Abstract
Clear cell renal cell carcinoma (ccRCC) is a common human malignancy. Despite numerous efforts, there is still no reliable biomarker or combination of biomarkers available for daily practice. Our study was designed to explore the expression profile of messenger RNA (mRNA) and long non-coding RNA (lncRNA) transcripts in ccRCC in order to identify potential diagnostic biomarkers for patients with ccRCC. Total RNA from corresponding normal and malignant tissue of 15 patients with ccRCC was isolated. Expression profiling was performed using a custom Agilent gene expression microarray which allowed the analysis of 34,144 mRNA and 32,183 lncRNA transcripts. We observed that a subset of mRNA (n = 1064; 3.1%) and lncRNA (n = 1308; 4.1%) transcripts are dysregulated (fold change > 2) in ccRCC tissue. The relative higher number of differentially expressed lncRNAs indicates that lncRNA profiling may be better suited for diagnostic purposes; a number of so far unknown RNAs with potential diagnostic interest in ccRCC are identified by our gene expression profiling study. The data are deposited in the Gene Expression Omnibus (GSE61763).
Keywords: Biomarker, lncRNA, mRNA, Gene expression, Renal cell carcinoma
| Specifications | |
|---|---|
| Organism/cell line/tissue | Homo sapiens, normal and clear cell renal carcinoma tissue |
| Sex | Male and female |
| Sequencer or array type | Agilent SurePrint G3 Human Gene Expression Microarray v1.1, custom designed 60 k (AMADID #041648) |
| Data format | Normalized data: SOFT, MINiML, TXT |
| Experimental features | Corresponding normal and malignant fresh-frozen tissue samples from 15 clear cell renal cell carcinoma patients; analysis of lncRNA and mRNA expression profile |
| Consent | All patients provided written informed consent |
| Sample source location | University Hospital Bonn, Bonn, Germany |
1. Direct link to deposited data
2. Experimental design, materials and methods
2.1. Tissue samples
Tissue samples were prospectively collected from patients undergoing partial or radical nephrectomy for renal tumors at the Department of Urology at the University Hospital Bonn. Tumor and normal renal tissue samples were fresh-frozen and stored at − 80 °C in the Biobank at the Centrum for Integrated Oncology at the University Hospital Bonn. Histology of the tissue specimen was confirmed by a uropathologist using hematoxylin and eosin staining. All patients provided written informed consent prior to surgery. The study was approved by the local ethic committee (280/12).
2.2. RNA preparation
The RNA isolation procedure was described in detail before [1]. In brief, total RNA was isolated from approximately 50 mg frozen renal tissue using the mirVana miRNA Isolation Kit (Ambion, Foster City, CA, USA). Residual DNA fragments were removed by DNase-treatment (Ambion DNA-free Kit). The RNA quality and quantity (A260/A280 ratio close to 2, A260/A230 close to 2) was first determined with the Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Additionally, agarose gel electrophoresis was performed to exclude RNA degradation. Finally, the samples were analyzed at the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) using the RNA 6000 Nano Kit; samples with a RNA Integrity Number (RIN) > 6 were used for expression profiling experiments.
2.3. Gene expression profiling
The RNA samples were shipped on dry ice to Biogazelle (Gent, Belgium), where the gene expression microarray experiments were performed as a contract service. A custom microarray based on the Agilent SurePrint G3 technology was used to determine the expression of 34,144 mRNA and 32,183 lncRNA transcripts. The lncRNA content is based on LNCpedia 2.1 [2] and allowed profiling of 17,5125 lncRNA genes.
The workflow at Biogazelle was as follows: 100 ng total RNA was labeled using the low input Quick Amp labeling Kit (Agilent Technologies). The labeled RNA was then hybridized to the custom gene expression microarray (AMADID #041648). The microarrays were scanned using an Agilent G2565CA Microarray Scanner with the Scan Control software v8.5. The Probe intensities were extracted using the Agilent Feature Extraction software, and probe intensities were background corrected and normalized (quantile normalization) using the LIMMA package in R. All procedures were performed according to the manufacturer's instructions.
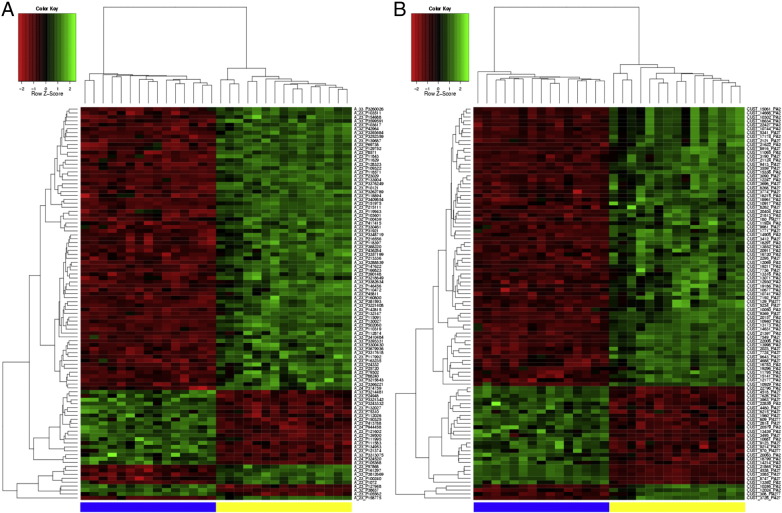
The background subtracted and normalized expression data were groupwise normalized deducting group mean values. Pairwise fold change, for each gene, was calculated by dividing ccRCC group mean of RNA expression by normal group mean RNA expression levels. Correlation analysis was performed using the R-Base Package v3.0.2 applying the Pearson's correlation coefficient. In order to identify clinically relevant dysregulated RNA transcripts, we considered differential expression as present if the fold-change was > 2, and the adjusted p-value (false discovery rate) was p < 0.05. A heatmap (see Fig. 1) was created using ArrayMining [3].
Fig. 1.
The heatmap is showing the 100 most differentially expressed mRNA (A) and lncRNA (B) transcripts in ccRCC and normal renal tissue. Expression levels are coded in colors and range from green (up-regulation) to red (down-regulation). The columns represent individual tissue samples; all ccRCC (blue) and normal (yellow) samples are located within a separate cluster. The rows represent individual genes.
3. Discussion
So far, there is no biomarker for ccRCC available for daily practice [4]. Our gene expression profile of 15 corresponding normal and malignant renal tissue samples from patients with ccRCC indicated dysregulation of 3.1% (368 upregulated and 696 downregulated transcripts) of mRNA and 4.1% (568 upregulated and 740 downregulated transcripts) of lncRNA transcripts in ccRCC tissue. As expected, the mRNA and lncRNA expression profiles allowed distinguishing normal and malignant tissue samples accurately based on the molecular signature. Interestingly, the expression changes seemed to be more pronounced within the subgroup of lncRNA than mRNA molecules, suggesting that lncRNA could be better suited as diagnostic biomarker. An earlier comparison of SAGE libraries of a broad spectrum of normal and tumor tissues also indicated a highly aberrant lncRNA expression profile in human cancers, as well as a tissue specific lncRNA expression profile [5]. Other researchers also reported lncRNA expression profiles in ccRCC tissue [6], [7], but the analyses were restricted to small cohorts thereby limiting powerful statistics. Nevertheless, it is obvious that unique lncRNA expression profiles exist in ccRCC, and the identification of novel differentially expressed lncRNA is an important step towards better understanding of ccRCC.
Conclusion
We have identified an expression profile of mRNA and lncRNA in ccRCC and normal renal tissue, and RNA expression levels allowed accurate identification of tumor tissue based on the molecular signature. Our study provides novel insights and thus a better understanding of ccRCC carcinogenesis.
Disclosures
The authors declare no conflicts of interest.
Acknowledgments
The tissue samples were collected within the framework of the Center for Integrated Oncology CIO Köln Bonn; the CIO is supported by the German Cancer Aid. This work was supported by a grant of the Rudolf Becker Foundation to Sven Perner.
References
- 1.Blondeau J.J., Deng M., Syring I. Identification of novel long non-coding rnas in clear cell renal cell carcinoma. Clin. Epigenetics. 2015;7:10. doi: 10.1186/s13148-015-0047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Volders P.J., Helsens K., Wang X. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 2013;41:D246–D251. doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glaab E., Garibaldi J.M., Krasnogor N. Arraymining: a modular web-application for microarray analysis combining ensemble and consensus methods with cross-study normalization. BMC Bioinformatics. 2009;10:358. doi: 10.1186/1471-2105-10-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellinger J., Muller S.C., Dietrich D. Epigenetic biomarkers in the blood of patients with urological malignancies. Expert. Rev. Mol. Diagn. 2015;15:505–516. doi: 10.1586/14737159.2015.1019477. [DOI] [PubMed] [Google Scholar]
- 5.Gibb E.A., Vucic E.A., Enfield K.S. Human cancer long non-coding RNA transcriptomes. PLoS One. 2011;6:e25915. doi: 10.1371/journal.pone.0025915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu G., Yao W., Wang J. Lncrnas expression signatures of renal clear cell carcinoma revealed by microarray. PLoS One. 2012;7:e42377. doi: 10.1371/journal.pone.0042377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin C., Han Z., Qian J. Expression pattern of long non-coding RNAs in renal cell carcinoma revealed by microarray. PLoS One. 2014;9:e99372. doi: 10.1371/journal.pone.0099372. [DOI] [PMC free article] [PubMed] [Google Scholar]