Abstract
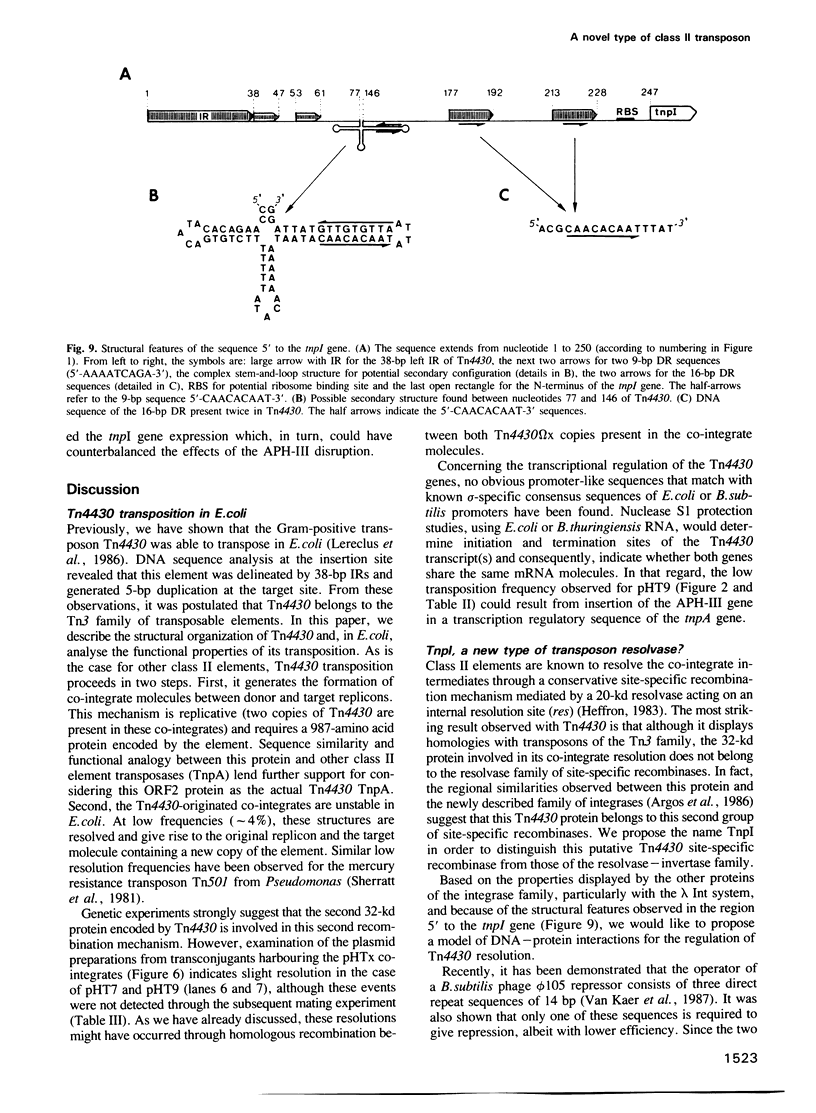
The 4149-bp transposon Tn4430 from Bacillus thuringiensis is delineated by 38-bp inverted repeats and codes for a 113-kd protein that shares homology with the transposases (TnpA) of Tn3, Tn21 and Tn501. Through transpositional recombination, this protein generates the formation of co-integrates between both donor and target replicons, with duplication of Tn4430 molecules. These features are characteristic of transposons of the Tn3 family (class II elements). The second step of the transposition process, the co-integrate resolution, is mediated by a 32-kd protein. This protein (TnpI) displays regional similarities with site-specific recombinases of the integrase family, such as Int of bacteriophage lambda, Cre of bacteriophage P1 or TnpA and TnpB of the Tn554 transposon. Moreover, the 250-bp sequence upstream to the tnpI gene contains several structural features that are reminiscent of the attP attachment site of phage lambda. This unique association between the integrase-like TnpI recombinase and the TnpA transposase qualifies Tn4430 as a member of a new group within the class II mobile genetic elements.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argos P., Landy A., Abremski K., Egan J. B., Haggard-Ljungquist E., Hoess R. H., Kahn M. L., Kalionis B., Narayana S. V., Pierson L. S., 3rd The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986 Feb;5(2):433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur A., Sherratt D. Dissection of the transposition process: a transposon-encoded site-specific recombination system. Mol Gen Genet. 1979 Oct 1;175(3):267–274. doi: 10.1007/BF00397226. [DOI] [PubMed] [Google Scholar]
- Botterman J., Zabeau M. A standardized vector system for manipulation and enhanced expression of genes in Escherichia coli. DNA. 1987 Dec;6(6):583–591. doi: 10.1089/dna.1987.6.583. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Brown N. L., Winnie J. N., Fritzinger D., Pridmore R. D. The nucleotide sequence of the tnpA gene completes the sequence of the Pseudomonas transposon Tn501. Nucleic Acids Res. 1985 Aug 12;13(15):5657–5669. doi: 10.1093/nar/13.15.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell A. Some general questions about movable elements and their implications. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):1–9. doi: 10.1101/sqb.1981.045.01.003. [DOI] [PubMed] [Google Scholar]
- Chandler M., Galas D. J. Cointegrate formation mediated by Tn9. II. Activity of IS1 is modulated by external DNA sequences. J Mol Biol. 1983 Oct 15;170(1):61–91. doi: 10.1016/s0022-2836(83)80227-7. [DOI] [PubMed] [Google Scholar]
- Evans L. R., Brown N. L. Construction of hybrid Tn501/Tn21 transposases in vivo: identification of a region of transposase conferring specificity of recognition of the 38-bp terminal inverted repeats. EMBO J. 1987 Sep;6(9):2849–2853. doi: 10.1002/j.1460-2075.1987.tb02582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel R. A., James A. A., Kolodner R. recA-independent general genetic recombination of plasmids. Nature. 1981 Nov 12;294(5837):184–186. doi: 10.1038/294184a0. [DOI] [PubMed] [Google Scholar]
- Grindley N. D., Reed R. R. Transpositional recombination in prokaryotes. Annu Rev Biochem. 1985;54:863–896. doi: 10.1146/annurev.bi.54.070185.004243. [DOI] [PubMed] [Google Scholar]
- Guyer M. S., Reed R. R., Steitz J. A., Low K. B. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- Heffron F., McCarthy B. J., Ohtsubo H., Ohtsubo E. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell. 1979 Dec;18(4):1153–1163. doi: 10.1016/0092-8674(79)90228-9. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Klier A., Fargette F., Ribier J., Rapoport G. Cloning and expression of the crystal protein genes from Bacillus thuringiensis strain berliner 1715. EMBO J. 1982;1(7):791–799. doi: 10.1002/j.1460-2075.1982.tb01249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad J. W., Schnepf H. E., Whiteley H. R. Diversity of locations for Bacillus thuringiensis crystal protein genes. J Bacteriol. 1983 Apr;154(1):419–428. doi: 10.1128/jb.154.1.419-428.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad J. W., Whiteley H. R. Inverted repeat sequences flank a Bacillus thuringiensis crystal protein gene. J Bacteriol. 1984 Oct;160(1):95–102. doi: 10.1128/jb.160.1.95-102.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad J. W., Whiteley H. R. Three classes of homologous Bacillus thuringiensis crystal-protein genes. Gene. 1986;43(1-2):29–40. doi: 10.1016/0378-1119(86)90005-3. [DOI] [PubMed] [Google Scholar]
- Laban A., Cohen A. Interplasmidic and intraplasmidic recombination in Escherichia coli K-12. Mol Gen Genet. 1981;184(2):200–207. doi: 10.1007/BF00272905. [DOI] [PubMed] [Google Scholar]
- Lane D., de Feyter R., Kennedy M., Phua S. H., Semon D. D protein of miniF plasmid acts as a repressor of transcription and as a site-specific resolvase. Nucleic Acids Res. 1986 Dec 22;14(24):9713–9728. [PMC free article] [PubMed] [Google Scholar]
- Lederberg E. M., Cohen S. N. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974 Sep;119(3):1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lereclus D., Lecadet M. M., Klier A., Ribier J., Rapoport G., Dedonder R. Recent aspects of genetic manipulation in Bacillus thuringiensis. Biochimie. 1985 Jan;67(1):91–99. doi: 10.1016/s0300-9084(85)80234-0. [DOI] [PubMed] [Google Scholar]
- Lereclus D., Menou G., Lecadet M. M. Isolation of a DNA sequence related to several plasmids from Bacillus thuringiensis after a mating involving the Streptococcus faecalis plasmid pAM beta 1. Mol Gen Genet. 1983;191(2):307–313. doi: 10.1007/BF00334831. [DOI] [PubMed] [Google Scholar]
- Lereclus D., Ribier J., Klier A., Menou G., Lecadet M. M. A transposon-like structure related to the delta-endotoxin gene of Bacillus thuringiensis. EMBO J. 1984 Nov;3(11):2561–2567. doi: 10.1002/j.1460-2075.1984.tb02174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Mahillon J., Seurinck J., van Rompuy L., Delcour J., Zabeau M. Nucleotide sequence and structural organization of an insertion sequence element (IS231) from Bacillus thuringiensis strain berliner 1715. EMBO J. 1985 Dec 30;4(13B):3895–3899. doi: 10.1002/j.1460-2075.1985.tb04163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McLaughlin J. R., Murray C. L., Rabinowitz J. C. Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus beta-lactamase gene. J Biol Chem. 1981 Nov 10;256(21):11283–11291. [PubMed] [Google Scholar]
- Michiels T., Cornelis G., Ellis K., Grinsted J. Tn2501, a component of the lactose transposon Tn951, is an example of a new category of class II transposable elements. J Bacteriol. 1987 Feb;169(2):624–631. doi: 10.1128/jb.169.2.624-631.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E., Huwyler L., de Freire Bastos M. do C. Transposon Tn554: complete nucleotide sequence and isolation of transposition-defective and antibiotic-sensitive mutants. EMBO J. 1985 Dec 1;4(12):3357–3365. doi: 10.1002/j.1460-2075.1985.tb04089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E., Löfdahl S. Transposition of Tn554 does not generate a target duplication. Nature. 1984 Jan 19;307(5948):292–294. doi: 10.1038/307292a0. [DOI] [PubMed] [Google Scholar]
- Primrose S. B., Ehrlich S. D. Isolation of plasmid deletion Mutants and study of their instability. Plasmid. 1981 Sep;6(2):193–201. doi: 10.1016/0147-619x(81)90066-4. [DOI] [PubMed] [Google Scholar]
- Rambach A., Hogness D. S. Translation of Drosophila melanogaster sequences in Escherichia coli. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5041–5045. doi: 10.1073/pnas.74.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve J. N., Mendelson N. H., Coyne S. I., Hallock L. L., Cole R. M. Minicells of Bacillus subtilis. J Bacteriol. 1973 May;114(2):860–873. doi: 10.1128/jb.114.2.860-873.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. H., Clewell D. B. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J Bacteriol. 1985 Nov;164(2):782–796. doi: 10.1128/jb.164.2.782-796.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherratt D., Arthur A., Burke M. Transposon-specified, site-specific recombination systems. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):275–281. doi: 10.1101/sqb.1981.045.01.040. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumar A. G., Hahn J., Dubnau D. Studies on the synthesis of plasmid-coded proteins and their control in Bacillus subtilis minicells. Plasmid. 1979 Apr;2(2):279–289. doi: 10.1016/0147-619x(79)90046-5. [DOI] [PubMed] [Google Scholar]
- Smith G. M., Jones P. Tn7 transposition: a multigene process. Identification of a regulatory gene product. Nucleic Acids Res. 1986 Oct 24;14(20):7915–7927. doi: 10.1093/nar/14.20.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3'5"-aminoglycoside phosphotransferase type III. Gene. 1983 Sep;23(3):331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]
- Trieu-Cuot P., Gerbaud G., Lambert T., Courvalin P. In vivo transfer of genetic information between gram-positive and gram-negative bacteria. EMBO J. 1985 Dec 16;4(13A):3583–3587. doi: 10.1002/j.1460-2075.1985.tb04120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Klier A., Courvalin P. DNA sequences specifying the transcription of the streptococcal kanamycin resistance gene in Escherichia coli and Bacillus subtilis. Mol Gen Genet. 1985;198(2):348–352. doi: 10.1007/BF00383017. [DOI] [PubMed] [Google Scholar]
- Van Kaer L., Van Montagu M., Dhaese P. Transcriptional control in the EcoRI-F immunity region of Bacillus subtilis phage phi 105. Identification and unusual structure of the operator. J Mol Biol. 1987 Sep 5;197(1):55–67. doi: 10.1016/0022-2836(87)90609-7. [DOI] [PubMed] [Google Scholar]
- Ward E., Grinsted J. The nucleotide sequence of the tnpA gene of Tn21. Nucleic Acids Res. 1987 Feb 25;15(4):1799–1806. doi: 10.1093/nar/15.4.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley H. R., Schnepf H. E. The molecular biology of parasporal crystal body formation in Bacillus thuringiensis. Annu Rev Microbiol. 1986;40:549–576. doi: 10.1146/annurev.mi.40.100186.003001. [DOI] [PubMed] [Google Scholar]
- Wishart W. L., Broach J. R., Ohtsubo E. ATP-dependent specific binding of Tn3 transposase to Tn3 inverted repeats. Nature. 1985 Apr 11;314(6011):556–558. doi: 10.1038/314556a0. [DOI] [PubMed] [Google Scholar]
- Zabeau M., Stanley K. K. Enhanced expression of cro-beta-galactosidase fusion proteins under the control of the PR promoter of bacteriophage lambda. EMBO J. 1982;1(10):1217–1224. doi: 10.1002/j.1460-2075.1982.tb00016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubay G. The isolation and properties of CAP, the catabolite gene activator. Methods Enzymol. 1980;65(1):856–877. doi: 10.1016/s0076-6879(80)65079-4. [DOI] [PubMed] [Google Scholar]