Abstract
The transcription factor p63 is a key regulator in epidermal keratinocyte proliferation and differentiation. However, the role of p63 in gene regulation during these processes is not well understood. To investigate this, we recently generated genome-wide profiles of gene expression, p63 binding sites and active regulatory regions with the H3K27ac histone mark (Kouwenhoven et al., 2015). We showed that only a subset of p63 binding sites are active in keratinocytes, and that differentiation-associated gene expression dynamics correlate with the activity of p63 binding sites rather than with their occurrence per se. Here we describe in detail the generation and processing of the ChIP-seq and RNA-seq datasets used in this study. These data sets are deposited in the Gene Expression Omnibus (GEO) repository under the accession number GSE59827.
Keywords: ChIP-seq, RNA-seq, p63, Epidermal differentiation, Gene regulation
| Specifications | |
|---|---|
| Organism/cell line/tissue | Homo sapiens |
| Sex | Female |
| Sequencer or array type | Illumina HiSeq 2000 |
| Data format | Raw ChIP-seq and RNA-seq: FASTQ files Processed RNA-seq: FPKM gene expression table Processed ChIP-seq: wiggle files, peaks files, RNAPII gene body RPKM table |
| Experimental factors | Cultured primary human epidermal keratinocytes derived from abdominal skin of healthy volunteers |
| Experimental features | ChIP-seq and RNA-seq in differentiating epidermal keratinocytes at days 0, 2, 4 and 7 to identify genes regulated by p63 |
| Consent | The healthy donors gave their informed consent |
| Sample source location | Nijmegen, The Netherlands |
1. Direct link to deposited data
2. Experimental design, materials and methods
2.1. Experimental design
In this study we elucidated the role of the transcription factor p63 in the regulation of the differentiation process of keratinocytes [1]. We differentiated primary human epidermal keratinocytes in two-dimensional culture, and collected cells at different time points after the initiation of differentiation. This process mimics the in vivo keratinocyte differentiation process [2]. We collected cells from two different keratinocyte cell lines at four time points: day 0 (proliferating keratinocytes), day 2 (early differentiation), day 4 (mid-differentiation) and day 7 (late differentiation).
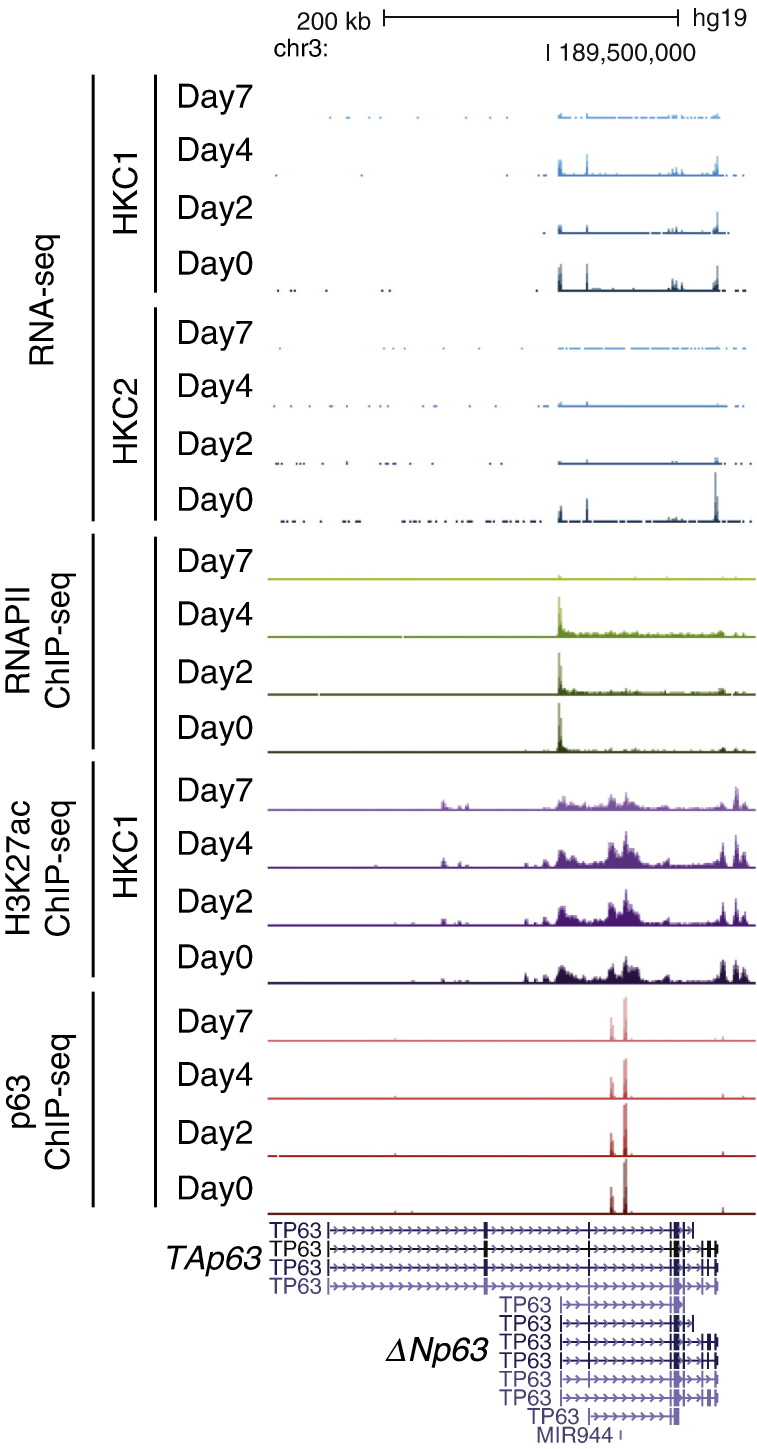
To investigate the role of p63 in this process, we performed chromatin immunoprecipitation of p63 followed by deep sequencing (ChIP-seq) analysis, in order to identify the genome-wide binding sites of p63 at each stage of differentiation. We similarly performed ChIP-seq of the H3K27ac histone modification, which marks active regulatory elements [3]. To measure gene activity we used two approaches: ChIP-seq of RNA polymerase II (RNAPII) in order to identify genes undergoing active transcription, and RNA sequencing (RNA-seq) in order to measure steady state RNA levels. These two approaches showed good genome-wide correlation of gene transcription and expression [1], but differed in specific cases. In particular, transcription of non-coding RNAs was more readily detectable by RNAPII ChIP-seq than by RNA-seq [1]. This combination of datasets allowed us to survey the genome-wide expression dynamics during keratinocyte differentiation, as well as the genome-wide changes in regulatory element activity, and to identify different sets of genes whose expression dynamics correlate with the activity of nearby p63-associated regulatory elements (Fig. 1).
Fig. 1.
Screenshot from genome browser (UCSC) to visualize RNA-seq, and ChIP-seq signal at the TP63 gene locus during keratinocyte differentiation.
2.2. Human primary keratinocyte culture and differentiation induction
Epidermal keratinocytes were obtained from skin biopsies taken from the abdomen of two healthy volunteers, labeled HKC1 and HKC2, and cultured in Keratinocyte Growth Medium (KGM). Differentiation was induced by cell contact inhibition and excluding several growth factor supplements from the medium: bovine pituitary extract (Bio Whittaker), EGF (Sigma), insulin (Sigma), and hydrocortisone (Calbiochem). The medium was changed every second day, and before harvesting of the RNA and chromatin. For both cell lines, cells were collected on day 0 before differentiation induction, and on days 2, 4 and 7 after induction of differentiation.
2.3. Chromatin immunoprecipitation
ChIP-seq data were only generated for cell line HKC1. Chromatin immunoprecipitation (ChIP) for p63 was performed using 2 μg of antibody per reaction that recognizes the alpha isoforms of p63 (H129, Santa Cruz). RNAPII ChIP was performed using 1.5 μg of RNAPII antibody (8WG16, Santa Cruz) per reaction and the antibody detects all forms of the large subunit of RNA polymerase II. H3K27ac ChIP was performed using 2 μg of the antibody (H3K27ac, C15410174, pAb-174–050, Diagenode) per reaction and the antibody recognizes acetylated lysine 27 residue on the H3 histone protein. RNAPII, p63, and H3K27ac ChIP experiments were performed as previously described [4], with a minor change of using magnetic beads (Novex by Life Technologies, 10008D and 10009D).
2.4. RNA extraction
RNA-seq data were generated for both keratinocyte cell lines, in order to facilitate differential expression calculations by providing replicates. RNA extraction was carried out using the NucleoSpin RNA II kit (740,955.50; MACHEREY-NAGEL, Düren, Germany). As a control for the RNA-seq data cDNA synthesis was carried out from 1 μg total RNA using the iScript cDNA synthesis kit (Bio-Rad; 170-8891, CA, Hercules, USA). For RNA-seq, a starting amount of 1 μg RNA per sample was used, and rRNA was depleted using the Ribo-Zero rRNA Removal Kit (Human/Mouse/Rat; MRZH11124, Epicenter, Madison, WI, USA) according to the manufacturer's instructions. The RNA fragmentation reactions were performed using fragmentation buffer (5 ×; 200 mM Tris-Ac, 500 mM Potassium-Ac, 150 mM Magnesium-Ac) in a final concentration of 1 × per reaction. Each 50 μl fragmentation reaction was incubated at 95 °C for 1.5 min on a thermal cycler, and placed on ice for 10 min. Ethanol precipitation was used to purify the reactions. The fragmented, rRNA depleted RNA was combined with 5 μg of random hexamers (Roche) in a final volume of 11 μl, incubated at 65 °C for 5 min, then immediately placed on ice. The remaining reagents were then added to the reaction: 4 μl 5 × First strand buffer (Invitrogen), 2 μl DTT (0.1 M Invitrogen), 1 μl dNTP mix (10 mM Invitrogen), 2 μl Actinomycin D (0.1 μg/μl Sigma), 1 μl of RNase H (40 U/ml Ambion), and 1 μl SuperScript III (200 U Invitrogen). The first strand reaction was incubated at 25 °C for 10 min, and 50 °C for 90 min, followed by deactivation at 70 °C for 15 min, after which the MinElute Reaction Cleanup Kit (Qiagen) was used according to the manufacturer's protocol. The second strand was synthesized by adding to the purified sample (100 μl total volume): 20 μl 5 × Second strand buffer (Invitrogen), 4 μl 5 × First strand buffer (Invitrogen), 2 μl DTT (0.1 M Invitrogen), 1 μl random hexamers (5 mg/ml Roche), dUTP mix (12.5 mM Invitrogen), 1 μl RNase H (8 U/ml Ambion), 1 μl Escherichia coli DNA polymerase I (10 U/μl Invitrogen), and 1 μl E. coli DNA ligase (10 U/μl NEB). After 2 h at 16 °C, 1 μl of T4 DNA polymerase (10 U/μl Promega) was added and the reaction was incubated at 16 °C for 10 min. The ds-cDNA was purified using the MinElute Reaction Cleanup Kit (Qiagen), according to the manufacturer's protocol.
2.5. Illumina library preparation and sequencing
DNA samples were prepared for sequencing starting with 6 ng total DNA for the p63 and RNAPII ChIP-seq samples, 10 ng total DNA for the H3K27ac ChIP-seq samples, and 5 ng total cDNA for the RNA-seq samples, as measured by Qubit (Invitrogen). Subsequently, the DNA fragments were prepared for adaptor ligation by adding an adenosine base to the 3′-end of the DNA using 5 μl of Klenow buffer, 10 μl of dATP (1 mM), and 1 μl Klenow exo (3′–5'exo) (5 U) (NEB, M0210S). The reaction was incubated for 30 min at 37 °C, purified using the MinElute Reaction Cleanup Kit (Qiagen), and the DNA was eluted in 10 μl of EB. The adaptors were ligated to the DNA fragments by addition of 15 μl 2 × T4 DNA Ligase buffer, 1 μl of NEXTflex adaptor (for appropriate adaptor dilutions see the Bio Scientific ChIP-seq barcodes protocol; 514,120), and 4 μl T4 DNA ligase (3 U) (NEB, M0203S). The reaction was incubated for 15 min at 21 °C, followed by purification using the MinElute Reaction Cleanup Kit (Qiagen), and eluted in 10 μl of EB. Preceding the Pre-PCR step, the RNA-seq samples were treated with the USER enzyme by adding 3 μl of USER enzyme and 10 μl 5 × Phusion buffer to the DNA sample in a total volume of 48 μl. This reaction was incubated at 37 °C for 15 min, 95 °C for 10 min, on ice for 5 min. This was followed by a pre-PCR reaction by adding 1.5 μl of dNTPs (10 mM), 2 μl NEXTflex primers, and 0.5 μl Phusion polymerase (2 U/ml) (Finnzymes, F-530 L) to a total volume of 52 μl. The pre-PCR reaction for the end repaired and adaptor ligated ChIP-samples was assembled in a total volume of 50 μl by adding 10 μl 5 × Phusion buffer, 1.5 μl dNTPs (10 mM), 2 μl NEXTflex primers, and 0.5 μl Phusion polymerase. Pre-PCR was performed using the following conditions: 20 s at 98 °C, (10 s at 98 °C, 30 s at 65 °C, 30 s at 72 °C) for 4 cycles, 5 min at 72 °C, and kept at 4 °C. The DNA was purified using the MinElute Reaction Cleanup Kit (Qiagen), eluted in 10 μl of EB and size selected using an E-gel system (Invitrogen catnr. G661002) according to the manufacturer's protocol. In the size range of 300–310 bp approximately 23 μl of DNA was collected (DNA fragments + adaptors) that was PCR amplified using 10 μl 5 × Phusion buffer, 1.5 μl dNTPs (10 mM), 2 μl NEXTflex primers, 0.5 μl Phusion polymerase (2 U/ml) mixture in a total volume of 50 μl with the following PCR conditions: 20 s at 98 °C, (10 s at 98 °C, 30 s at 65 °C, 30 s at 72 °C) for 11 cycles, 5 min at 72 °C, and kept at 4 °C. The PCR product was purified using AMPure XP beads (Beckman Coulter, A63881) according to the manufacturer's protocol and eluted in 20 μl of EB. DNA size and integrity were confirmed on a Bio-Rad Experion apparatus. Cluster generation and single end sequencing of 50 cycles was performed with the Illumina HiSeq 2000 sequencer according to standard Illumina protocols. Samples were barcoded using NEXTflex ChIP-seq 6 barcodes, and multiple samples were pooled and sequenced together in the same lane. After sequencing reads were demultiplexed based on their barcodes using standard Illumina bcl2fastq software and the resulting 42 bp reads were used for all further downstream processing. These raw read datasets are available for all ChIP-seq and RNA-seq samples under GEO accession number GSE59827.
2.6. ChIP-seq data processing
The 42 bp single end reads were uniquely mapped to the human genome NCBI build 37 (hg19) using bwa 0.6.1 with standard parameters [5]. All p63, H3K27ac and RNAPII ChIP-seq datasets were normalized to the same sequencing depth by randomly removing aligned reads. This normalization was performed separately for all datasets associated with each target protein. Duplicated reads were removed before normalization. Wiggle tracks of these normalized samples were generated for visualization purposes in the UCSC genome browser using our in-house “sum_bed.pl.” Perl script (Supplementary materials). The MACS2 program was used for peak recognition for the p63 and RNAPII ChIP-seq datasets. This is an updated version of the MACS program [6] that is specifically designed to process mixed signal types (URL https://github.com/taoliu/MACS). It was run with default settings and a P value threshold of 1E − 9, using input genomic DNA as a background control. Peak recognition for the H3K27ac ChIP-seq dataset was performed using MACS2 with default settings for broad peak calling and a P value threshold of 1E − 9, using input genomic DNA as a background control. Active p63 binding sites were identified by overlapping the p63 peaks with the H3K27ac peaks using the intersect tool from the BEDTools suite [7]. Gene transcription rates were quantified as RNAPII ChIP-seq reads per kilobase per million mapped reads (RPKM) falling within gene bodies. Genes were taken from the RefSeq transcriptome [8], and their genomic locations were retrieved from the UCSC genome browser [9]. For downstream analyses the longest genomic transcript variant was used per gene. For more detailed microRNA analysis, the occupancy of RNAPII ChIP-seq datasets for transcribed pri-miRNAs was quantified by RPKM calculation of the detected sequenced reads for all miRNA genes that were found to co-localize with the detected RNAPII MACS2 peaks using the intersect tool from the BEDTools suite [7]. A cutoff of RPKM > 1 was set, and only miRNAs, smaller than 200 bp, intersecting these RNAPII peak regions were considered. All wiggle tracks, the p63 and H3K27ac MACS2 peak files, and the RefSeq gene body RNAPII RPKM table for all days are available under GEO accession number GSE59827.
2.7. RNA-seq data processing
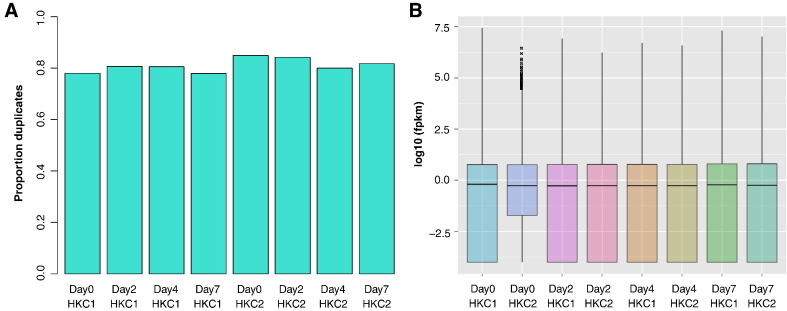
The 42 bp single end reads were mapped to the human genome NCBI build 37 (hg19) using gsnap version 2013-03-31 [10], mapping the spliced reads against the Ensembl 68 reference transcriptome [11]. Resulting mapped reads were stored in BAM files. Duplicate rates were relatively high at around 80% (Fig. 2A), which is common in RNA-seq data where highly expressed genes can affect duplication rates. Additionally, we used relatively short single-end reads and a SNP-tolerant read mapper. Wiggle tracks of these samples were generated for visualization in the UCSC genome browser using our in-house “sum_bed.pl.” Perl script (Supplementary materials). To make the signal heights comparable in the browser view, these wiggle tracks were generated from normalized BAM files in which excess reads were randomly discarded from all but the smallest BAM file using the Picard Tools DownsampleSam program (http://broadinstitute.github.io/picard/), in order to use a similar number of reads from all samples. Transcript quantification was performed with cufflinks and cuffdiff v2.1.1 [12], using the RefSeq transcriptome [8] which was downloaded from the UCSC genome browser website [9] in June 2013. Expression levels were quantified as fragments per kilobase per million mapped reads (FPKM) and normalized across all samples by the cuffdiff program, which computes expression levels at both the individual transcript level and the gene level, the latter by combining the data from all gene-associated transcripts per gene. Quality control of the expression data was performed using the R package cummeRbund (http://compbio.mit.edu/cummeRbund/), and no batch effect was observed despite the fact that HKC2 day2 had fewer lowly expressed genes (Fig. 2B). All wiggle tracks and the transcript- and gene-level FPKM tables are available under GEO accession number GSE59827.
Fig. 2.
Overall properties of RNA-seq datasets. (A) Proportion of uniquely mapped reads that are duplicates in RNA-seq samples. (B) Box plot of expression levels (log10 (FPKM)) for RNA-seq samples.
Acknowledgments
This work is supported by NWO/ALW/MEERVOUD/836.12.010 (HZ), NGI/BioRange program of the NBIC (MO), Radboud University fellowship (HZ) and NCMLS PhD fellowship (ENK). We would like to thank Simon van Heeringen for allowing us to provide his sum_bed.pl. script as a supplementary file.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.gdata.2015.06.002.
Appendix A. Supplementary data
Supplementary material.
References
- 1.Kouwenhoven E.N., Oti M., Niehues H., Van Heeringen S.J., Schalkwijk J., Stunnenberg H.G., Van Bokhoven H., Zhou H. Transcription Factor p63 Bookmarks and Regulates Dynamic Enhancers During Epidermal Differentiation. EMBO Rep. 2015 doi: 10.15252/embr.201439941. pii: e201439941, http://dx.doi.org/10.15252/embr.201439941. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Ruissen F., de Jongh G.J., Zeeuwen P.L., Van Erp P.E., Madsen P., Schalkwijk J. Induction of normal and psoriatic phenotypes in submerged keratinocyte cultures. J. Cell. Physiol. 1996;168(2):442–452. doi: 10.1002/(SICI)1097-4652(199608)168:2<442::AID-JCP23>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 3.Karlic R., Chung H.R., Lasserre J., Vlahovicek K., Vingron M. Histone modification levels are predictive for gene expression. Proc. Natl. Acad. Sci. U. S. A. 2010;107(7):2926–2931. doi: 10.1073/pnas.0909344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smeenk L., van Heeringen S.J., Koeppel M., van Driel M.A., Bartels S.J., Akkers R.C., Denissov S., Stunnenberg H.G., Lohrum M. Characterization of genome-wide p53-binding sites upon stress response. Nucleic Acids Res. 2008;36(11):3639–3654. doi: 10.1093/nar/gkn232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H., Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng J., Liu T., Qin B., Zhang Y., Liu X.S. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012;7(9):1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pruitt K.D., Brown G.R., Hiatt S.M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C.M., Hart J., Landrum M.J., McGarvey K.M., Murphy M.R., O'Leary N.A., Pujar S., Rajput B., Rangwala S.H., Riddick L.D., Shkeda A., Sun H., Tamez P., Tully R.E., Wallin C., Webb D., Weber J., Wu W., DiCuccio M., Kitts P., Maglott D.R., Murphy T.D., Ostell J.M. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42(Database issue):D756–D763. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer L.R., Zweig A.S., Hinrichs A.S., Karolchik D., Kuhn R.M., Wong M., Sloan C.A., Rosenbloom K.R., Roe G., Rhead B., Raney B.J., Pohl A., Malladi V.S., Li C.H., Lee B.T., Learned K., Kirkup V., Hsu F., Heitner S., Harte R.A., Haeussler M., Guruvadoo L., Goldman M., Giardine B.M., Fujita P.A., Dreszer T.R., Diekhans M., Cline M.S., Clawson H., Barber G.P., Haussler D., Kent W.J. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41(Database issue):D64–D69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu T.D., Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26(7):873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flicek P., Ahmed I., Amode M.R., Barrell D., Beal K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fairley S., Fitzgerald S., Gil L., Garcia-Giron C., Gordon L., Hourlier T., Hunt S., Juettemann T., Kahari A.K., Keenan S., Komorowska M., Kulesha E., Longden I., Maurel T., McLaren W.M., Muffato M., Nag R., Overduin B., Pignatelli M., Pritchard B., Pritchard E., Riat H.S., Ritchie G.R., Ruffier M., Schuster M., Sheppard D., Sobral D., Taylor K., Thormann A., Trevanion S., White S., Wilder S.P., Aken B.L., Birney E., Cunningham F., Dunham I., Harrow J., Herrero J., Hubbard T.J., Johnson N., Kinsella R., Parker A., Spudich G., Yates A., Zadissa A., Searle S.M. Ensembl 2013. Nucleic Acids Res. 2013;41(Database issue):D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.