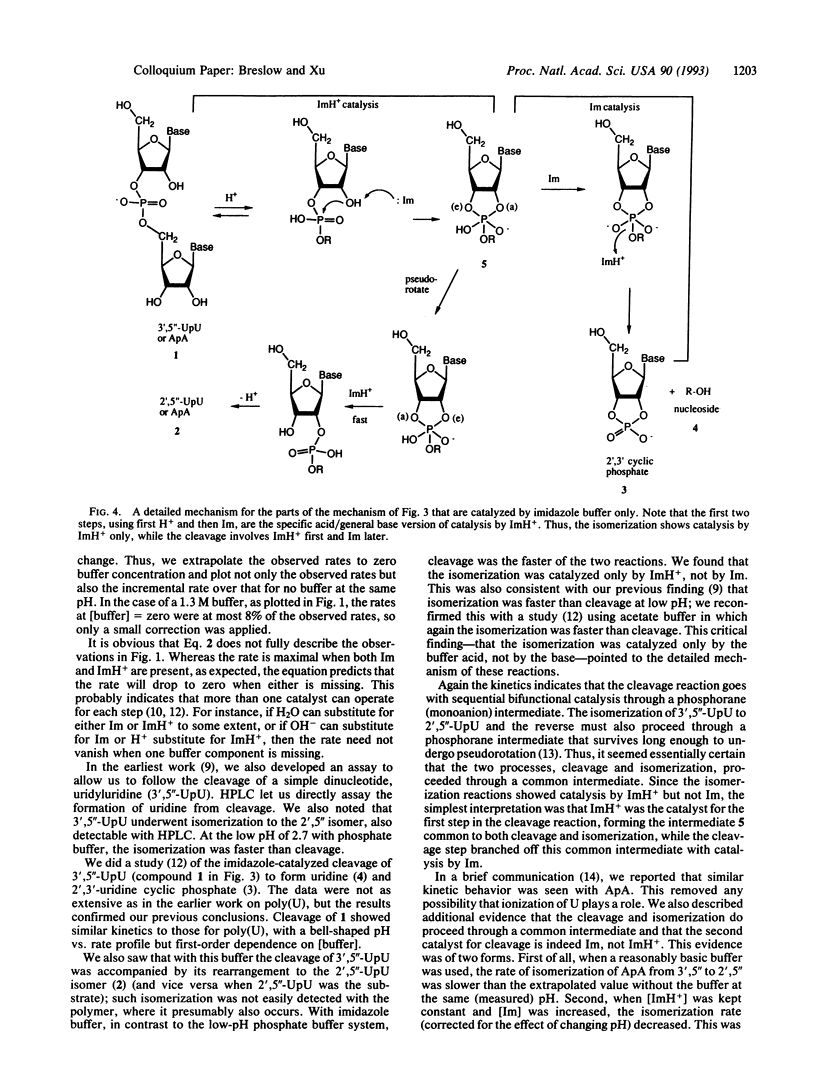
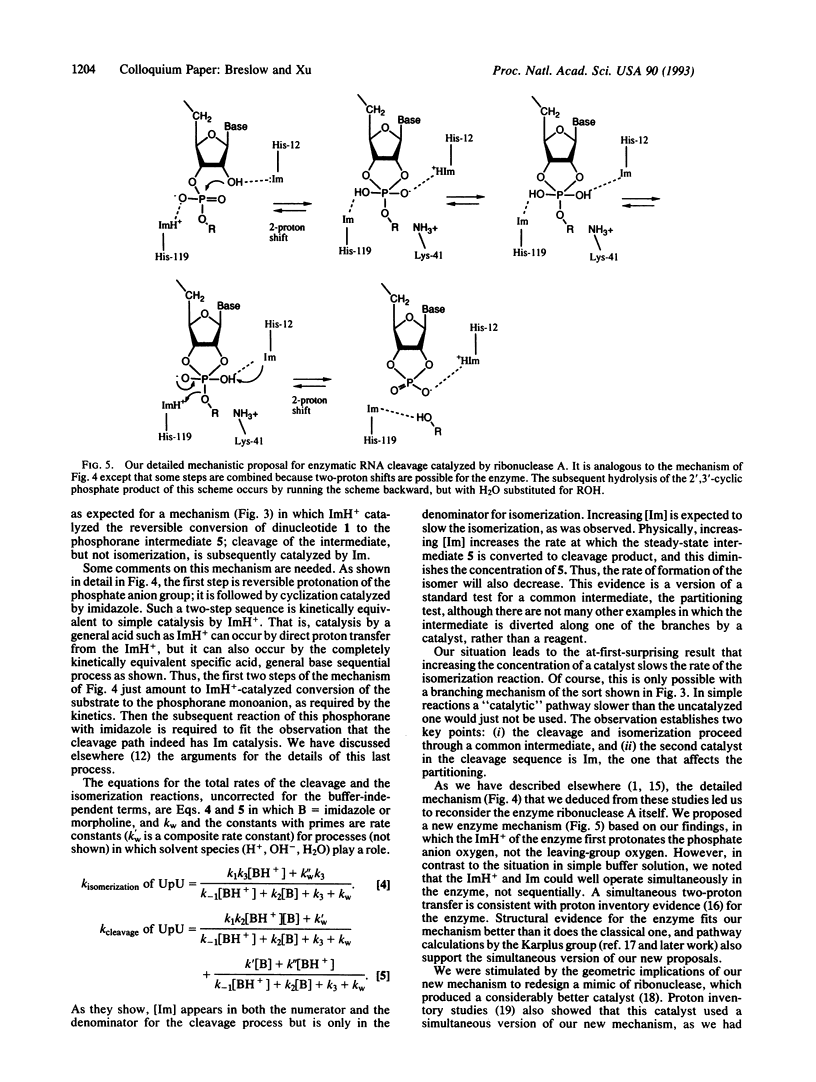
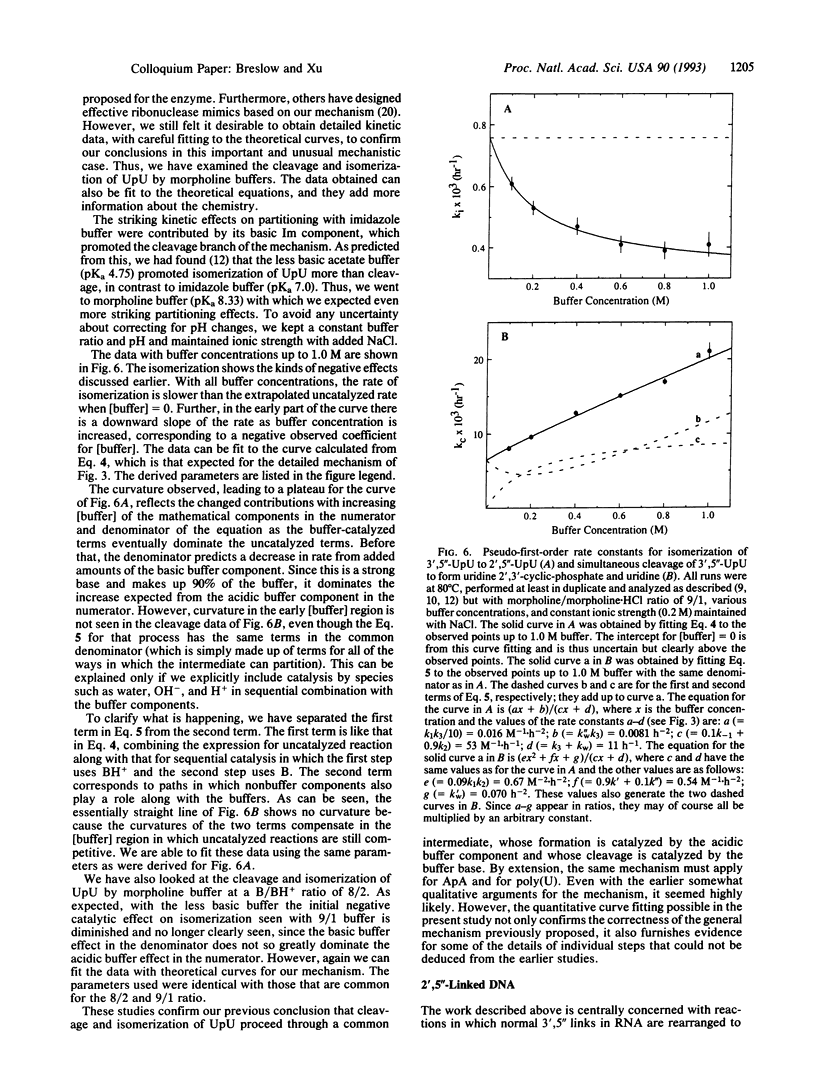
The enzyme ribonuclease A catalyzes the cleavage of RNA, using the imidazole groups of histidine-12 and histidine-119 as its principal catalytic groups. Model studies show that RNA can be cleaved by imidazole buffer itself and that, as in the enzyme, a bell-shaped pH vs. rate profile is seen. This indicates that one imidazole functions as a base, while the other, as the imidazolium ion, functions as an acid. However, in contrast to the enzymatic case, the simple model uses the imidazoles in sequential, rather than simultaneous, bifunctional catalysis. Mechanistic studies on this reaction and on the reactions of simple dinucleotides catalyzed by imidazole and other buffers establish the details of the process. The results let us propose a mechanism for the enzymatic process different from the standard one; they also stimulated us to design an improved mimic of the enzyme that uses a mechanism like that proposed for the enzyme. Critical to the mechanistic studies is observation of the rearrangement of normal 3',5'' RNA nucleotides to the 2',5'' isomers. This led us to investigate the properties of DNA isomers in which a 2',5'' link also replaces the normal 3',5'' one. The results indicate that poor base stacking in a double helix with such links makes them less suitable as genetic units.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anukanth A., Ponnuswamy P. K. 2'5'-linked polynucleotides do form a double-stranded helical structure: a result from the energy minimization study of A2'p5'A. Biopolymers. 1986 Apr;25(4):729–752. doi: 10.1002/bip.360250414. [DOI] [PubMed] [Google Scholar]
- Breslow R., Huang D. L., Anslyn E. On the mechanism of action of ribonucleases: dinucleotide cleavage catalyzed by imidazole and Zn2+. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1746–1750. doi: 10.1073/pnas.86.6.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran R., Labelle M., Czarnik A. W., Breslow R. An assay to determine the kinetics of RNA cleavage. Anal Biochem. 1985 Feb 1;144(2):563–568. doi: 10.1016/0003-2697(85)90154-x. [DOI] [PubMed] [Google Scholar]
- Dhingra M. M., Sarma R. H. Why do nucleic acids have 3'5' phosphodiester bonds? Nature. 1978 Apr 27;272(5656):798–801. doi: 10.1038/272798a0. [DOI] [PubMed] [Google Scholar]
- Eftink M. R., Biltonen R. L. Energetics of ribonuclease A catalysis. 1. pH, ionic strength, and solvent isotope dependence of the hydrolysis of cytidine cyclic 2',3'-phosphate. Biochemistry. 1983 Oct 25;22(22):5123–5134. doi: 10.1021/bi00291a011. [DOI] [PubMed] [Google Scholar]
- Kierzek R., He L., Turner D. H. Association of 2'-5' oligoribonucleotides. Nucleic Acids Res. 1992 Apr 11;20(7):1685–1690. doi: 10.1093/nar/20.7.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirshnan R., Seshadri T. P., Viswamitra M. A. Visualisation of a 2'-5' parallel stranded double helix at atomic resolution: crystal structure of cytidylyl-2',5'-adenosine. Nucleic Acids Res. 1991 Jan 25;19(2):379–384. doi: 10.1093/nar/19.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakenfull D. G., Richardson D. I., Jr, Usher D. A. Models of ribonuclease action. I. General species catalysis in the hydrolysis of a nucleotide diester analog. J Am Chem Soc. 1967 Oct 11;89(21):5491–5492. doi: 10.1021/ja00997a055. [DOI] [PubMed] [Google Scholar]
- Parthasarathy R., Malik M., Fridey S. M. X-ray structure of a dinucleoside monophosphate A2'p5'C that contains a 2'-5' link found in (2'-5')oligo(A)s induced by interferons: single-stranded helical conformation of 2'-5'-linked oligonucleotides. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7292–7296. doi: 10.1073/pnas.79.23.7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan A. R., Olson W. K. Conformational studies of (2'-5') polynucleotides: theoretical computations of energy, base morphology, helical structure, and duplex formation. Nucleic Acids Res. 1986 Jul 11;14(13):5461–5479. doi: 10.1093/nar/14.13.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J., Lohrmann R., Orgel L. E., Schneider-Bernloehr H., Weimann B. J., Miles H. T. Non-enzymic oligonucleotide synthesis on a polycytidylate template. J Mol Biol. 1969 Mar 14;40(2):227–234. doi: 10.1016/0022-2836(69)90471-9. [DOI] [PubMed] [Google Scholar]
- Tung C. H., Wei Z., Leibowitz M. J., Stein S. Design of peptide-acridine mimics of ribonuclease activity. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):7114–7118. doi: 10.1073/pnas.89.15.7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usher D. A., McHale A. H. Hydrolytic stability of helical RNA: a selective advantage for the natural 3',5'-bond. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1149–1153. doi: 10.1073/pnas.73.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]



