Abstract
The modeling and analysis of networks and network data has seen an explosion of interest in recent years and represents an exciting direction for potential growth in statistics. Despite the already substantial amount of work done in this area to date by researchers from various disciplines, however, there remain many questions of a decidedly foundational nature — natural analogues of standard questions already posed and addressed in more classical areas of statistics — that have yet to even be posed, much less addressed. Here we raise and consider one such question in connection with network modeling. Specifically, we ask, “Given an observed network, what is the sample size?” Using simple, illustrative examples from the class of exponential random graph models, we show that the answer to this question can very much depend on basic properties of the networks expected under the model, as the number of vertices nV in the network grows. In particular, adopting the (asymptotic) scaling of the variance of the maximum likelihood parameter estimates as a notion of effective sample size, say neff, we show that whether the networks are sparse or not under our model (i.e., having relatively few or many edges between vertices, respectively) is sufficient to yield an order of magnitude difference in neff, from O(nV) to . We then explore some practical implications of this result, using both simulation and data on food-sharing from Lamalera, Indonesia.
Keywords: Asymptotic normality, Consistency, Exponential random graph model, Maximum likelihood
1 Introduction
Since roughly the mid-1990s, the study of networks has increased dramatically. Researchers from across the sciences — including biology, bioinformatics, computer science, economics, engineering, mathematics, physics, sociology, and statistics — are more and more involved with the collection and statistical analysis of data associated with networks. As a result, statistical methods and models are being developed in this area at a furious pace, with contributions coming from a wide spectrum of disciplines. See, for example, the work of Jackson (2008), Kolaczyk (2009), and Newman (2010) for recent overviews from the perspective of economics, statistics, and statistical physics, respectively.
A cross-sectional network is typically represented mathematically by a graph, say, G = (V, E), where V is a set of nV vertices (commonly written V = {1, …, nV}) and E is a set of |E| edges (represented as vertex pairs (u, υ) ∈ E). Edges can be either directed (wherein (u, υ) is distinct from (υ, u)) or undirected. Prominent examples of networks represented in this fashion include the World Wide Web graph (with vertices representing web-pages and directed edges representing hyper-links pointing from one page to another), protein-protein interaction networks in biology (with vertices representing proteins and undirected edges representing an affinity for two proteins to bind physically), and friendship networks (with vertices representing people and edges representing friendship nominations in a social survey).
A great deal of attention in the literature has been focused on the natural problem of modeling networks—of the presence and absence of their edges in particular. There is by now a wide variety of network models that have been proposed, ranging from models of largely mathematical interest to models designed to be fit statistically to data. See, for example, the sources cited above or, for a shorter treatment, the review paper by Airoldi et al. (2009). The derivation and study of network models is a unique endeavor, due to a number of factors. First, the defining aspect of networks is their relational nature, and hence the task is effectively one of modeling complex dependencies among the vertices. Second, quite often there is no convenient space associated with the network, and so the type of distance and geometry that can be exploited in modeling other dependent phenomena, like time series and spatial processes, generally are not available when modeling networks. Finally, network problems frequently are quite large, involving hundreds if not thousands or hundreds of thousands of vertices and their edges. Since a network of nV vertices can in principle have on the order of edges, in network modeling and analysis — particularly statistical analysis of network data — the sheer magnitude of the network can be a critical factor in this area.
Suppose that we observe a network, in the form of a directed graph G = (V, E), where V is a set of nV = |V| vertices and E is a set of ordered vertex pairs, indicating edges. We will focus on graphs with no self-loops: (u, u) ∉ E for any u ∈ V. Alternatively, we may think of G in terms of its nV × nV adjacency matrix Y, where Yij = 1, if (i, j) ∈ E, and 0, otherwise, with Yii ≡ 0. What is our sample size in this setting? At the opening workshop of the recent Program on Complex Networks, held in August of 2010 at the Statistical and Applied Mathematical Sciences Institute (SAMSI), in North Carolina, USA, this question in fact evoked three different responses:
it is the number of unique entries in Y, i.e., nV(nV − 1);
it is the number of vertices, i.e., nV ; or
it is the number of networks, i.e., one.
Which answer is correct? And, why should it matter?
Despite the already vast literature on network modeling, to the best of our knowledge this question has yet to be formally posed much less answered. Closest to doing so are, perhaps, Frank and Snijders (1994) and Snijders and Borgatti (1999), who offer some discussion of this issue in the context of jackknife and bootstrap estimation of variance in network contexts. That this should be so is particularly curious given that the analogous questions have been asked and answered in other areas involving dependent data. In particular, the notion of an effective sample size has been found to be useful in various contexts involving dependent data, including survey sampling, time series analysis, spatial analysis, and even genetic case-control studies (Thibaux and Zwiers, 1984; Yang et al., 2011). Given a sample of size n in such contexts, an effective sample size — say, neff — typically is defined in connection with the variance of an estimator of interest. An understanding of neff, as a function of n, can help lend important insight into a variety of fundamental and inter-related concerns, including the precision with which inference can be done, the amount of information contributed by the data towards learning a parameter(s), and, more practically, the resources needed for data collection.
For example, in survey sampling, where nontrivial dependencies can arise through the use of complex sampling designs, neff generally is taken to be the sample size necessary under simple random sampling with replacement to obtain a variance equal to that resulting from the actual design used (e.g., Lavrakas (2008)). Alternatively, consider a simple AR(1) time series model, where Xt = μ + ϕXt−1 + Zt, for |ϕ| < 1 and Zt independent and identically distributed normal random variables, with mean zero and variance σ2. For a sample of size n, the sample mean X̄n, the natural and unbiased estimator of μ, has a variance that behaves asymptotically in n like σ2/[n(1 − ϕ)2]. Contrasting this expression with σ2/n, corresponding to the case of independent and identically distributed Xt (i.e., equivalent to the case where ϕ ≡ 0), the value neff = n(1 − ϕ)2 is sometimes interpreted as an effective sample size.
In these and similar contexts, it is often possible to show that, whereas nominally the relevant (asymptotic) variance scales inversely with the sample size n, under dependency a different scaling obtains, reflecting a combination of (a) the nominal sample size n, and, importantly, (b) the dependency structure in the data. Since networks are defined by relational data and, hence, consist of random variables that are inherently dependent, it seems not unreasonable to hope that we might similarly gain insight into the above question ‘What is the sample size?’ in a network setting, with the corresponding neff expected to be some function of the number of vertices nV, modified by characteristics of the network structure itself.
Following similar practice in these other fields, therefore, we will interpret the scaling of the asymptotic variances of maximum likelihood estimates in a network model as an effective sample size. In this paper we provide some initial insight into the question of what is the effective sample size in network modeling, focusing on the impact of what is arguably the most fundamental of network characteristics — sparsity. A now commonly acknowledged characteristic of real-world networks is that the actual number of edges tends to scale much more like the number of vertices (i.e., O(nV)) than the number of potential edges (i.e., ). Here we demonstrate that two very different regimes of asymptotics, corresponding to responses 1 and 2 above, obtain for maximum likelihood estimates in the context of a simple case of the popular exponential random graph models, under non-sparse and sparse variants of the models. Response 3 suggests no meaningful asymptotics other than via independent replication. These may arise in some unexpected settings, such as with discrete-time Markov models for evolution of networks over time (Hanneke et al., 2010; Krivitsky and Handcock, 2014, for example). However, we do not explore this direction here.
We will also show that the notion of regime of asymptotics relates to the notion of consistency, as it applies to networks. Krivitsky et al. (2011) showed, informally, that their offset model was consistent, in the sense that if the network’s asymptotic regime agreed with the model, the coefficients of the non-offset terms would converge to some asymptotic value. Although the results of Shalizi and Rinaldo (2013) suggest that consistency may be meaningless for linear ERGMs with nontrivial dependence structure, our results, both theoretical and simulated, suggest that offsets that control the asymptotic regime of the network model can produce consistency-like properties.
As a technical aside, we note that the exponential random graph models we consider here are only relatively simple versions of those commonly used in practice. We choose to work with these models because (i) they are amenable to relatively standard tools in producing the theoretical results we require, while, nevertheless, (ii) they are sufficient in allowing us to highlight in a straightforward and illustrative manner our key finding – that the question of effective sample size in network settings can in fact be expected to be non-trivial and that the answer in general is likely to be subtle, depending substantially on basic model assumptions. That such insight may be obtained already for the simplest models in this class not only speaks to the fundamental nature of our results, but also appears to be fortunate, in that it would appear that theoretical analysis of the key quantity involved in our calculations becomes decidedly more delicate when even moderately more sophisticated models are considered. We provide further comments in this direction at the end of this paper.
The rest of this paper is organized as follows. Some background and definitions are provided in Section 2. Our main results are presented in Section 3, first for the case where edges arise as independent coin flips; second, for the case in which flips corresponding to edges to and from a given pair of vertices are dependent; and, third, for the case of triadic (friend-of-a-friend) effects, which we study via simulation. We then illustrate some practical implications of our results, through a simulation study in Section 4, exploring coverage of confidence intervals associated with our asymptotic arguments, and through application to food-sharing networks in Section 5, where we examine the extent to which real-world data can be found to support non-sparse versus sparse variants of our models. Finally, some additional discussion may be found in Section 6.
2 Background
There are many models for networks. See Kolaczyk (2009), Chapter 6, or the review paper by Airoldi et al. (2009). The class of exponential random graph models has a history going back roughly 30 years and is particularly popular with practitioners in social network analysis. This class of models specifies that the distribution of the adjacency matrix Y follow an exponential family form, i.e., pθ(Y = y) ∝ exp (θ⊤g(y)), for vectors θ of parameters and g(·) of sufficient statistics. However, despite this seemingly appealing feature, work in the last five years has shown that exponential random graph models must be handled with some care, as both their theoretical properties and computational tractability can be rather sensitive to model specification. See Robins et al. (2007), for example, and Chatterjee and Diaconis (2013), for a more theoretical treatment.
Here we concern ourselves only with certain examples of the simplest type of exponential random graph models, wherein the dyads (Yij, Yji) and (Ykℓ, Yℓk) are assumed independent, for (i, j) ≠ (k, ℓ), and identically distributed. These independent dyad models arguably have the smallest amount of dependency to still be interesting as network models. A variant of the models introduced by Holland and Leinhardt (1981), they are in fact too simple to be appropriate for modeling in most situations of practical interest. However, they are ideal for our purposes, as they allow us to quickly obtain non-trivial insight into the question of effective sampling size in network modeling, using relatively standard tools and arguments.
Outside of Section 3.3, the models we consider are all variations of the form
| (1) |
with sufficient statistics
| (2) |
a so-called Bernoulli model with reciprocity. The parameter α governs the propensity of pairs of vertices i and j to form an edge (i, j), and the parameter β governs the tendency towards reciprocity, forming an edge (j, i) that reciprocates (i, j). This model can be motivated from the independence and homogeneity assumptions given above by an argument analogous to that of Frank and Strauss (1986) using the Hammersley–Clifford Theorem (Besag, 1974), with dependence graph being D = {{(i, j), (j, i)} : (i, j) ∈ V2 ∧ i < j}, the set of cliques of D being {{(i, j)} : (i, j) ∈ V2 ∧ i ≠ j} ∪ {{(i, j), (j, i)} : (i, j) ∈ V2 ∧ i < j}, and simplifying for homogeneity.
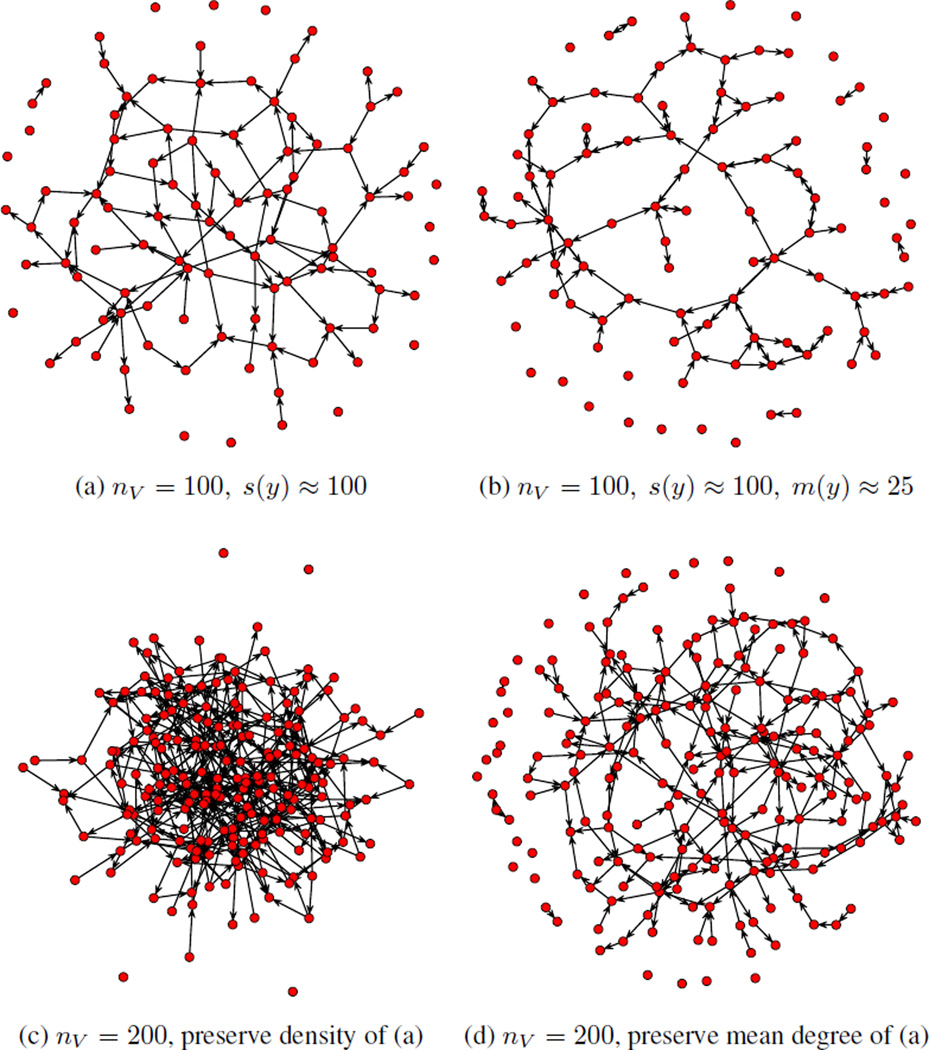
Of interest will be both this general model and the restricted model pα ≡ pα, 0, wherein β = 0 and there is no reciprocity, and not just dyads, but individual potential ties within dyads are independent. We will refer to this latter model simply as the Bernoulli model. Realizations of networks from this model without and with reciprocity (holding expected edge count s(y) fixed) are given in Figure 1(a) and (b), respectively.
Figure 1.
Sampled networks drawn from four configurations of (1). (a) shows a realization from a model with expected mean degree 1 on 100 vertices, and no reciprocity effect. (b) shows a realization from model with the same network size and mean degree as (a), but with reciprocity parameter β set such that the expected number of mutual ties is 25. (c) is a realization of the model from (a), scaled to 200 vertices, preserving density; while (d) preserves mean degree.
Importantly, in both the Bernoulli model and the Bernoulli model with reciprocity, we will examine the question of effective sample size under both the original model parameterization and a reparameterisation in which parameter(s) are shifted by a value log nV. Krivitsky et al. (2011) introduced such shifts in an undirected context as a way of adjusting models like (1) for network size such that realizations with fixed α and β would produce network distributions with asymptotically constant expected mean degree, Eα,β[2s(Y)/nV], for varying nV. That is, a configuration (α, β) that would produce a typical nV = 100 realization like that in Figure 1a would produce an nV = 200 realization like that in Figure 1d. The model’s baseline asymptotic behavior is to have a constant expected density, Eα,β[2s(Y)/{nV(nV − 1)], such that a parameter configuration that would produce a network like 1a for nV = 100 would produce a network like 1c for nV = 200.
In a directed context, “degree” of a given vertex i is ambiguous, as it can refer to the number of ties that vertex makes to others (∑j≠i Yij, “outdegree”), number of ties others make to that vertex (∑j≠i Yji, “indegree”), the number of others to whom that vertex has at least one connection of either type (∑j≠i max(Yij, Yji)), and the number of connections that vertex has (∑j≠i (Yij + Yji)). In this work, we use either of the first two. Then, “mean degree” of Y is s(Y)/nV, with mean outdegree and mean indegree trivially equal; and density is s(Y)/{nV (nV − 1)}.
Motivated by similar concerns, we use the presence or absence of such shifts to produce two different types of asymptotic behavior in our network model classes, corresponding to sparse (asymptotically finite mean degree) and non-sparse (asymptotically infinite mean degree) networks, respectively. Because it is widely recognized that most large real-world networks are sparse networks, this distinction is critical, and, as we show below, it has fundamental implications on effective sample size.
3 Main Results
3.1 Bernoulli Model
We first present our results for the Bernoulli model. Let pα denote the model pα,0, as defined above, and let denote the same model, but under the mapping α ↦ α − log nV of the density parameter. Then, it is easy to show that under pα the mean vertex in- and out-degree tends to infinity and the network density stays at logit−1(α) as nV → ∞, while under , the mean degree tend to eα while the density tends to zero. In fact, the limiting in- and out-degree distributions tend to a Poisson law with the stated mean.
From the perspective of traditional random graph theory, the offset model of Krivitsky et al. (2011) is asymptotically equivalent to the standard formulation of an Erdős-Rényi random graph, in which the probability of an edge scales like eα/nV. Alternatively, from the perspective of social network theory, it is use-ful to examine the log-odds that Yij = 1, conditional on the status of all other potential edges. Defining Y[−ij] to be the network Y with edge (i, j) removed if present, this can be expressed as
This quantity goes from being a constant value α under p = pα to a value α − log nV under . This reflects the intuition that as long as there is a cost associated with forming and maintaining a network tie, an individual will be able to maintain ties with a shrinking fraction of the network as the network grows, with the average number of maintained ties being unaffected by the growth of the network beyond a certain point (Krivitsky et al., 2011).
Given the observation of a network Y randomly generated with respect to either of these models, initial insight into the effective sample size can be obtained by studying the asymptotic behavior of the Fisher information, which we denote ℐ(α) and ℐ†(α) under pα and , respectively. Straightforward calculation shows that while
in contrast,
So , while ℐ(α)† = O(nV), a difference by an order of magnitude.
The implications of this difference are immediately apparent when we consider the asymptotic behavior of the maximum likelihood estimates of α under the two models.
Theorem 1
Let α̂ and α̂† denote the maximum likelihood estimates of the parameter α0 under the models pα0 and , respectively, where α0 ∈ [αmin, αmax], for finite α̂min, αmax. Then under the model pα0, the estimator α̂ is -consistent for α0, and
while under the model , the estimator α̂† is -consistent for α0, and
The proof of these results uses largely standard techniques for asymptotics of estimating equations, but with a few interesting twists. Note that, for fixed nV, the dyads (Yij, Yji) constitute nV (nV − 1)/2 independent and identically distributed bivariate random variables under both pα and . Consistency of the estimators in both cases can be argued by verifying, for example, the conditions of Theorem 5.9 of van der Vaart (2000) for consistency of estimating equations. Similarly, the proof of asymptotic normality of the estimators can be based on the usual technique of a Taylor series expansion of the log-likelihood and, due to the fact that we have assumed an exponential family distribution, the asymptotic normality of the sufficient statistic s(y) in (2). However, in the case of the sparse model , the dyads {(Yij, Yji)}i<j follow a different distribution for each nV, and therefore an array-based central limit theorem is required to show the asymptotic normality of s(y). But since increasing the number of vertices from, say, nV − 1 to nV, as nV → ∞, increases the number of dyads in our model by nV − 1, a standard triangular array central limit theorem is not appropriate here. Rather, a double array central limit theorem is needed, such as Theorem 7.1.2 of Chung (2001). A full derivation is provided in the supplementary materials.
3.2 Bernoulli Model with Reciprocity
From Theorem 1 we see that the effective sample size neff in this context can be either on the order of nV or of , depending on the scaling of the assumed model, i.e., on whether the model is sparse or not. From a non-network perspective, these results can be largely anticipated by the rescaling involved, in that the transformation α ↦ α − log nV induces a rescaling of the expected number of edges by . Now, however, consider the full Bernoulli model with reciprocity, pα, β, defined in (1). Even with just two parameters the situation becomes notably more subtle.
Let ℐ(α, β) be the 2 × 2 Fisher information matrix under this model. Then calculations (not shown) completely analogous to those required for our previous results show that and, similarly, asymptotic properties of the maximum likelihood estimate of (α, β) analogous to those for pα hold.
Let us focus then on sparse versions of pα,β. The offset used previously, i.e., mapping α to α − log nV, is not by itself satisfactory. Call the resulting model . Standard arguments show that the limiting in- and out-degree distributions under this model will be Poisson with mean parameter eα. On the other hand, the expected number of reciprocated out-ties a vertex has, , behaves like e2α+β/nV, and therefore tends to zero as nV → ∞. Thus, β plays no role in the limiting behavior of the model, and, indeed, reciprocity vanishes. This fact can also be understood through examination of the Fisher information matrix, say ℐ†(α, β), in that direct calculation shows
That is, only the information on α grows with the network. Under , only the affinity parameter α can be inferred in a reliable manner.
However, the same intuition that suggests that as the network becomes larger, a given vertex i will have an opportunity for contact with a smaller and smaller fraction of it also suggests that if there is a preexisting relationship in the form of a tie from j to i, such an opportunity likely exists regardless of how large the network may be. This, as well as direct examination of the exact expression for the information matrix ℐ†(α, β), suggests that the − log nV penalty on tie logprobability should not apply to reciprocating ties, which may be implemented by mapping β ↦ β + log nV. Call this model, in which is augmented with this additional offset for β, the model . The corresponding conditional log-odds of an edge now have the form
which exactly captures the intuition described.
It can be shown that under we have ℐ‡(α, β) = O(nV), indicating that information on both parameters grows at the same rate in nV. It can also be shown that the limiting in-and out-degree distribution is now Poisson with mean parameter eα + e2α+β, and that tends to e2α+β. So, both parameters play a role in the limiting behavior of the model and the additional offset induces an asymptotically constant expected per-vertex reciprocity in addition to asymptotically constant expected mean degree.
Finally, we have the following analogue of Theorem 1.
Theorem 2
Let (α̂‡, β̂‡) denote the maximum likelihood estimate of the parameter (α0, β0) under the model , where (α0, β0) ∈ [αmin, αmax] × [βmin, βmax], for finite αmin, αmax, βmin, βmax. Then (α̂‡, β̂‡) is -consistent for (α0, β0), and
Proof of this theorem, using arguments directly analogous to those of Theorem 1, may be found in the supplementary materials. From the theorem we see that under the sparse model , as under , the effective sample size neff is nV.
3.3 Triadic effects
Although there has been some work on obtaining closed-form asymptotics for ERGMs with triadic — friend-of-a-friend — effects (Chatterjee et al., 2011) or showing that they might not exist (Shalizi and Rinaldo, 2013), these results do not appear to be directly applicable to the per-capita asymptotic regimes that we consider in this work. In this section, we thus use simulation in an attempt to extend the intuition developed in Section 3.2 — that reciprocating ties should not be “penalized” for the network size — to these triadic effects. For the sake of simplicity, we will consider undirected networks only.
A tie between i and k and a tie between k and j — i.e., that i knows k and k knows j — should create a preexisting relationship between i and j. That is, k can “introduce” i and j regardless of how large the network is otherwise. Thus, given i − k − j relationships, a potential relationship between i and j should not be penalized for network size (though i − k − j themselves are); and more such two-paths (i.e., i − k′ − j) should have no further effect on this penalty. This suggests an offset on the statistic called the transitive ties (Snijders et al., 2010, eq. 8), or, equivalently, Geometrically-Weighted Edgewise Shared Partners (GWESP) (Morris et al., 2008) with parameter α fixed at 0, i.e.,
| (3) |
Unlike the more familiar count of the number of triangles (∑i<j<k yijyikyjk), t(y) only considers whether a two-path exists, not how many of them there are. (This also makes it far less prone to ERGM degeneracy. (Schweinberger, 2011))
Consider the following model, with edge count and transitive tie count (3):
| (4) |
As with , the coefficient on s is penalized by network size, in the form of log(nV)α⋆, with α⋆ being 1 in . However, the penalty is then partially negated by increasing the coefficient on t by log(nV)γ⋆. This means that, on a sparse network,
The reason that this is an approximation is that, on an otherwise empty network having ties (i, k) and (k, j), adding a tie (i, j) creates not one but three transitive ties, by making all three of the ties transitive (hence the coefficient 3 on the γs). However, as the network becomes more dense, (i, k) and/or (k, j) may already be transitive when (i, j) is added, so only two or one transitive ties might be created.
Therefore, on a sufficiently sparse network (i.e., sufficiently large nV for a given mean degree), in order to cancel the network size penalty for a tie (i, j) but retain it for (i, k) and (k, j), γ⋆ = α⋆/3. With α⋆ = 1 per the same reasoning as before, this means that our heuristic suggests that γ⋆ ≈ 1/3. We verify this empirically as follows. Define t′(y)— per-capita transitive ties — as
In other words, for each vertex i, the number of its neighbors who have ties to at least one other neighbor of i is counted, and the resulting measures averaged over all actors in the network. It can be shown easily that t′(y) ≡ 2t(y)/nV.
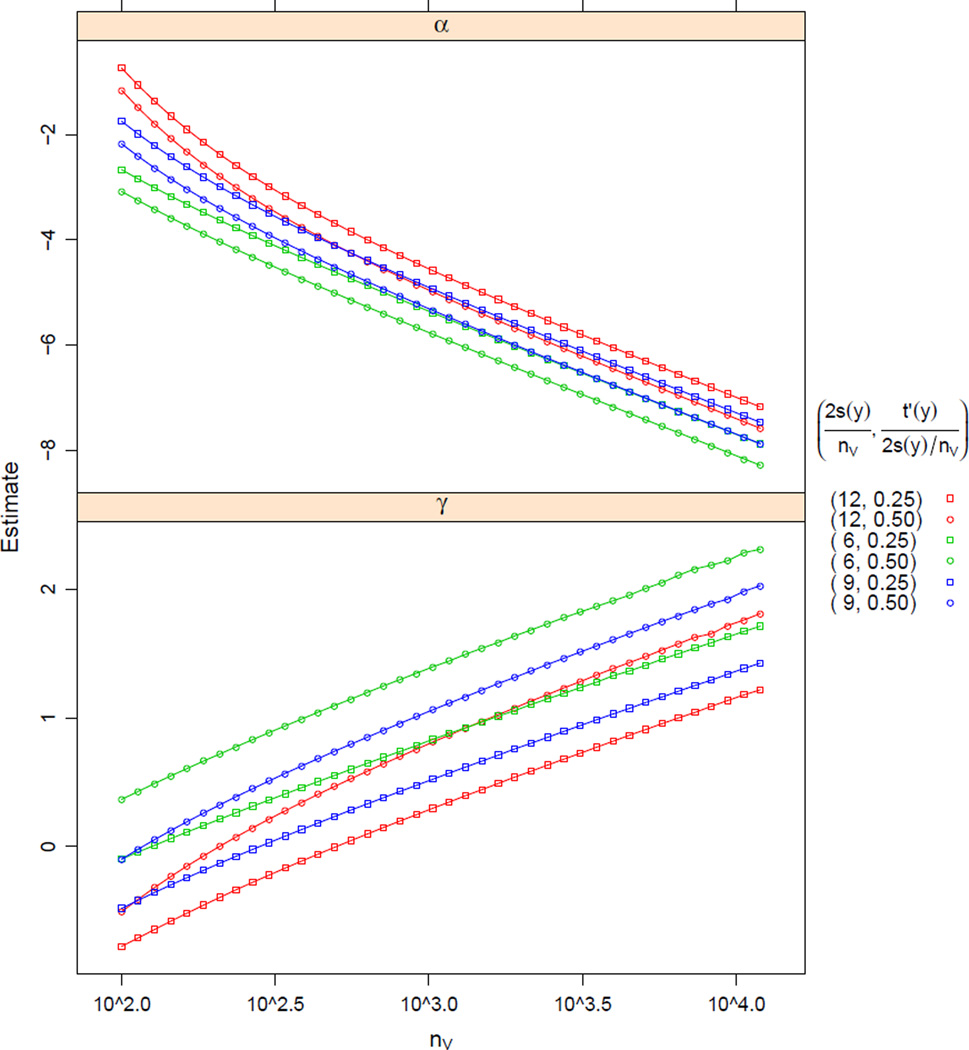
We can then ask if there exist constant values of α⋆ and γ⋆ that produce stable mean degree (2s(y)/nV) and stable per-capita transitive ties (t′(y)). We constructed a series of 40 networks, sized from 100 to 12,000, logarithmically spaced, for each of three configurations of mean degree, 6, 9, and 12; and two levels of per-capita transitivity for each: 1/2 of the mean degree and 1/4 of the mean degree. (This was done because per-capita transitivity cannot exceed the mean degree.) For each combination of nV, 2s/nV, and t′/(2s/nV), we used simulated annealing to construct a network y with these statistics, and then fit an ERGM pα,γ (without offsets) to it to obtain point estimates for what is effectively (−log(nV)α⋆ + α0, log(nV)γ⋆ + γ0). The calculations were performed using the ergm package (Handcock et al., 2014; Hunter et al., 2008) for the R computing environment (R Core Team, 2013).
We show the results in Figure 2. Our intuition seems to be confirmed, to the extent that our network sizes are sufficiently large to confirm or disconfirm it. The trend in both parameter estimates appears to be more linear (in log nV) as nV increases, suggesting that unique α⋆ and γ⋆ exist. For α̂, the asymptotic slope (i.e., −α⋆) is very close to −1 regardless of the mean degree and the amount of transitivity, and for α̂, the slope (i.e., γ⋆) decreases as log nV increases, though it does not quite obtain the exact value of 1/3 for the network sizes considered. (E.g., considering only nV > 5,000, 2s(y)/nV = 6, and t′(y)/(2s(y)/nV) = 1/4 — the fastest-converging configuration — gave the slope of 0.35.)
Figure 2.
Maximum likelihood estimates from fitting pα,γ to networks with a variety of sizes, densities (distinguished by color), and levels of transitivity (distinguished by plotting symbol). Note that the horizontal axis is plotted on the logarithmic scale.
Notably, even though given a particular value of the sufficient statistic (s(y), t(y)), the natural parameters (α, γ) would be determined exactly, we have to use Monte Carlo MLE (Hunter and Handcock, 2006) to estimate them, so there is some noise in the point estimates.
Overall, it appears that the coefficients of sparser networks with weaker transitivity tend to approach linearity faster. Thus, we performed a follow-up simulation study, this one with mean degree 2, transitivity proportion 1/8, and 40 values of nV between 10,000 and 40,000, logarithmically spaced.
Based on all of the values of nV considered, α̂⋆ = 1.00037 (95% CI: (1.00029, 1.00044)) and γ̂⋆ = 0.3377 (95% CI: (0.3369, 0.3386)), closer to the theoretical values of 1 and 1/3 than the smaller network sizes. The confidence intervals do not include the theoretical values, but we would not expect the asymptotic values to be attained for any finite network size. Indeed, there is evidence of nonlinearity in that range (P-value of predictor log(nV)2 term is < 0.0001 for the α⋆ response and 0.04 for the γ⋆ response, with negative coefficient for both). Furthermore, fitting only the 20 data points with nV > 20,000 produces (α̂⋆, γ̂⋆) = (1.000072, 0.3347), and fitting only the 10 data points with nV > 29,000, (α̂⋆, γ̂⋆) = (1.00030, 0.3334).
This very strongly suggests meaningful and interpretable asymptotic behavior for triadic closure ERGM terms as well. In particular, the asymptotic linearity with a known coefficient suggests a form of consistency for “intercepts” α0 and γ0, as it is they that control the asymptotic mean degree and per-vertex amount of triadic closure in (4).
To relate this to the notion of effective sample size neff used earlier, defined through the scaling of the information matrix ℐ(α, γ), we simulated the sufficient statistics from the above-described fits. For an exponential family, the variance covariance matrix of sufficient statistics under the MLE approximates the information matrix (Hunter and Handcock, 2006, eq. 3.5, for example). We find that the entries of ℐ̂(α̂, γ̂)/nV = Varα̂, γ̂ ([s(Y), t(Y)]) /nV do not exhibit any trend at all as a function of nV, for fixed mean degree and per-vertex transitivity. (In particular, for a linear trend, P-values are 0.31, 0.49, and 0.41 for Var(s(Y))/nV, Var(t(Y))/nV, and Cov(s(Y), t(Y))/nV, respectively. Exploratory plots do not show any pattern, except for greater variability in estimates of variance for higher nV.) This strongly suggests that the asymptotics of the model (4) have an effective sample size neff of nV as well.
4 Coverage of Wald Confidence Intervals
Our asymptotic arguments in Section 3 were developed primarily for the purpose of establishing the scaling associated with the asymptotic variance, so as to provide insight into the question of effective sample size — our main focus here. However, the asymptotically normal distributions we have derived are of no little independent interest themselves, as they serve as as a foundation for doing formal inference on the model parameters in practice. By way of illustration, here we explore their use for constructing confidence intervals, particularly those based on Theorem 2: under a model , the Wald confidence intervals using plug-in estimators for the standard errors are for α and .
Because our asymptotics are in Nυ = |V|, we examine a variety of network sizes. The desired asymptotic properties of the network are expressed in terms of the per-capita mean value parameters— and . We study two configurations:
and
.
In other words, the expected mean outdegree is set to 1, and expected numbers of out-ties that are reciprocated are 0.25 × 2 = 0.5 and 0.40 × 2 = 0.8 per vertex, respectively. These represent two levels of mutuality, though note that even (1) reprsents substantial mutuality, especially for larger networks.
For each nV = 10, 15, 20, …, 200, we estimate the natural parameters of the model corresponding to the desired mean value parameters, and then simulate 100,000 networks from each configuration, evaluating the MLE and constructing a Wald confidence interval at each of level of the customary 80%, 90%, 95%, and 99%, for α and for β (individually), checking the coverage.
For some of the smaller sample sizes, the simulated network statistics for some realizations were not in the interior of their convex hull. (Barndorff-Nielsen, 1978, Thm. 9.13, p. 151) That is, their values were the maximal or minimal possible: s(y) = 0 (empty graph), s(y) = nV(nV − 1) (complete graph), m(y) = 0 (no edges reciprocated), and/or m(y) = s(y)/2 (every extant edge reciprocated). For those, the MLE did not exist. (For (1), the fraction was 8.2% for nV = 10 and none of the 100,000 realizations had no MLE for nV ≥ 55. For (2), it was 14.2% for nV = 10 and none of the realizations had no MLE for nV ≥ 65.)
Our results are conditional on the MLE existing. From the frequentist perspective, one might argue that if the MLE did not exist for a real dataset, we would not have reported that type of confidence interval, so it should be excluded from the simulation as well.
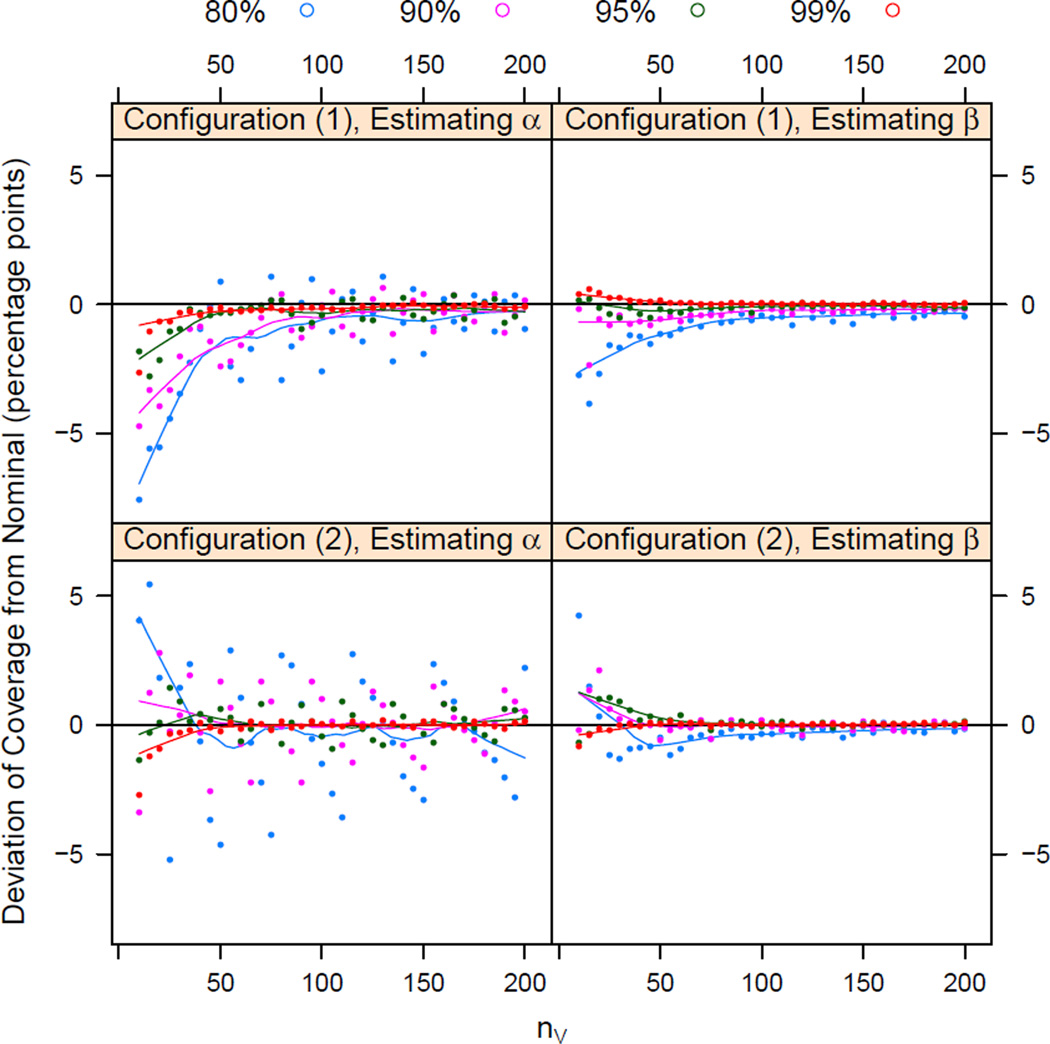
We report coverages for selected network sizes in Table 1 and provide a visualization in Figure 3. Overall, the 80% coverage appears to be varied — and not very conservative—while higher levels of confidence appear to be more consistently conservative, particularly for estimates of β. Coverage for α appears to oscillate as a function of network size. This is particularly noticeable for the lower confidence levels and stronger mutuality (2). Tendency of a confidence interval for a binomial proportion to oscillate around the nominal level is a known phenomenon (Brown et al., 2001, 2002, and others), though it is interesting to note that it appears to be more prominent for the density, rather than mutuality, parameter and that it appears to be stronger for stronger mutuality.
Table 1.
Simulated Theorem 2 confidence interval coverage levels for selected network sizes and two levels of reciprocity: lower (1) and higher (2).
| Coverage | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 80.0% | 90.0% | 95.0% | 99.0% | ||||||
| nV | α | β | α | β | α | β | α | β | |
| (1) | 10 | 72.4% | 77.3% | 85.3% | 89.8% | 93.2% | 95.2% | 96.4% | 99.4% |
| 20 | 74.5% | 77.3% | 86.0% | 89.4% | 92.9% | 94.9% | 98.3% | 99.5% | |
| 50 | 80.9% | 78.8% | 87.6% | 89.4% | 94.7% | 94.8% | 98.9% | 99.2% | |
| 100 | 77.4% | 79.6% | 90.0% | 90.0% | 94.6% | 94.9% | 98.9% | 99.1% | |
| 200 | 79.0% | 79.5% | 90.1% | 89.8% | 94.9% | 94.9% | 98.9% | 99.0% | |
| (2) | 10 | 84.0% | 84.2% | 86.6% | 89.8% | 93.6% | 94.3% | 96.3% | 98.2% |
| 20 | 81.8% | 80.3% | 92.8% | 92.1% | 95.1% | 96.0% | 98.1% | 98.8% | |
| 50 | 75.3% | 79.5% | 91.7% | 89.4% | 95.6% | 95.1% | 98.8% | 99.0% | |
| 100 | 78.5% | 79.7% | 91.0% | 90.2% | 94.5% | 94.9% | 99.0% | 99.1% | |
| 200 | 82.2% | 79.9% | 90.5% | 89.9% | 95.3% | 95.1% | 99.2% | 99.1% | |
Figure 3.
Scatterplot of differences between simulated coverage and nominal coverage for the two configuration studied, as a function of network size nV. Color denotes the nominal coverage levels, and smoothing lines have been added. Note that the differences are differences in percentage points (simulated % − nominal %), not percent differences .
5 Example: Food-Sharing Networks in Lamalera
While the results of Section 3 are important in establishing how closely the question of effective sample size in network modeling is tied to the structural property of (non)sparseness expected of the networks modeled, there remains the important practical question of establishing in applications just which model (i.e., sparse or non-sparse) is most appropriate. While a full and detailed study of this question is beyond the scope of this work, we present here an initial exploration.
Note that, in exploring this question, we face a problem similar to that pointed out by Krivitsky et al. (2011): it requires a collection of closed networks of a variety of sizes yet substantively similar social structure. Furthermore, our results are limited to modeling density and reciprocity, so the networks should be well-approximated by this model. Here, we use data collected by Nolin (2010), in which each of 317 households in Lamalera, Indonesia was asked to list the households to whom they have given and households from whom they have received food in the preceding season. Lamalera is split, administratively, into two villages, which are further subdivided into wards, and then into neighborhoods. Nolin (2010) fit several ERGMs to the network, finding that distance between households had a significant effect on the propensity to share, as did kinship between members of the households involved. Nolin also found a significant positive mutuality effect.
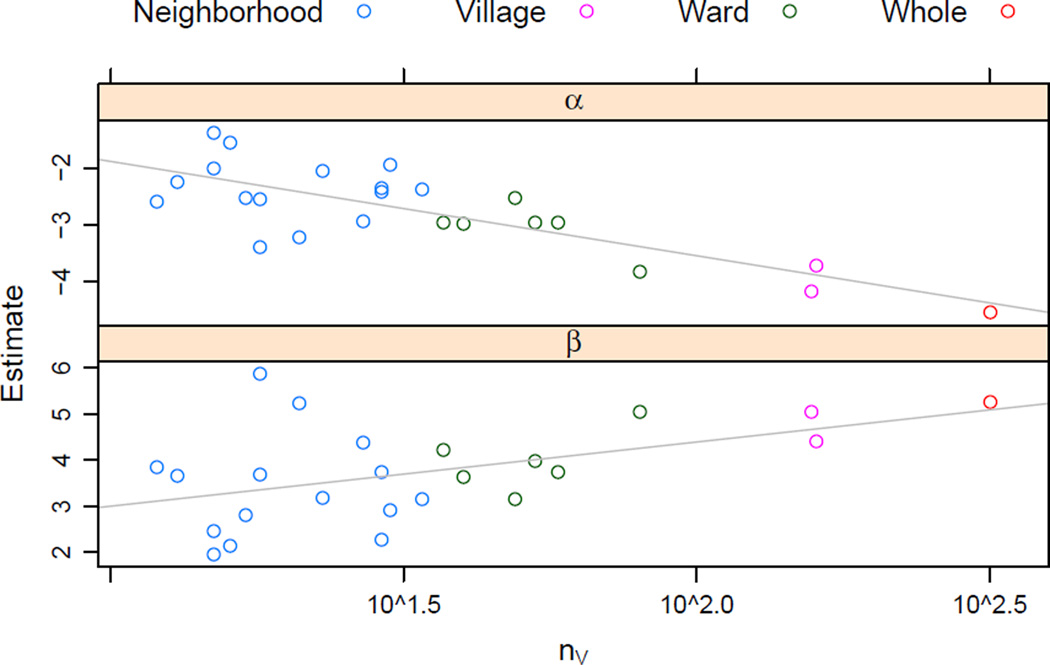
In our study, we make use of the geographic effect by constructing a series of 24 overlapping subnetworks, consisting of Lamalera itself, its 2 constituent villages, 6 wards, and 15 neighborhoods, with network sizes ranging from 12 to 317. We then fit the baseline model pα,β to each network. If pα,β is the most realistic asymptotic regime for these data, we would expect estimates α̂ and β̂ to have no relationship to log nV for the corresponding network. If is the most realistic, we would expect no relationship between log nV and β̂, but an approximately linear relationship with α̂, with slope around − 1. Lastly, if is the most realistic, we would expect the slope of the relationship between log nV and α̂ to be around − 1 and between log nV and β̂ to be around +1.
The estimated coefficients and the slopes are given in Figure 4. The results are suggestive. The relationship between β̂ and log nV is clearly negative, while the relationship between β̂ and log nV is clearly positive, and the magnitudes of both slopes are closer to 1 than to 0 (although both are far from equaling 1). Overlap between the subnetworks induces dependence among the coefficients, so it is not possible to formally test or estimate how significant this difference is. Nevertheless, the preponderance of evidence is that is the best of the three considered. That is, a sparse model that does not enforce sparsity on reciprocating ties appears to be preferable here.
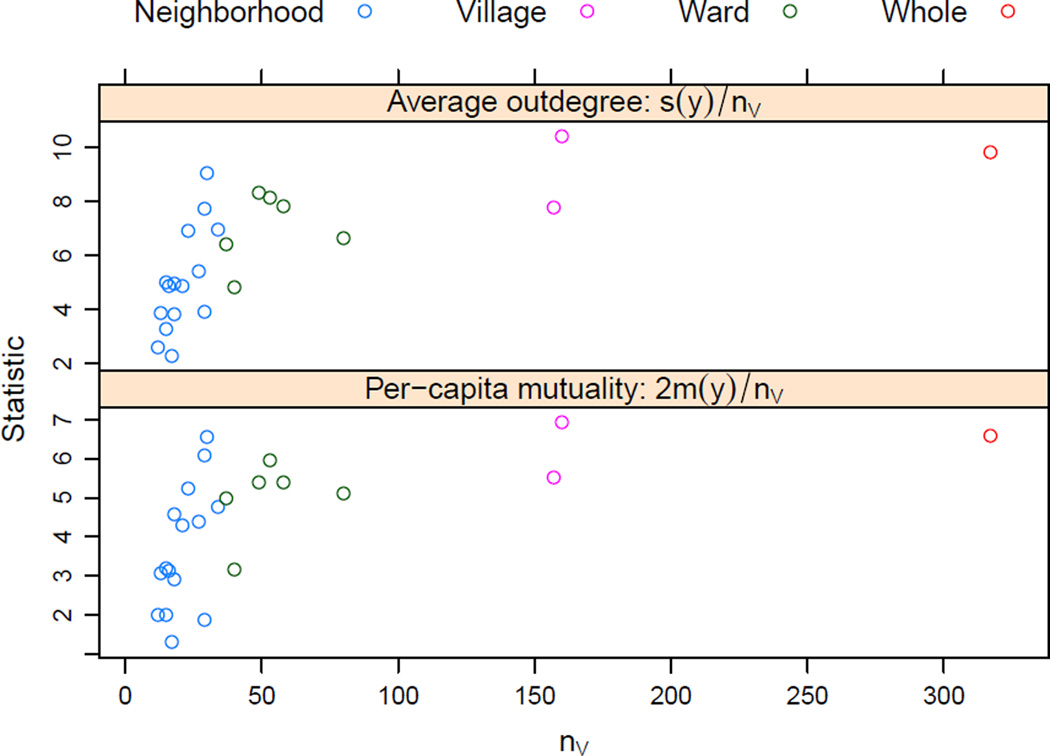
Figure 4.
Maximum likelihood estimates from fitting pα,β to each subdivision of the Lamalera food-sharing network. Note that the horizontal axis is plotted on the logarithmic scale. Colors indicate subdivision type. The least-squares coefficients from regressing α̂ and β̂ on log nV are −0.72 and +0.60, respectively.
A possible explanation for why the magnitudes of the slopes are substantially less than 1 is that both the argument of Krivitsky et al. (2011) and our argument in Section 3.2 rely on the assumption that the network is closed — no relationships of interest are to or from actors outside of the network observed — or, at least, that the stable mean degree and per-capita reciprocity are for the ties within it. However, while there is likely to be very little food sharing out of or into Lamalera, and relatively little between the two villages it comprises (7% of all food-sharing ties in the network are between villages), there is more sharing between the wards (28% are between wards), and even more between neighborhoods (44%). Thus, the closed-network assumption is violated. (The respective between-subdivision percentages for reciprocated ties are 6%, 22%, and 39%.) When each of the subdivisions of the network is considered in isolation, these ties are lost, so the smaller subdivisions appear, to the model, to have smaller mean degree and per-capita mutuality. (See Figure 5.) This, in turn, means that smaller subdivisions have a decreased α̂ (increasing the slope for it in Fig. 4) and, because mutual ties suffer less of this “attrition” than ties do overall, the β, after adjusting for the decreased α̂, is increased for smaller networks, thus reducing the slope for β̂ in Fig. 4. It is not unlikely that this pattern will hold in any network with an unobserved spatial structure, whose subnetworks of interest are contiguous regions in this space.
Figure 5.
Per-capita network statistics as a function of nV. Colors indicate subdivision type. Note that the larger subdivisions have more within-subdivision ties.
6 Discussion
Unlike conventional data, network data typically do not have an unambiguous notion of sample size. The theoretical developments and the examples we have presented show that the effective sample size neff associated with a network depends strongly on the model assumed for how the network scales. In particular, in the case of reciprocity, whether or not the model for scaling takes into account the notion of preexisting relationship affects whether reciprocity is even meaningful for large networks. In the case of triadic effects, a similar notion — along with the inutition that as the network size changes each individual’s view of triadic closure should not — implies a specific scaling regime which, in turn, implies a specific notion of the effective sample size.
The models we study here are relatively simple examples of network models. However, with reciprocity, our work includes an important aspect that already allows us a glimpse beyond the treatments of, say, Chatterjee et al. (2011) and Rinaldo et al. (2011), for so-called beta models, where the dependency induced here by reciprocity is absent. In addition, the results for reciprocity suggest that the effective modeling of triadic (e.g., friend of a friend of a friend) effects in a manner indexed to network size requires a more complex treatment yet. However, our simulation shows, perhaps somewhat surprisingly, that if triadic closure is considered on a per-capita basis, effective sample size ultimately behaves similar to the way it does in the simpler cases.
We note that asymptotic theory supporting methods for the construction of confidence intervals for network parameters is only beginning to emerge. The most traction appears to have been gained in the context of stochastic block models (e.g., Bickel and Chen (2009), Choi et al. (2012), Celisse et al. (2012), and Rohe et al. (2011)), although progress is beginning to be had with exponential random graph models as well (e.g., Chatterjee et al. (2011), Chatterjee and Diaconis (2013), and Rinaldo et al. (2011)). Most of these works present consistency results for maximum likelihood and related estimators, with the exception of Bickel and Chen (2009), which also includes results on asymptotic normality of estimators. Our work contributes to this important but nascent area with both our theoretical developments and our simulation studies. In particular, the asymptotic regime of pα,γ; is one that appears to neither become degenerate nor approaches Erdős-Rényi.
The lack of an established understanding of the distributional properties of parameter estimates in commonly used network models is particularly unfortunate given that a number of software packages now allow for the easy computation of such estimates. For example, packages for computing estimates of parameters in fairly general formulations of exponential random graph models routinely report both estimates and, ostensibly, standard errors, where the latter are based on standard arguments for exponential families. Unfortunately, practitioners do not always seem to be aware that the use of these standard errors for constructing normal-theory confidence intervals and tests is lacking fully formal justification. From that perspective, our work appears to be one of the first to begin laying the necessary theoretical foundation to justify practical confidence interval procedures in exponential random graph models. See Haberman (1981) for another contribution in this direction, proposed as part of the discussion of the original paper of (Holland and Leinhardt, 1981).
In order to successfully build upon our work, and extend our results to more sophisticated instances of exponential random graph models, certain technical challenges must be overcome. First, we note that our notion of effective sample size is tied directly to the asymptotic behavior of the Fisher information matrix of our model (denoted ℐ(θ) in the supplementary materials). Given that exponential random graph models are, by definition, of exponential family form, this information matrix is in principle given by the matrix of partial second derivatives of the cumulant generating function (denoted ψ in the supplementary materials, so that ℐ = ∂2ψ(θ)/∂θ∂θT). Due to the use of independent dyads in our theoretical work (i.e., our models are variations on Bernoulli models), the corresponding likelihoods factor over dyads, and hence the information matrices are simply proportional to powers of Nυ (i.e., linear or quadratic). This is in analogy to the canonical setting of independent and identically distributed observations. However, in more general settings beyond the case of independent dyads — including even the models with triadic effects we studied in simulation, the likelihood cannot be expected to factor in such a simple manner. Hence, the analysis of the Fisher information promises to be decidedly more subtle. In fact, there appears to be almost no work to date studying this matrix in any detail. To the best of our knowledge the only such work is the recent manuscript by Pu et al. (2013), introducing a deterministic approach to approximating this matrix (stochastic approximations may, of course, be produced using MCMC) based on a lower-bound of the cumulant generating function. This bound, however, has only an implicit representation.
Second, in the case of more general exponential random graph models than those studied here, there will be a need for a correspondingly more sophisticated central limit theorem, in order to produce results on asymptotic normality analogous to those we present for the simpler models we study. Even for our models, the tool we used was somewhat nonstandard, in that we required a double-array central limit theorem. The more general case will require a central limit theorem capable of handling the nontrivial global dependencies induced by effects even as seemingly simple as triadic closure or the like. Progress on the first point above is a likely prerequisite to understanding the nature of these dependencies sufficiently well to know just what sort of central limit theorem is required.
Finally, there is, as always with exponential random graph models, the issue of instability and degeneracy that must be kept in mind (e.g., Handcock (2003) and Chatterjee et al. (2011)). It has been discovered only relatively recently that substantial care must be taken in specifying network effects in exponential random graph models. Without such care, it is possible to produce models for which the corresponding distributions turn out to be near-degenerate and, in turn, the estimation of parameters, highly unstable. Schweinberger (2011) has recently shed important light on this issue, showing that instability and degeneracy are related to the scaling of the linear term in exponential family distributions generally and, more specifically, in exponential random graph models. These scaling results can be expected to have implications on the role that scaling necessarily plays in the types of calculations we have presented here.
Acknowledgments
This work was begun during the 2010–2011 Program on Complex Networks at SAMSI and was partially supported by ONR award N000140910654 (EDK), ONR award N000140811015 (PNK), NIH award 1R01HD068395-01 (PNK), and Portuguese Foundation for Science and Technology Ciência 2009 Program (PNK). Krivitsky also wishes to thank David Nolin for the Lamalera food-sharing network dataset.
Contributor Information
Eric D. Kolaczyk, Department of Mathematics and Statistics, Boston University, Boston, MA 02215, USA.
Pavel N. Krivitsky, School of Mathematics and Applied Statistics, University of Wollongong, Wollongong, NSW 2500, Australia.
References
- Airoldi Edoardo M, Blei David M, Fienberg Stephen E, Xing Eric P. A survey of statistical network models. Foundations and Trends in Machine Learning. 2009;2:129–233. [Google Scholar]
- Barndorff-Nielsen Ole E. Information and Exponential Families in Statistical Theory. New York: John Wiley & Sons, Inc.; 1978. ISBN 0471995452. [Google Scholar]
- Besag Julian. Spatial interaction and the statistical analysis of lattice systems (with discussion) Journal of the Royal Statistical Society, Series B. 1974;36:192–236. ISSN 0035-9246. [Google Scholar]
- Bickel Peter J, Chen Aiyou. A nonparametric view of network models and Newman–Girvan and other modularities. Proceedings of the National Academy of Sciences. 2009;106(50):21068–21073. doi: 10.1073/pnas.0907096106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown Lawrence D, Tony Cai T, DasGupta Anirban. Interval estimation for a binomial proportion. Statistical Science. 2001 May;16(2):101–117. ISSN 0883-4237. [Google Scholar]
- Brown Lawrence D, Tony Cai T, DasGupta Anirban. Confidence intervals for a binomial proportion and asymptotic expansions. The Annals of Statistics. 2002;30(1):160–201. [Google Scholar]
- Celisse Alain, Daudin Jean-Jacques, Pierre Laurent. Consistency of maximum-likelihood and variational estimators in the stochastic block model. Electronic Journal of Statistics. 2012;6:1847–1899. [Google Scholar]
- Chatterjee Sourav, Diaconis Persi. Estimating and understanding exponential random graph models. The Annals of Statistics. 2013;41(5):2428–2461. [Google Scholar]
- Chatterjee Sourav, Diaconis Persi, Sly Allan. Random graphs with a given degree sequence. The Annals of Applied Probability. 2011;21(4):1400–1435. [Google Scholar]
- Choi David S, Wolfe Patrick J, Airoldi Edoardo M. Stochastic blockmodels with a growing number of classes. Biometrika. 2012;99(2):273–284. doi: 10.1093/biomet/asr053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Kai L. A course in probability theory. Academic Press; 2001. [Google Scholar]
- Frank Ove, Snijders Tom AB. Estimating the size of hidden populations using snowball sampling. Journal of Official Statistics. 1994;10:53–67. [Google Scholar]
- Frank Ove, Strauss David. Markov graphs. Journal of the American Statistical Association. 1986;81(395):832–842. ISSN 0162-1459. [Google Scholar]
- Haberman Shelby J. An exponential family of probability distributions for directed graphs: Comment. Journal of the American Statistical Association. 1981;76(373):60–61. [Google Scholar]
- Handcock Mark S. Technical Report No. 39. University of Washington: Center for Statistics and the Social Sciences; 2003. Assessing degeneracy in statistical models of social networks. [Google Scholar]
- Handcock Mark S, Hunter David R, Butts Carter T, Goodreau Steven M, Krivitsky Pavel N, Morris Martina. ergm: Fit, Simulate and Diagnose Exponential-Family Models for Networks. 2014 doi: 10.18637/jss.v024.i03. The Statnet Project ( http://www.statnet.org), URL CRAN.R-project.org/package=ergm. R package version 3.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanneke Steve, Fu Wenjie, Xing Eric P. Discrete temporal models of social networks. Electronic Journal of Statistics. 2010;4:585–605. ISSN 1935-7524. [Google Scholar]
- Holland Paul W, Leinhardt Samuel. An exponential family of probability distributions for directed graphs. Journal of the American Statistical Association. 1981:33–50. [Google Scholar]
- Hunter David R, Handcock Mark S. Inference in curved exponential family models for networks. Journal of Computational and Graphical Statistics. 2006;15(3):565–583. ISSN 1061-8600. [Google Scholar]
- Hunter David R, Handcock Mark S, Butts Carter T, Goodreau Steven M, Morris Martina. ergm: A package to fit, simulate and diagnose exponential-family models for networks. Journal of Statistical Software. 2008;24(3):1–29. doi: 10.18637/jss.v024.i03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson Matthew O. Social and economic networks. Princeton University Press; 2008. [Google Scholar]
- Kolaczyk Eric D. Statistical analysis of network data: methods and models. Springer Verlag; 2009. [Google Scholar]
- Krivitsky Pavel N, Handcock Mark S. A separable model for dynamic networks. Journal of the Royal Statistical Society, Series B. 2014;76(1):29–46. doi: 10.1111/rssb.12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivitsky Pavel N, Handcock Mark S, Morris Martina. Adjusting for network size and composition effects in exponential-family random graph models. Statistical Methodology. 2011 doi: 10.1016/j.stamet.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrakas Paul J. Encyclopedia of survey research methods. SAGE Publications, Incorporated; 2008. [Google Scholar]
- Morris Martina, Handcock Mark S, Hunter David R. Specification of exponential-family random graph models: Terms and computational aspects. Journal of Statistical Software. 2008 May;24(4):1–24. doi: 10.18637/jss.v024.i04. ISSN 1548-7660. URL http://www.jstatsoft.org/v24/i04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman Mark EJ. Networks: an introduction. Oxford University Press; 2010. [Google Scholar]
- Nolin David A. Food-sharing networks in lamalera, indonesia: Reciprocity, kinship, and distance. Human Nature. 2010;21(3):243–268. doi: 10.1007/s12110-010-9091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Wen, Choi Jaesik, Amir Eyal, Espelage Dorothy L. Learning exponential random graph models. 2013 Unpublished manuscript, available at https://www.ideals.illinois.edu/handle/2142/45098. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. URL http://www.R-project.org/. [Google Scholar]
- Rinaldo Alessandro, Petrovic Sonja, Fienberg Stephen E. Maximum likelihood estimation in network models. Arxiv preprint arXiv:1105.6145. 2011 [Google Scholar]
- Robins Garry, Snijders Tom AB, Wang Peng, Handcock Mark S, Pattison Philippa. Recent developments in exponential random graph (p*) models for social networks. Social Networks. 2007;29(2):192–215. [Google Scholar]
- Rohe Karl, Chatterjee Sourav, Yu Bin. Spectral clustering and the high-dimensional stochastic blockmodel. The Annals of Statistics. 2011;39(4):1878–1915. [Google Scholar]
- Schweinberger Michael. Instability, sensitivity, and degeneracy of discrete exponential families. Journal of the American Statistical Association. 2011;106(496):1361–1370. doi: 10.1198/jasa.2011.tm10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalizi Cosma Rohilla, Rinaldo Alessandro. Consistency under sampling of exponential random graph models. Annals of Statistics. 2013;41(2):508–535. doi: 10.1214/12-AOS1044. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders Tom AB, Borgatti Stephen P. Non-parametric standard errors and tests for network statistics. Connections. 1999;22(2):161–170. [Google Scholar]
- Snijders Tom AB, van de Bunt Gerhard G, Steglich Christian EG. Introduction to stochastic actor-based models for network dynamics. Social Networks. 2010;32(1):44–60. ISSN 0378-8733. [Google Scholar]
- Thibaux Jean H, Zwiers Francis W. The interpretation and estimation of effective sample size. Journal of Climate and Applied Meteorology. 1984 May;23(5):800–811. ISSN 0733-3021. [Google Scholar]
- van der Vaart Aad W. Asymptotic statistics. Number 3 in Cambridge series in statistical and probabilistic mathematics: Cambridge University Press; 2000. [Google Scholar]
- Yang Yaning, Remmers Elaine F, Ogunwole Chukwuma B, Kastner Daniel L, Gregersen Peter K, Li Wentian. Effective sample size: Quick estimation of the effect of related samples in genetic case–control association analyses. Computational Biology and Chemistry. 2011;35(1):40–49. doi: 10.1016/j.compbiolchem.2010.12.006. ISSN 1476-9271. doi: http://dx.doi.org/10.1016/j.compbiolchem.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]