Abstract
Aging, also called senescence, is thought to be a physiological phenomenon that commonly occurs in various organs and tissues (Enoki et al., 2007 [1]). Many older adults experience dysfunction in their salivary glands, for example xerostomia, which is defined as dry mouth resulting from reduced or absent saliva flow (Nagler et al., 2004 [2]). In the present study, we investigated gene expression in submandibular glands of young (8 weeks old) and adult (50 weeks old) mice to analyze association of aging with gene expression profiling in mouse submandibular glands. Whole-genome gene expression profiles were analyzed using an Illumina Sentrix system with Mouse-WG-6 v.2 Expression BeadChips (Illumina). Of the genes screened, 284 showed detection values at a significance level of P < 0.01. Among those, the expression of 94 genes (33%) showed a greater decrease in adult mice as compared to young mice. On the other hand, that of 190 genes (77%) was increased in the adults more than in young mice. The data obtained in this study are publicly available in the Gene Expression Omnibus (GEO) database (accession number GSE66857).
Keywords: Submandibular gland, Aging, Microarray
| Specifications | |
|---|---|
| Organism/cell line/tissue | Mus musculus/submandibular glands of young (8 weeks old) and adult (50 weeks old) mice |
| Sex | Male |
| Sequencer or array type | Illumina Mouse-WG-6 v.2 expression bead chip |
| Data format | Raw and analyzed |
| Experimental factors | Expression patterns in submandibular glands of young (8 weeks old) and adult (50 weeks old) mice were analyzed to determine aging-dependent gene expression. |
| Experimental features | Microarray analysis of gene expression associated with aging in mouse submandibular glands. |
| Consent | N/A |
| Sample source location | 1-5-8 Hatanodai, Shinagawa, Tokyo 142-8555, Japan |
1. Direct link to deposited data
2. Experimental design, materials, and methods
2.1. Introduction
The submandibular glands (SMGs) participate as major salivary secreting organs to secrete fluids rich in proteins that are critical for maintenance of oral health [1], [2], [3]. The SMGs have been reported to increase in proportional volume of fat and connective tissues with a reduction in that of acini with aging, though without any remarkable change in the volume of the duct system [4]. For investigation of age-dependent changes in the expression of genes in SMGs, a gene expression array can provide a comprehensive view of the expression pattern. The Illumina Sentrix system using Mouse-WG-6 v.2 Expression BeadChips (Illumina) reflects the latest advancements in mouse genomics and provides biologically relevant information for gene expression studies, while GenomeStudio Data Analysis Software is useful for visualizing and analyzing data obtained with the Illumina array platform. In the present study, we used cDNA microarray analysis to detect age-associated changes in gene expression of mouse SMGs.
2.2. Animal treatment
All animal experiments were conducted in accordance with the guidelines of Showa University. C57BL/6J mice were obtained from Sankyo Laboratory and housed in the Animal Facility at Showa University. We used 8- and 50-week-old mice as the young and adult, respectively, groups.
2.3. Tissue preparation
For general histopathological examinations, all samples were fixed in 4% paraformaldehyde and processed into frozen sections using routine procedures, then stained with hematoxylin–eosin (H–E).
2.4. Whole-genome expression assay and microarray data analysis
Whole-genome gene expression profiles in the SMGs of each mouse were analyzed using an Illumina Sentrix system with Mouse-WG-6 v.2 Expression BeadChips (Illumina), which includes 45,281 Illumina probes to detect transcriptants covering 30,854 genes, using a previously reported method [5]. First, total RNA was extracted with TRIzol reagent (Life Technologies) from whole SMGs, then 500 ng was subjected to RNA amplification, which was performed with an Illumina TotalPrep RNA Amplification Kit (Ambion), according to the manufacturer's instructions. Biotinylated cRNA was then hybridized to Mouse-WG-6 v.2 Expression BeadChips and reacted with streptavidin-cy3 (GE Healthcare). Finally, the expression intensity of the transcripts on the BeadChips was detected using an Illumina iScan reader. Raw BeadChip image data were subjected to expression analyses using the manufacturer's software (GenomeStudio v.2011.1, Gene Expression Module v.1.9.0). A heat map was generated by hierarchical clustering of the selected transcripts based on the gene expression profiles (expression ratio: AVG_signal of adult mice/AVG_signal of young mice).
2.5. Quantitative real-time PCR
Total RNA was extracted using TRIzol reagent (Life Technologies), then reverse transcribed using ReverTra Ace® qPCR RT Master Mix (TOYOBO). Quantitative real-time PCR was performed using a SYBR green Fast PCR system (GE Healthcare), with the following primer sequences shown in Table 1.
Table 1.
Sequences of primers used for quantitative PCR.
| Gene | Primer | Sequence |
|---|---|---|
| Pdcd4 | Forward | 5′-GGATGAGACCGCATTTGAGAA-3′ |
| Reverse | 5′-AGGCTAAGGACACTGCCAACAC-3′ | |
| Ttr | Forward | 5′-GGTCAAAGTCCTGGATGCTGTC-3′ |
| Reverse | 5′-CCAGTACGATTTGGTGTCCAGTTC-3′ | |
| Pdk4 | Forward | 5′-CAGGTTATGGGACAGACGCTATCA-3′ |
| Reverse | 5′-TGCTTGGGATACACCAGTCATCA-3′ | |
| Ly6d | Forward | 5′-CCAGCAGGGCCATGTCA-3′ |
| Reverse | 5′-AGGTCAGTCTGGCAGCATTGT-3′ | |
| Kik1 | Forward | 5′-TGACAGATGACATGTTGTGTGCAG-3′ |
| Reverse | 5′-GATACCCGGCACATTGGGTTTA-3′ | |
| Creld2 | Forward | 5′-GCGATGGCCAGTACTGTGAGAA-3′ |
| Reverse | 5′-CTGTACAGCCCACGCAGGTAGA-3′ | |
| Igfbp2 | Forward | 5′-GGCCGGTACAACCTTAAGCA-3′ |
| Reverse | 5′-GGGTTCACACACCAGCACTC-3′ | |
| Sdf2l1 | Forward | 5′-GCTGCACTCACACGACATCAA-3′ |
| Reverse | 5′-CGCGAATCCGCCAGTAACTA-3′ | |
| Tgm2 | Forward | 5′-CAACCTGACCCTGGATCCCTA-3′ |
| Reverse | 5′-TCAGGCACCCGCTGTACTTC-3′ |
3. Results and discussion
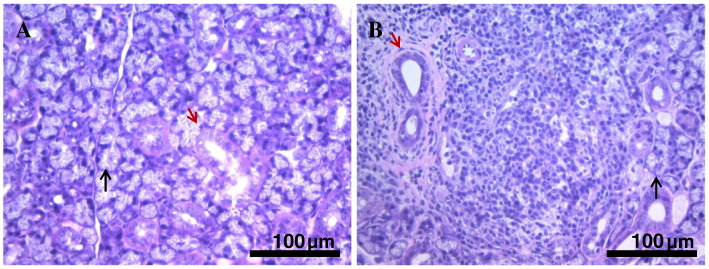
SMGs in both the young and adult mice consisted of acini, a duct system, and interstitial connective tissues. The nuclei of the acinar cells were restricted to the basal side, where cytoplasm filled clear granules in both phenotypes. In the young mice, duct cells as well as intercalated, granular, striated, and excretory cells were clearly identified in the duct system of the SMGs (Fig. 1A). On the other hand, histological findings of those from adult mice showed mainly periductal inflammation of lymphocytes in the SMGs and inflammatory cell infiltration leading to destruction of acini (Fig. 1B).
Fig. 1.
Representative sections from submandibular glands of young and adult mice (H & E, scale bars: 100 μm). Black arrows, acinar cell; red arrows, duct.
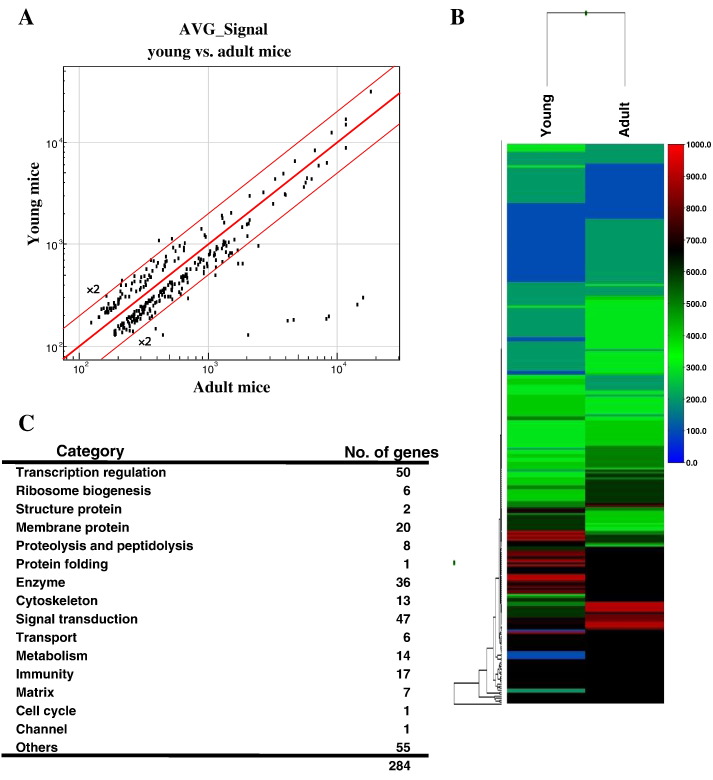
To elucidate the molecular mechanisms of changes associated with age in the mice, we performed whole-genome expression assays using the Illumina Sentrix platform with Mouse WG-6 v.2 Expression BeadChips. Obtained raw signal intensity data were analyzed using GenomeStudio Software. Following baseline subtraction and normalization, among the 45,281 transcripts of all detected genes, 284 showed an expression detection significance of P < 0.01. The expression levels of those 284 transcripts differed between the young and adult mice. Scatterplots for both groups displayed a symmetrical AVG_Signal distribution around linear identity lines to form a 45° angle. The expression levels of 3 genes in the adult mice showed a greater than two-fold reduction, while those of 12 genes showed a greater than two-fold increase (Fig. 2A). Of the 284 transcripts, 94 exhibited lower and 190 exhibited higher expression in adults as compared to the young mice (Fig. 2B). The categories of the altered genes are shown in Fig. 2C. Of the 284 genes, 50 (17.6%) encode transcription regulation, 47 (16.5%) encode signal transduction, 36 (12.7%) encode enzymes, and 17 (6%) encode immunity. Table 2 shows the 30 genes with lower (< 0.69) and 32 genes with higher (> 1.63) fold changes in expression level in the adults as compared to the young mice. To verify the microarray data, 9 genes (red in Table 2) were randomly selected and quantitative PCR was performed (Fig. 3), with the results consistent with the microarray findings.
Fig. 2.
Genome wide gene expression profiling of submandibular glands from C57BL/6J mice at 8 (young) and 50 (adult) weeks old. Biotinylated cRNA was synthesized from total RNA and hybridized to Illumina MouseWG-6 v.2 Expression BeadChips. Raw expression intensity data were analyzed using GenomeStudio Software. (A) Scatterplots of the AVE_Signal for young and adult mice were generated. (B) Heat map illustration of gene expression profiles in young and adult mice. Hierarchical clustering was based on relative gene expression levels (AVG_Signal/AVG_Signal of C57BL/6J mouse). (C) Selected 284 transcript expression changes among young and adult mice based on whole-genome expression analysis.
Table 2.
Highly differentially expressed gene profiles in submandibular glands from young and adult mice. Genes noted by redfont were subjected to quantitative real-time PCR analysis.
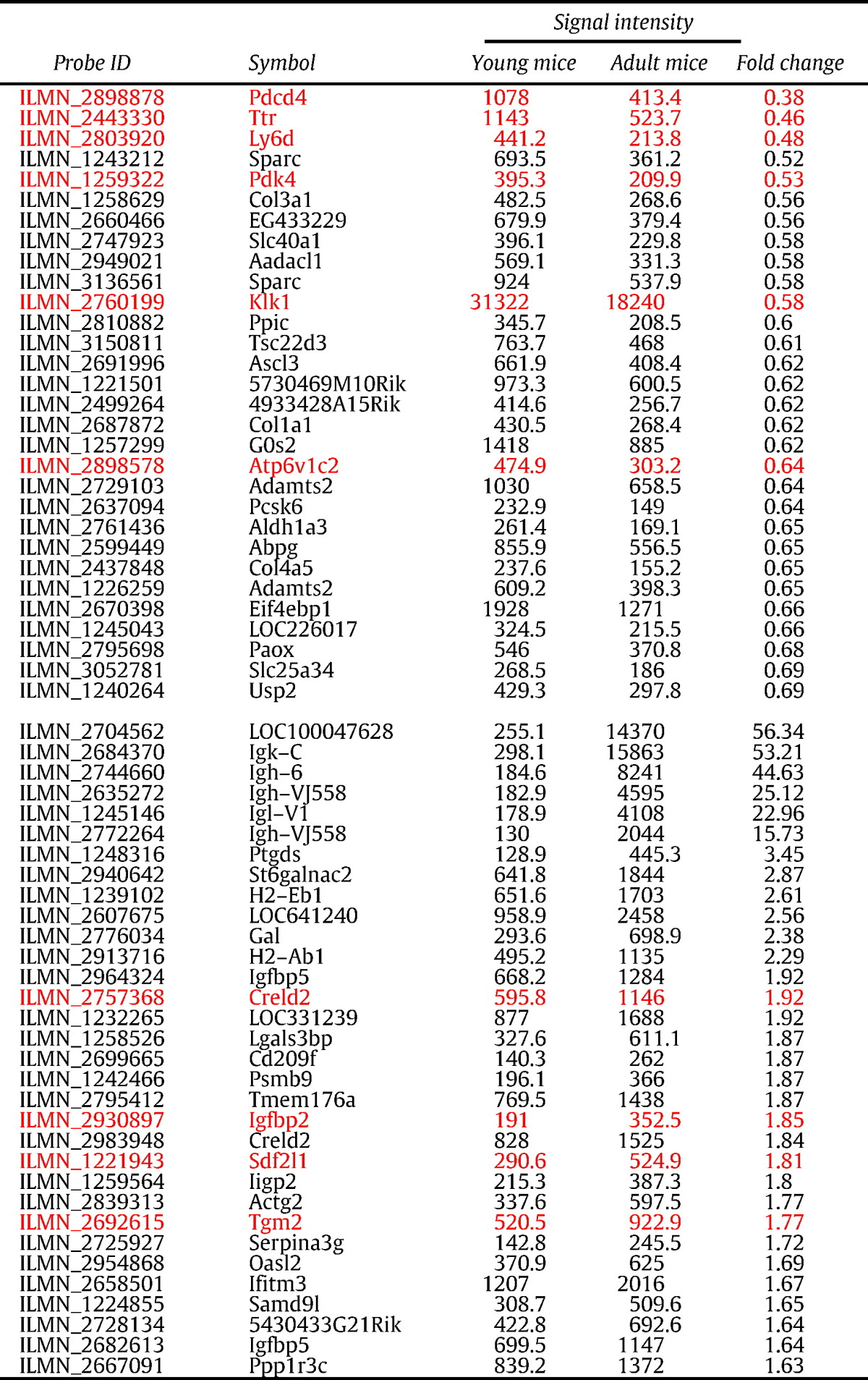
Fig. 3.
Quantitative PCR analysis. Results are shown as the mean ± SEM of 6 samples. *P < 0.05, **P < 0.0; Student's t test.
Among the 9 picked-up genes, Pdcd4 (Programmed cell death 4) is characterized as a potent tumor suppressor and known to inhibit the function of transcription factors, such as AP-1 transactivation [6], [7], [8]. Furthermore, Hayashi et al. demonstrated that miR-21, whose target gene is Pdcd4 and regulates branching morphogenesis in the submandibular glands [8]. In the present study, Pdcd4 was decreased in the SMGs of adult mice. Additional experiments are required to fully elucidate the mechanism involved in that decrease.
Conflict of interest
The authors declare that there are no potential conflicts of interest with respect to the authorship and/or publication of this article.
Acknowledgments
This work was supported in part by the Grants-in-Aid for scientific research from the Japan Society for the Promotion of Science (24592813 to AY).
References
- 1.Enoki N., Kiyoshima T., Sakai T., Kobayashi I., Takahashi K., Terada Y., Sakai H. Age-dependent changes in cell proliferation and cell death in the periodontal tissue and the submandibular gland in mice: a comparison with other tissues and organs. J. Mol. Histol. 2007;38:321–332. doi: 10.1007/s10735-007-9105-6. [DOI] [PubMed] [Google Scholar]
- 2.Nagler R.M. Salivary glands and the aging process: mechanistic aspects, health-status and medicinal-efficacy monitoring. Biogerontology. 2004;5:223–233. doi: 10.1023/B:BGEN.0000038023.36727.50. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen C.Q., Sharma A., Lee B.H., She J.X., McIndoe R.A., Peck A.B. Differential gene expression in the salivary gland during development and onset of xerostomia in Sjogren's syndrome-like disease of the C57BL/6.NOD-Aec1Aec2 mouse. Arthritis Res. Ther. 2009;11:R56. doi: 10.1186/ar2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki H., Amizuka N., Noda M., Amano O., Maeda T. Histological and immunohistochemical changes in the submandibular gland in klotho-deficient mice. Arch. Histol. Cytol. 2006;69:119–128. doi: 10.1679/aohc.69.119. [DOI] [PubMed] [Google Scholar]
- 5.Kasai S., Ikeda K. Reduced supraspinal nociceptive responses and distinct gene expression profile in CXBH recombinant inbred mice. J. Pain. 2013;14:648–661. doi: 10.1016/j.jpain.2013.01.773. [DOI] [PubMed] [Google Scholar]
- 6.Krichevsky A.M., Gabriely G. miR-21: a small multi-faceted RNA. J. Cell. Mol. Med. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang H.S., Jansen A.P., Nair R., Shibahara K., Verma A.K., Cmarik J.L., Colburn N.H. A novel transformation suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-kappaB or ODC transactivation. Oncogene. 2001;20:669–676. doi: 10.1038/sj.onc.1204137. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi T., Koyama N., Azuma Y., Kashimata M. Mesenchymal miR-21 regulates branching morphogenesis in murine submandibular gland in vitro. Dev. Biol. 2011;352:299–307. doi: 10.1016/j.ydbio.2011.01.030. [DOI] [PubMed] [Google Scholar]