Abstract
To date, only a few conserved miRNAs have been predicted in hexaploid (AABBDD) bread wheat and till now community behavior among miRNA is still in dark. Analysis of publically available 1287279 ESTs from NCBI resulted 262 putative pre-miRNAs and 39 novel mature miRNAs. A total 22,468 targets were identified on 21 chromosomes. MiRNA target community was identified for genomes with different levels of cross talks. Gene ontology of these community targets suggests their differential involvement in different metabolisms along with common and stringent involvement in nitrogen metabolism.
Abbreviations: miRNAs, microRNAs; GRN, Gene Regulatory Network; FDR, false discovery rate
Keywords: MicroRNAs, EST, MiRNA community, Nitrogen metabolism
| Specifications | |
|---|---|
| Organism/cell line/tissue | Triticum aestivum |
| Sex | N/A |
| Sequencer or array type | N/A |
| Data format | Raw ESTs (Fasta files), CAP3 assembled contigs (Fasta files) |
| Experimental factors | Pooled ESTs from multiple experiments majorly belongs to biotic and abiotic stress |
| Experimental features | Novel miRNAs were identified from assembled ESTs and targets were identified on the draft genome sequence of wheat. |
| Consent | N/A |
| Sample source location | NCBI EST database |
1. Direct link to deposited data
2. Introduction
MicroRNAs (miRNAs) play important roles in plant growth regulations, development and adaptation to abiotic stresses [1], [2], [3]. It is an important class of small RNAs, a non-protein coding segment of genome, originates from fold back precursors and functions as negative regulators of gene expression in plant [1], [4], [5]. A large number of miRNAs have been discovered and functionally identified in plant kingdom. Currently, 8496 mature miRNAs have been identified from Viridiplantae and deposited in a publicly available database (miRbase, Release 21, June 2014) [6]. In agriculturally important monocot species, miRNAs have been reported in large number for fully sequenced genome but fewer in partially or incomplete sequenced genomes. Though, wheat (Triticum aestivum) is one of the most extensively cultivated crop among monocots but only 115 mature miRNAs have been reported [7]. Whereas in other monocots viz. Oryza sativa, Zea mays, Sorghum bicolor and Hordeum vulgare, 713, 321, 241 and 71 mature miRNAs have been reported respectively. The regulatory role of many of these miRNAs under drought, salinity and low temperature stress has been demonstrated [8], [9], [10]. Moreover, the homeostasis of the nutrients such as sulfur, copper and phosphate, which are critical for growth and development, has been found to be regulated by miRNAs [11], [12], [13]. A number of putative miRNAs in wheat have been identified [14], [15], [16], [17] yet the association of miRNA with different metabolic processes is still in dark. Therefore, identification of novel miRNAs as well as their association with metabolic pathways in wheat may facilitate its improvement and production.
The most challenging problem in understanding plant miRNAs is the identification of novel miRNAs. EST based computational analysis is a powerful technique for identifying conserved miRNAs, especially for plants with partially or un-sequenced genomes [18]. Thus in the present study, EST based computational technique was employed in search of potential miRNA in wheat. Available miRNAs in miRBase and downloaded wheat EST dataset were used for detection of potential miRNAs. Altogether 39 novel miRNAs were detected and 22,468 target genes were identified in wheat. Further, the mechanism of miRNA-target crosstalk was deciphered by employing a meta-analysis for chromosomal mapping, gene ontology, and miRNA-target community network. It was observed that most of these miRNAs in wheat were primarily involved in regulating nitrogen metabolism that needs to be further validate.
3. Materials and methods
3.1. MiRNA reference set
For prediction of potential miRNAs, previously known miRNAs (8496) of whole Viridiplantae were obtained from miRNA Registry database (Release 21, June 2014). To avoid the overlap of miRNAs, the redundant miRNA sequences were removed using an in-house Perl script and the remaining sequences were used as a reference set.
3.2. Software used
Initially, software BLAST-2.2.14 was used from NCBI GenBank. CAP3 was used for assembly in the form of contigs and singlets. Triplet SVM classifier was used to predict potential miRNA precursor [19]. These precursor sequences were used for BLASTx analysis for removing the protein-coding sequences and retained only non-protein encoding sequences. RNAfold was used online to analyze secondary structure of RNAs. BLASTn from NCBI (http://www.ncbi.nlm.nih.gov) was used to analyze the potential targets of miRNAs. psRNATarget web server (http://plantgrn.noble.org/psRNATarget/) was used for identification of the potential targets.
3.3. Prediction of miRNAs
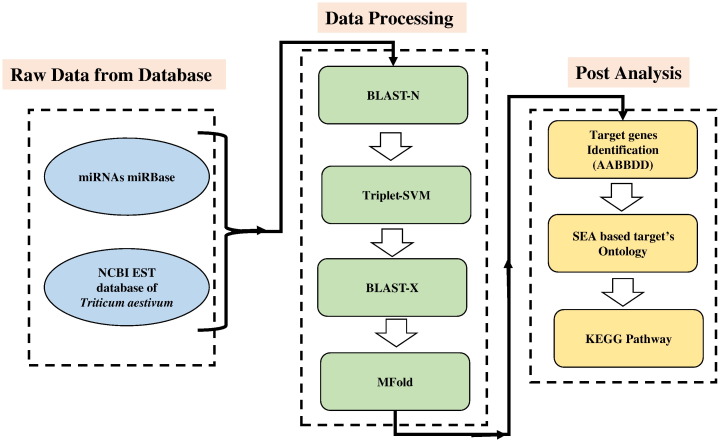
The overall workflow followed in the present study is represented as schematic diagram (Fig. 1). Firstly, 1,287,279 ESTs of T. aestivum from dbEST (http://www.ncbi.nlm.nih.gov/nucest/) and 8496 known mature miRNAs of Viridiplantae along with their precursor sequences from miRBase (http://www.mirbase.org/cgi-bin/browse.pl) were downloaded. All, the downloaded EST sequences were trimmed using EST trimmer. The trimmed EST sequences were then assembled using CAP3 results in contigs and singlets. Only unique miRNA was aligned pairwise by BLASTn program with a threshold (e-value at 10) and the word-match size between the query and the database was kept at 7 bp. The following two criteria were applied to select candidate miRNA from the homolog search: a) Candidate miRNAs must contain at least 18 ntd length with no gaps within alignment and b) maximum mismatch between known miRNA and assembled EST sequences should not be more than 3. These aligned ESTs were used as precursors with flanking regions of 50 nucleotides. Candidate hairpin forming precursors were filtered out by Triplet-SVM classifier. Filtered precursors were subjected to BLASTx and Rfam search [20] and non-protein coding precursors were finally obtained.
Fig. 1.
Workflow for the identification of miRNAs and their targets in Triticum aestivum (AABBDD).
3.4. Prediction of secondary structure
Potentially stable secondary structures from the precursors were generated by RNAfold algorithm. The parameters used for the secondary structure prediction using RNAfold were: minimum free energy and partition function; avoid isolated base pairs; dangling energy on both sides of the helix in any case; RNA parameters; rescale energy parameters at a given temperature 37 °C; interactive RNA secondary structure plot; RNA secondary structure plots with reliability annotation (partition function folding only) and mountain plot (Supplementary Table 1) [21]. Finally, these structures of precursors were manually curated using rules of Zhang et al. [22].
3.5. Prediction of mRNA targets of miRNAs
The draft genome sequences of T. aestivum for all 21 (3 ∗ 7) chromosomes were downloaded from URGI database (http://wheat-urgi.versailles.inra.fr/Seq-Repository). Chromosomes sequences were used for miRNA targets search using psRNATarget web server (http://plantgrn.noble.org/psRNATarget/) [23]. Parameters used were maximum expectation = 3.0, length for complementarity scoring (hspsize) = 20 bp, number of top target genes for each small RNA = 200, target accessibility-allowed maximum energy to unpair the target site (UPE) = 25, flanking length around target site for target accessibility analysis = 17 bp in upstream/13 bp in downstream and range of central mismatch leading to translational inhibition = 9–11 ntd.
3.6. Gene ontology analysis
To better understand the function of T. aestivum miRNAs, AgriGO tool (http://bioinfo.cau.edu.cn/agriGO/) [24] was employed to investigate the predicted target genes. For enrichment analysis, a hypergeometric distribution based statistical test (level of significance at 0.05%) was used to reject the chances of randomness in association to target genes with their corresponding ontology term. To moderate the false positives in multiple hypothesis testing procedure Benjamini and Hochberg false discovery rate (FDR) correction [25] was applied.
4. Results
4.1. Potential miRNAs in T.aestivum
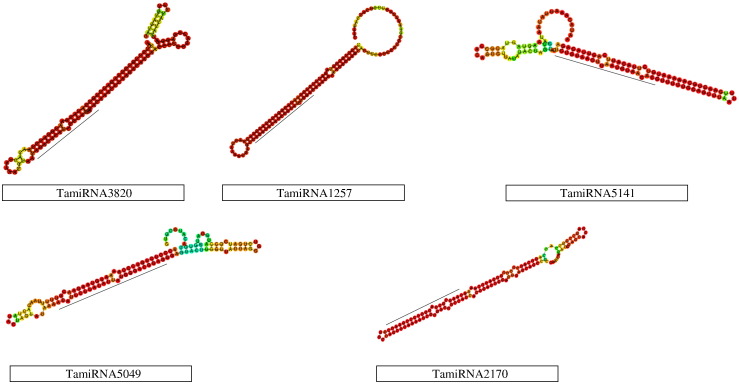
Overall schema of the present study is given in Fig. 1. Here, total 1,287,279 wheat ESTs were downloaded and pre-processed for poly-A/T tail removals and then assembled into 36,140 contigs and 414,041 singlets. These assembled sequences were used for homolog search against 3999 unique, Viridiplantae miRNAs (out of total 8496 downloaded miRNAs). 13,815 candidate precursors with the flanking region of 50 nucleotides were selected using an in-house Perl script. As per the standard rule, a precursor must form a hairpin structure [22] therefore all precursors were processed with Triplet-SVM [19] resulting in 1196 hairpin precursors. These precursors having similarity with non-redundant protein database were discarded. Stable secondary structure of the remaining 262 precursor sequences was obtained using RNAfold algorithm [26]. Following Zhang et al. [27], precursors were manually curated and 39 novel miRNAs were obtained (Supplementary file 1). Top five stable secondary structures (based on MFEI value) are given in Fig. 2 (Table 1 and Supplementary file 1).
Fig. 2.
Stable secondary structures of top five novel miRNAs of Triticum aestivum.
Table 1.
Newly identified top five miRNAs from ESTs of Triticum aestivum.
| S. no | Reference species | miRNA header | miRNA | EST/contig | Length mature miRNA | Length precursor | Strand | A + U (%) | MFE (kcal/mol) | MFEI | AMFEI |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hvu-miR1120 | > TamiRNA5062 | ACACUCUUAUAUUAUGGGACGGAG | CJ539616.1 | 24 | 124 | + | 60.48 | − 63.60 | − 1.30 | − 51.29 |
| 2 | bdi-miR5180a | > TamiRNA2170 | UAAGUGUCACGGUUUUGAACU | DY742248.1 | 21 | 121 | + | 53.72 | − 58.10 | − 1.04 | − 48.02 |
| 3 | hvu-miR5049c | > TamiRNA5374 | AGACACUUAUUUUGGGACGGAGG | Contig18580 | 23 | 123 | − | 60.98 | − 56.40 | − 1.18 | − 45.85 |
| 4 | bdi-miR1122 | > TamiRNA1257 | UAGAUACAUCCGUAUCUGGA | Contig17189 | 20 | 120 | − | 64.17 | − 51.20 | − 1.19 | − 42.67 |
| 5 | bdi-miR5181b | > TamiRNA2186 | UCCGAUCCAUAAUAAGUGUCG | CJ958667.1 | 21 | 121 | + | 56.20 | − 49.90 | − 0.94 | − 41.24 |
5. Result and discussion
5.1. Chromosomal distribution of targets
In the present study using a bioinformatics approach, 39 new miRNAs were identified from the EST libraries of T. aestivum and their genome wide (using draft genome sequence) target was searched using psRNATarget. None of the predicted miRNAs showed identity with the previously reported miRNAs in wheat and therefore these are addition into the existing wheat miRNA dataset. Further, we identified 22,468 non-redundant potential targets on 21 chromosome reported recently as draft genome sequence. The observed distribution of the miRNA targets is given in Table 2 Supplementary file 2.
Table 2.
Chromosomal distribution of the miRNA targets in on chromosomes of Triticum aestivum (AABBDD).
| Wheat chromosomes | miRNA target counts |
|---|---|
| Chr1 | 2783 |
| Chr2 | 3112 |
| Chr3 | 5947 |
| Chr4 | 2740 |
| Chr5 | 2808 |
| Chr6 | 3437 |
| Chr7 | 4753 |
5.2. MiRNA target community
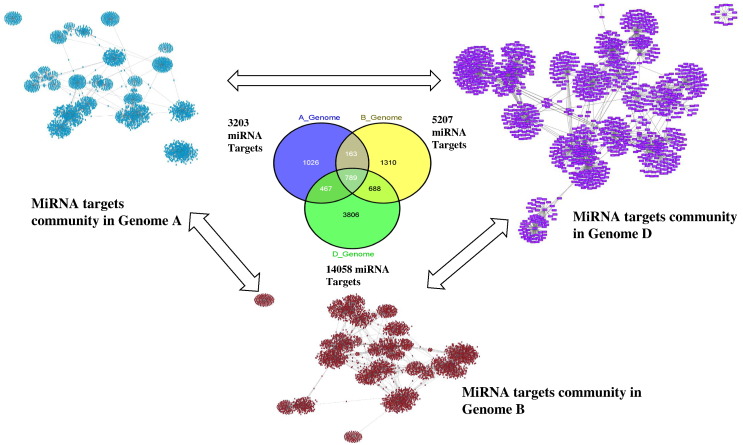
Gene Regulatory Network (GRN) in the present study explores the dimension of miRNA and their target multiplicity and cooperative in the sense of their complex genome organization and duplication event. The miRNA target gene identified for three genomes was used for the network generation using Cytoscape 3.0.2 [28]. Here we have used the UPE energy information in between the miRNA and their targets for network generation. There were three different networks formed as depicted in Fig. 3. The GRN represented in blue color is from A genome for 39 miRNAs regulating 3203 mRNA targets (Fig. 3a). Moreover, GRN represented in red color is from B genome that contributes 5207 mRNA targets, captured by 39 miRNAs (Fig. 3b), whereas from D genome, the largest number i.e., 14,058 of miRNA targets was identified for 39 miRNAs (Fig. 3c). This network graph of miRNA and their targets thus explore the actual contribution from all three genomes (AABBDD).
Fig. 3.
MiRNA target community within three genomes (AABBDD) of wheat revealing the within and in between crosstalks.
5.3. T.aestivum miRNA targets and their functions
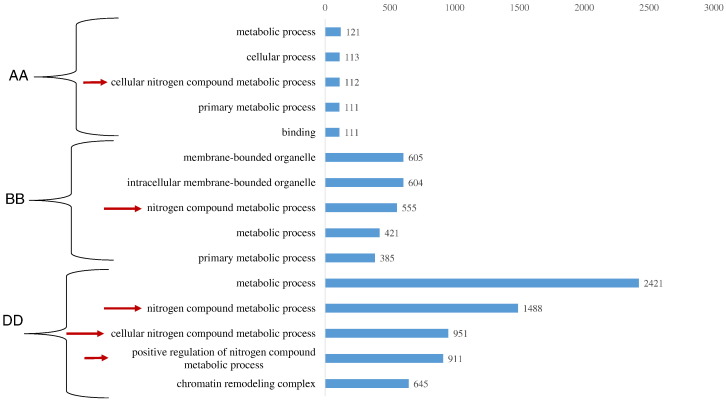
With the ongoing efforts in the field of wheat genomics it is logical to identify the contribution of three genomes governing polyploidy in wheat. Separately, genome contribution analysis for miRNA targeted shown that three genomes (AA), (BB) and (DD) contribute in different ratios. In result we got a total of 22,486 targets of 39 miRNAs in wheat genome. For the identification of biological process separately governed by three genomes (AABBDD), gene ontology was performed using agriGO tool with the parameter of p value of < 0.05 (Supplementary file 3). Due to the huge number of targets we performed a comparative analysis of gene ontology, where only top five activities were shown in Fig. 4 and Supplementary file 1. A genome suggests their involvement in different metabolic and cellular processes along with nitrogen metabolic process. Whereas, in case of B genome miRNA targets we found that membrane bound organelle and primary metabolic process, along with nitrogen metabolic process was found to be enriched. However with the case of D genome, similar type of activities were observed along with highly enriched process related to chromatin remodeling complex.
Fig. 4.
Genome wise enriched biological activities in Triticum aestivum (AABBDD) on the basis of SEA analysis. Only top five GO terms has been plotted here.
6. Conclusion
However miRNA-related research is one of the most common research topics in biological areas but miRNA research on wheat is still very far from the other plant species. Till now, only 115 known mature miRNAs have been deposited in mirBASE for T. aestivum. In the present study, we have tried to identify potential miRNAs in this highly economical plant species and we are able to identify 39 potential miRNAs along with 22,468 potential target genes in three genomes (AABBDD). Our analysis therefore added novel as well as potential miRNAs into the existed database. These miRNA were shown to regulate their targets with different ratios in all three genomes (AABBDD). Further they have found to exist as a “miRNA community” with different levels of crosstalks. Interestingly we found “nitrogen metabolism” as a common activity within miRNA community of three genomes suggesting their strong as well as common regulation by them.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgement
We acknowledge ICAR-Indian Agricultural Statistics Research Institute, PUSA, and New Delhi for providing all facility to conduct this study.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.gdata.2015.04.028.
Appendix A. Supplementary data
Supplementary file 1.
Supplementary file 2.
Supplementary file 3.
Supplementary Table 1.
References
- 1.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Chuck G., Candela H., Hake S. Big impacts by small RNAs in plant development. Curr. Opin. Plant Biol. 2009;12(1):81–86. doi: 10.1016/j.pbi.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Moldovan D., Spriggs A., Yang J., Pogson B.J., Dennis E.S., Wilson I.W. Hypoxia-responsive microRNAs and trans-acting small interfering RNAs in Arabidopsis. J. Exp. Bot. 2009:erp296. doi: 10.1093/jxb/erp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen E., Xie Z., Gustafson A.M., Carrington J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121(2):207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Carrington J.C., Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301(5631):336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 6.Griffiths-Jones S. MicroRNA Protocols. Springer; 2006. miRBase: the microRNA sequence database; pp. 129–138. [Google Scholar]
- 7.Sun F., Guo G., Du J., Guo W., Peng H., Ni Z., Sun Q., Yao Y. Whole-genome discovery of miRNAs and their targets in wheat (Triticum aestivum L.) BMC Plant Biol. 2014;14(1):142. doi: 10.1186/1471-2229-14-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sunkar R., Zhu J.-K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell Online. 2004;16(8):2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Y., Srivastava D. A developmental view of microRNA function. Trends Biochem. Sci. 2007;32(4):189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Arenas-Huertero C., Pérez B., Rabanal F., Blanco-Melo D., De la Rosa C., Estrada-Navarrete G., Sanchez F., Covarrubias A.A., Reyes J.L. Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant Mol. Biol. 2009;70(4):385–401. doi: 10.1007/s11103-009-9480-3. [DOI] [PubMed] [Google Scholar]
- 11.Yamasaki S., Ivanov P., Hu G.-f., Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009;185(1):35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones-Rhoades M.W., Bartel D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14(6):787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 13.Yang X.J., Finnegan P.M. Regulation of phosphate starvation responses in higher plants. Ann. Bot. 2010;105(4):513–526. doi: 10.1093/aob/mcq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao Y., Guo G., Ni Z., Sunkar R., Du J., Zhu J.-K., Sun Q. Cloning and characterization of microRNAs from wheat (Triticum aestivum L.) Genome Biol. 2007;8(6):R96. doi: 10.1186/gb-2007-8-6-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin W., Li N., Zhang B., Wu F., Li W., Guo A., Deng Z. Identification and verification of microRNA in wheat (Triticum aestivum) J. Plant Res. 2008;121(3):351–355. doi: 10.1007/s10265-007-0139-3. [DOI] [PubMed] [Google Scholar]
- 16.Yin Z., Shen F. Identification and characterization of conserved microRNAs and their target genes in wheat (Triticum aestivum) Genet. Mol. Res. 2010;9(2):1186–1196. doi: 10.4238/vol9-2gmr805. [DOI] [PubMed] [Google Scholar]
- 17.Li Y.-F., Zheng Y., Jagadeeswaran G., Sunkar R. Characterization of small RNAs and their target genes in wheat seedlings using sequencing-based approaches. Plant Sci. 2013;203:17–24. doi: 10.1016/j.plantsci.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 18.Zhang B., Pan X., Cannon C.H., Cobb G.P., Anderson T.A. Conservation and divergence of plant microRNA genes. Plant J. 2006;46(2):243–259. doi: 10.1111/j.1365-313X.2006.02697.x. [DOI] [PubMed] [Google Scholar]
- 19.Xue C., Li F., He T., Liu G.-P., Li Y., Zhang X. Classification of real and pseudo microRNA precursors using local structure-sequence features and support vector machine. BMC Bioinforma. 2005;6(1):310. doi: 10.1186/1471-2105-6-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffiths-Jones S., Moxon S., Marshall M., Khanna A., Eddy S.R., Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33(Suppl. 1):D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turner D.H., Mathews D.H. NNDB: the nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp892. gkp892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang B.H., Xiao Ping P., Qing Lian W., Cobb G.P., Anderson T.A. Identification and characterization of new plant microRNAs using EST analysis. Cell Res. 2005;15(5):336–360. doi: 10.1038/sj.cr.7290302. [DOI] [PubMed] [Google Scholar]
- 23.Dai X. Zhao PX: psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011;39(Suppl. 2):W155–W159. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du Z., Zhou X., Ling Y., Zhang Z., Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq310. (gkq310) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995:289–300. [Google Scholar]
- 26.Lorenz R., Bernhart S.H., Zu Siederdissen C.H., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA package 2.0. Algorithms Meol. Biol. 2011;6(1):26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang B.H., Pan X.P., Wang Q.L., George P.C., Anderson T.A. Identification and characterization of new plant microRNAs using EST analysis. Cell Res. 2005;15(5):336–360. doi: 10.1038/sj.cr.7290302. [DOI] [PubMed] [Google Scholar]
- 28.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file 1.
Supplementary file 2.
Supplementary file 3.
Supplementary Table 1.