Abstract
Genomic annotation of unique and combinatorial epigenetic modifications along with transcription factor occupancy is having a profound impact on our understanding of the genome. These studies have led to a better appreciation of the dynamic nature of the epigenetic and transcription factor binding components that reveal overarching principles of the genome as well as tissue specificity. In this minireview, we discuss the presence and potential functions of several of these features across the genome in osteoblast lineage cells. We examine how these features are modulated during cellular maturation, affect transcriptional output and phenotype, and how they alter the ability of cells to respond to systemic signals directed by calcemic hormones such as 1,25-dihydroxyvitamin D3 and PTH. In particular, we describe recent experiments which indicate that progressive stages of bone cell differentiation affect RUNX2 binding to the genome, modify and restrict patterns of gene expression, and dramatically alter cellular response to the vitamin D hormone. These studies expand our understanding of mechanisms that govern steroid hormone regulation of gene expression, while highlighting the increasing complexity that is evident relative to these basic cellular processes. The results also have profound implications with respect to the impact of skeletal diseases on transcriptional outcomes as well.
Keywords: Genome-wide analysis, transcription, osteoblast lineage cells, epigenetics
1. INTRODUCTION
Chromatin immunoprecipitation (ChIP), coupled initially to tiled oligonucleotide microarrays (ChIP-chip) and subsequently to massively parallel deep sequencing methods (ChIP-seq), together with numerous additional genome-scale techniques, has enabled investigators to annotate cellular genomes in ways fully unappreciated less than a decade earlier (1). While it has long been known that histones represent a particular focus of post translational modification, efforts by numerous groups including those of the ENCODE (Encyclopedia of DNA Elements) Consortium have focused upon both the identification of such modifications (marks) and elucidation of their structural and functional significance relative to the control of gene expression and other processes (2). These and additional efforts have led to an appreciation of the cell-type specific nature of histone modifications, the dynamic nature of their appearance during cellular development and differentiation, and the realization that their presence frequently denote specific functional attributes. Perhaps most importantly, the presence of many of these histone modifications or combinations thereof have been found to be enriched at sites of specific regulatory significance, whether as indicators of nucleosome presence, the location of promoters or of active enhancers (3–5). In addition, many of these marks provide insight into the functional state of transcription at specific genes, indicating whether genes are silenced, poised for activation or are actively being transcribed. Aside from the importance of these marks as predictors of the unique activities at genes of interest, their presence has accelerated an already emerging field of transcriptional and genomic enzymology associated with the exploration of chromatin active regulators and their capacity to dynamically impose or erase these marks, or to recognize and interpret these marks, presumably to affect downstream functions associated with the genome (6,7). Perhaps most importantly, the presence of many of these marks can now be overlaid across regions with small nucleotide polymorphisms (SNPs) associated with human disease, further supporting the idea that small changes in sequence, often times highly remote relative to neighboring target genes, can be associated with altered transcriptional output (8,9). The evidence for these functional linkages to minor changes in DNA sequence is rapidly accumulating.
In this brief review, we explore the ability of certain histone modifications to identify site-specific structural and functional features of genes expressed within the osteoblast lineage and to efficiently highlight regions that either contain pre-bound transcription factors or contain binding sites to which conditional transcription factors can be recruited. We also explore the dynamic nature by which these marks are not only altered during osteoblast differentiation, but influence the binding of key factors such as the master regulator RUNX2 and the inducible receptor for vitamin D (VDR) thus altering the transcriptome of cells as they become differentiated. The dynamic nature of these modifications suggests that the primary determinants of cellular response are not limited to changes in expression of transcription factors and their interaction with the genome, but also include changes to the target genome as well. The importance of this issue is highlighted by the fact that dynamic changes similarly occur to cellular genomes as a function of progression of diseases such as in cancer as well as throughout differentiation in the skeleton (10,11).
1. HISTONE MARKS AND DIFFERENTIATION
The Structural/Functional Significance of Known Histone Marks
A considerable effort over the past few years has led to the observation that epigenetic marks, whether at histones or on DNA, are dynamic and highly cell-specific, indicating that they impose strong functional consequences on gene expression profiles and are thus linked directly to differences in cellular phenotype (12). These can be seen, for example, in the unique transcriptomes of cells of the osteoblast lineage relative to those of many other lineages. However, these marks also identify common structural/functional features of genes in all cell types, highlighting their utility in defining domains at individual gene loci that represent regulatory regions (enhancers and repressors), transcriptional start sites, promoters, transcription and elongation functions, and marks that are indicators of overall chromatin condensation and activity (12). In many cases, the assignation of a particular common feature such as an enhancer at a gene requires the integration of more than one mark (or the absence of a mark), particularly as it relates to regions of regulatory significance (13,14). Using ChIP-seq analysis, we and others have shown that the presence of individual histone marks is particularly evident across genes that are expressed uniquely in the osteoblast lineage (15–17). Practically speaking, however, the presence of these marks as well as that of RNA polymerase II (RNA pol II) occupancy has been used to identify the regulatory regions of genes such as Tnfsf11 in T cells when the transcription factor(s) that controls expression of this gene is unknown (18). Additionally, a change in the content of many of these marks and their associations with specific genes can either presage ongoing developmental transitions or highlight the presence of clinical disease not only in bone but in other tissues as well.
The Dynamic Nature of Epigenetic Histone Marks During Osteoblast Differentiation
There is considerable evidence that cellular differentiation is accompanied by progressive changes to the epigenome, as measured by both qualitative and quantitative changes in histone marks across the genome (19). As the chromatin regulators that impose these marks do not have DNA sequence binding specificity per se, it seems likely that these factors are recruited to sites both early in development of the lineage and then later to additional sites during either differentiation or in mature cells, at least in part, through changes in the level of occupancy and/or activity of resident DNA sequence-specific transcription factors, a subject that is currently an active area of interest. Indeed, the underlying mechanisms for these changes have been explored in detail using developing T cells and macrophages (20–24). In these cases, it is clear that small collections of lineage determining factors play paramount roles in establishing the cell-specific enhancer landscapes de novo whereupon additional regulatory factors are then recruited in a selective fashion. We hypothesized early on that the differentiation of early osteoblast precursors to mineralizing osteoblasts and then further to osteocytes might follow this pattern as well. Accordingly, we contrasted the profiles of multiple histone modification across the mouse pre-osteoblastic MC3T3-E1 genome (POB) and in these same cells following 15 days of differentiation in vitro in osteogenic medium (OB) (15). We also examined these histone profiles in both IDG-SW3 osteoblasts (day 3) and differentiated osteocytes (day 35) (17). Of initial interest, we discovered that despite phenotypic differences in the MC3T3-E1 and IDG-SW3 mouse cell lines and dissimilarities in their differentiation states, the epigenetic histone landscapes on a genome-wide scale relative to a number of histone modifications were quite similar, suggesting that these marks were highly diagnostic for cells of the osteoblast lineage. We also discovered that while changes in the majority of histone modifications we examined were generally unaltered as a function of differentiation when linked to the transcriptome, they were highly correlated when contrasted exclusively with genes whose expression levels were changed as a result of differentiation. Of particular importance were the changes observed at H3K4me1, H3K4me2, H3K9ac, H3K27ac and H4K5ac, modifications that denote the locations of enhancers or that highlight variations in chromatin decondensation. Numerous changes were also noted at H3K4me3, a mark that specifies the location of gene promoters, and at H3K36me3, H4K20me1 and H4K5ac, marks that identify genomic regions spanning the transcription units (exons and introns) of genes. Interestingly, a bioinformatic examination of enhancer marks revealed that while quantitative changes in the levels of these signature marks were apparent at osteoblast regulated genes, locations where histone marks were newly commissioned or where existing marks were decommissioned were infrequently observed (15,17). Although the appearance of new marks is relatively subjective (relative to what is deemed the residual level of a basal histone mark versus background noise), the results do suggest that the programmed creation of the vast majority of regulatory enhancers in cells of the osteoblast lineage likely occurs early in the mesenchymal lineage. Moreover, current thought suggests a hierarchical model in which small collections of early transcription factors responsible for lineage development and differentiation conspire to establish an appropriate repertoire of regulatory enhancers (25). With respect to osteoblast lineage cells, it seems likely that the majority of these sites are established as a result of the expression of master regulators of early osteoblastogenesis such as RUNX2, OSTERIX and perhaps C/EBPβ (26–28).
Recent results suggest that local microenvironments can influence not only the epigenomic profile of specific cell types, but can result in the reprogramming of these cell types regardless of the apparent differences in their enhancer landscape. It seems likely that this high degree of cellular dynamism in macrophages portends the possibility for an increase in phenotype complexity in bone cells at different skeletal sites as well. Regardless of this speculation, it is clear that the striking changes to the epigenome and transcriptome that occur in bone cells during the differentiation process already have profound implications for skeletal homeostasis, endocrine function, adaptation to external stimuli, and communication with adjacent tissues such as fat, muscle, and the vascular system. In short, progressive changes to the genome during differentiation, a requirement for skeletal maintenance and function, suggests that this process represents not just a mechanism for the replenishment of skeletal cells with short half-lives but the availability of a continuum of cells with variable phenotypes and functions that likely function in unique and tissue site-specific ways.
2. VITAMIN D HORMONE ACTION IN OSTEOBLAST-LINEAGE CELLS AND THE IMPACT OF DIFFERENTIATION
A broad array of steroid and lipophilic hormones are active in the skeleton including the sex steroids, the glucocorticoids, thyroid hormone, the retinoids, PPAR ligands and the vitamin D hormone 1,25-dihydroxyvitamin D3 (1,25(OH)2D3). While recent studies have shown that a number of chromatin regulatory factors play significant roles in bone cells, thus altering skeletal dynamics related to either osteoblast lineage cells (29,30), little is known of the impact of most nuclear receptors on epigenetic histone modifications. As a result, we focus in the following section on our recent studies which examined the actions 1,25(OH)2D3 on osteoblast lineage cells.
The VDR Cistrome in Osteoblasts
Many genes are now known to represent direct mechanistic targets for the VDR. Recently, however, we and others have defined the nature of the VDR cistrome on a genome-wide scale in osteoblast precursors, their derivatives and in additional cell types as well (17,31–36) using ChIP-seq based analyses. Based largely upon our own summarized work, we have compiled a set of overarching principles through which the VDR acts to regulate the expression of genes as summarized in Table 1. These principles include both a genome-wide confirmation of previously held concepts as well as new principles/features identifiable through the advent of ChIP-seq analyses. Although all findings are interesting, perhaps one of the most important observations to emerge is the discovery that many, if not most, regulatory enhancers for the VDR, and indeed for almost all transcription factors, are located distal to gene promoters, frequently intronic or intergenic and often 10’s if not 100’s of kilobases from a target gene’s promoter. This particular finding suggests that a re-examination of many of the genes that were characterized early on by traditional methods may be necessary to determine the full complement and complexity of their regulatory components. Interestingly, the observation that regulatory regions can be found in remote locations relative to specific genes is important not only from a basic but also from a clinical perspective, as it is now clear that altered expression patterns for many disease-related genes are due to the presence of SNPs located in or near binding sites for key transcription factors that control their expression.
TABLE 1.
Overarching principles of 1,25(OH)2D3-mediated gene regulation in target cells. Principles in black represent those previously defined and now confirmed be genome-wide analysis. Principles in red represent newly defined genome-wide features of vitamin D action.
| VDR Binding Sites (The Cistrome): 2,000–8,000 1,25(OH)2D3-sensitive binding sites/genome whose number and location are determined by cell-type |
| Active Transcription Unit: The VDR/RXR heterodimer |
| Distal Binding Site Locations: Dispersed in cis-regulatory modules (CRMs or enhancers) across the genome; located in a cell-type specific manner near promoters, but predominantly within introns and distal intergenic regions; frequently located in clusters of elements |
| VDR/RXR Binding Site Sequence (VDRE): Induction mediated by classic hexameric half-sites (AGGTCA) separated by 3 base pairs; Repression mediated by divergent sites |
| Mode of DNA Binding: Predominantly, but not exclusively, 1,25(OH)2D3-dependent |
| Modular Features: CRMs contain binding sites for multiple transcription factors that facilitate both independent and synergistic interaction |
| Epigenetic CRM Signatures: Defined by the dynamically regulated post-translational histone H3 and H4 modifications and selectively regulated by 1,25(OH)2D3 |
| VDR Cistromes are highly dynamic: Cistromes change during cell differentiation, maturation, and disease activation and thus have consequential effects on gene expression |
VDR Regulation of Histone Acetylation in Bone Cells
Our initial studies of vitamin D action using ChIP-qPCR analysis revealed that VDR binding at the proximal elements associated with Spp1 and Cyp24a1 gene regulation resulted in a differential increase in the level histone H3 and H4 acetylation at these genes, suggesting the existence of a chromatin response to the actions of 1,25(OH)2D3 that might be gene-selective (37). Subsequent studies of the genes for Vdr, Tnfsf11 and others support this view (38,39). Consistent with these observations, we subsequently discovered that the effects of VDR binding on a genome-wide scale in osteoblasts and osteocytes also reflects this premise (17,35). Accordingly, while acetylation levels of H3K9, H4K5 and H3K27 were increased at sites of VDR action near many genes, sites in other genes were unaffected. It has long been known that one of the functions of the VDR in gene activation is to initiate the recruitment of coregulatory factors that include CBP, p300, and the SRC family of histone acetyltransferases (HATs) as well as histone deacetyltransferases (HDACs) (40). It is clear that the actions of these enzymes at histones associated with many genes likely account for the changes in acetylation that are observed, although the mechanism that underlies this site-selectivity is not understood. It seems likely that the requirement for gene activation differs among individual genes, perhaps based upon the nature of the residual expression level of the gene in question and the presence of additional transcription factors that contribute to this level of expression.
Acetylation levels represent a hallmark of chromatin decondensation and transcription factor accessibility to binding sites on DNA, particularly if access to those sites is restricted due to nucleosome positioning (41). Alternative explanations as to the role of increased acetylation as well as methylation include the possibility that site-specific increases lead to the recruitment of additional chromatin regulators that are necessary for nucleosomal redistribution, eviction or exchange, thereby enabling enhancer/promoter engagement through DNA reorganization (42). Separate studies using chromosome conformation capture (3C) have shown, for example, that the presence of estrogen and the estrogen receptor (ER) at distal enhancers facilitates this type of DNA reorganization (43). Interestingly, while our studies of this event at the Tnfsf11 (RANKL) and Cyp24a1 genes have shown linkage between the promoters for these genes and their associated distal enhancers, they do not appear to be influenced by 1,25(OH)2D3 (18,44). Increased methylation at specific sites on histones likewise precipitates changes in gene output, likely due in this case to the selective recruitment of chromatin regulators known as “readers” whose downstream actions are currently being characterized (45,46). Future studies will be required to delineate the consequence of increased acetylation and methylation by VDR at the molecular level and identify the specific players that are involved. Nevertheless, the observation that activated VDR initiates enhanced expression of specific chromatin regulators as well as their recruitment to genes provides an initial starting point.
The Role of RUNX2 and C/EBPβ as a Basal Regulator of Gene Expression in Osteoblasts and as both a Determinant and Facilitator of Vitamin D Action
Recent studies have shown that the distribution of RUNX2 across the osteoblast genome is altered as a result of differentiation, resulting in a contraction of the number of binding sites that is accompanied by a gross reduction in the osteoblast transcriptome (15,47). Surprisingly, approximately 25% of RUNX2 binding sites in both immature and mineralizing osteoblasts also contain C/EBPβ (15), supporting the dominant role for these two chromatin regulatory factors in the establishing the osteoblast phenotype. Interestingly, further examination revealed that RUNX2 could be found at 70% of the sites that bound the VDR/RXR heterodimer and that both RUNX2 and C/EBPβ could be found at 42% of these sites (35). A more detailed examination identified an even closer physical relationship wherein RUNX2 and C/EBPβ were found to bind 8 and 9 bp bi-directionally, respectively, from VDR/RXR peak centers, prompting its description as an “osteoblast enhancer complex”. As RUNX2 and C/EBPβ are independently active in the regulation of gene expression in osteoblast lineage cells, these findings suggest that enhancers of this type are likely capable of mediating both the independent actions of these transcription factors including the VDR, and perhaps integrating the actions of all three. Interestingly, other transcription factor arrangements for VDR/RXR, RUNX2 and C/EBPβ are also apparent as well. Thus, many genes including Spp1 and Mmp13 retain multiple enhancers each capable of independently binding RUNX2, C/EBPβ or the VDR (35). Given the linear distances between individual enhancers in these examples, we speculate that the activities of each regulatory module is manifested either independently or integrated collectively at target gene promoters via complex DNA looping in a manner that is reminiscent of that seen for the osteoblast enhancer complex. Interestingly, in view of the chromatin remodeling and complex transcriptional regulatory properties of the two master regulators RUNX2 and C/EBPβ (26,48,49), the role of these factors at enhancers is likely to be several fold. First, the prebound nature of these two factors and their broad master regulatory properties suggest that they may play a key role as enablers in establishing and maintaining a set of functional enhancers that are not only relevant to the osteoblast lineage, but essential for facilitating the availability of sites to which the VDR and other secondary regulators can be recruited. Second, the complex actions of these two factors in mediating direct regulatory actions in response to a wide variety of signaling pathways suggests that they may also contribute directly to gene regulation by 1,25(OH)2D3, in some cases potentiating and in others suppressing the hormone’s activity. If this hypothesis is correct, while the VDR is a primary determinant of vitamin D action, these two factors and likely others are also determinants of the quantitative and qualitative nature of the response, in part by contributing to processes such as histone modification that controls the output of gene expression.
The Impact of Osteoblast Differentiation on Cellular Response to 1,25(OH)2D3
The biological effects of 1,25(OH)2D3 on osteoblast lineage cells differ significantly and are clearly dependent upon the cellular state of maturation (50,51). In immature POBs, for example, 1,25(OH)2D3 is known to downregulate both RUNX2 and OSX and upregulate C/EBPβ expression, manifesting a negative impact on differentiation. The hormone also controls the expression of a number of additional genes including those that encode regulatory factors involved in the selective modulation of cellular function, regulators such as Spp1, Enpp1, Enpp3, Ank, and Alpl that control mineralization (52), and bone remodeling regulators such as Tnfsf11b and Tnfsf11 that control bone resorption (53). These findings raise the important underlying mechanistic question of how osteoblast differentiation can impact response to 1,25(OH)2D3. The discussion earlier in this minireview described the significant changes that occur to the RUNX2 and C/EBPβ cistromes as a result of osteoblast differentiation as well as the significant epigenetic modifications that occur to histones in a gene-selective fashion. These changes are likely responsible for the striking alteration in the transcriptome that is observed in the differentiated cell. We hypothesize that these transcription factor changes and the epigenomic alterations that are observed following differentiation are likely to alter cellular response to 1,25(OH)2D3. Transcriptomic response to 1,25(OH)2D3 is indeed changed as a function of differentiation, with a restriction in target gene regulation the major consequence (35). We also noted that these transcriptional changes were accompanied by a significant modification to the VDR cistrome, due, in part, to a differentiation-induced reduction in VDR expression that resulted in the absence of the VDR at numerous sites and a reduction at others. We also observed that 1,25(OH)2D3 caused a modest redistribution of RUNX2 and C/EBPβ binding across the genome in early osteoblasts suggesting that in addition to 1,25(OH)2D3’s ability to affect RUNX2 expression it was also capable of affecting the presence of RUNX2 at selected sites as well. Surprisingly, however, we found that despite the fact that 1,25(OH)2D3 no longer regulated the expression of many genes, a large cohort not only retained sensitive to 1,25(OH)2D3, but exhibited increased responsiveness to the hormone. Indeed, some genes manifested a response that was opposite that seen in immature cells. Interestingly, examination of the RUNX2 and C/EBPβ binding sites near these genes revealed frequent changes in the levels of these factors despite only limited differentiation-induced effect on the levels of RUNX2 and C/EBPβ protein. These changes suggest that transcription factor occupancy and activity can be altered during the course of differentiation, that these parameters can be affected by 1,25(OH)2D3, and that the accumulated changes are able to impact the qualitative nature of the genetic response to 1,25(OH)2D3. Accordingly, these results support the idea that the changing genome of the differentiated cell plays an important role in determining where and how 1,25(OH)2D3 acts to regulate transcription.
Biological Roles of 1,25(OH)2D3 in Osteocytogenesis and Similarities and Differences with PTH Action
The biological activity of 1,25(OH)2D3 is both to inhibit osteoblast lineage cell differentiation, perhaps through its actions to inhibit RUNX2 expression and/or to suppress Sost expression from mature osteocytes (17), and to stimulate mature cell function as measured by the induction of expression of osteocalcin, RANKL, and inhibitors of mineralization as well as other factors responsible for the osteoblast/osteocyte phenotype (51). We therefore explored whether this process was evident on a genome-wide scale during the osteoblast to osteocyte transition. As stated earlier, this transition resulted in a dramatic change in the transcriptome, silencing the expression of numerous osteoblastic genes, altering the overall expression levels (both up and down) of a large gene cohort common to both osteoblasts and osteocytes, and inducing the expression of numerous osteocyte-specific genes. Perhaps most importantly, we noted that 1,25(OH)2D3 reversed the expression of a large subset of genes that while active in both osteoblasts and osteocytes were either up- or down-regulated during the differentiation process. In contrast, 1,25(OH)2D3 reinforced the expression of a subset of the gene cohort that was uniquely upregulated in osteocytes. We conclude that in addition to its ability to impede osteoblast differentiation from its earlier precursors, 1,25(OH)2D3 retains its ability to retard osteoblast progression to the osteocyte phenotype. Unexpectedly, the actions of PTH on osteocyte differentiation are similar to those of 1,25(OH)2D3. Thus, PTH also opposes the modulation of genes that are both expressed and regulated in the two cell types during the osteoblast to osteocyte transition while reinforcing the expression of genes that are upregulated specifically in the osteocyte (54). Interestingly, the actions of PTH on the former cohort of genes appears to be mediated through the PKA/CREB signaling pathway while actions on the latter appear independent of CREB activation, as assessed by the general presence or absence of CREB at sites near genes belonging to each cohort (55). Surprisingly, while both 1,25(OH)2D3 and PTH reinforce the expression of genes specific to the osteocyte, the targets of the two hormones were generally independent of each other. These results suggest that while the biological impact of PTH and 1,25(OH)2D3 on changes in gene expression that occur during osteocyte differentiation are similar, their effects on the functional activity of mature osteocytes may be different. They also suggest that the response of these cells to PTH and 1,25(OH)2D3 may be cell transition-specific and thus sensitive to hormonal changes brought on during development and homeostasis and as a result of pathophysiologic states that may alter the levels of these two hormones.
3. SUMMARY
Cellular differentiation results in progressive changes in cellular phenotype and overall function due largely to significant underlying changes in the patterns of gene expression. These changes in gene expression are likely influenced by not only programmed changes in the expression of key transcription factors and other regulatory molecules, but also by pervasive alterations that occur to the genome in the form of diverse epigenetic modifications both to DNA and to histones. In this minireview, we identified many of the changes that occur to both the transcriptome and the epigenome of osteoblast lineage cells during the differentiation process from early osteoblast precursors to terminally differentiated osteocytes. We also described the impact of these changes on cellular response to secondary regulators using the hormone 1,25(OH)2D3 and its receptor as a paradigm. Accordingly, we show that differentiation alters the cistrome for the VDR, likely due to changes in both transcription factor occupancy at VDR target enhancers and selected changes in epigenomic modifications at histones. These changes affect the complex profile of genetic response to 1,25(OH)2D3, eliminating some responses, inducing others, and in some cases qualitatively and quantitatively altering the nature of the response. These findings indicate a profound ability of factors other than the VDR to act as major determinants of cellular response to 1,25(OH)2D3 and thus the hormone’s ability to modulate differentiation and mature cell function. Collectively, these overall findings add a new dimension to our understanding of the role of both the genome and the transcription factor regulome in establishing the cellular context that influences response to hormones such as 1,25(OH)2D3. This molecular and regulatory complexity, and the potential consequence of disease on these events is likely to be significant in vivo.
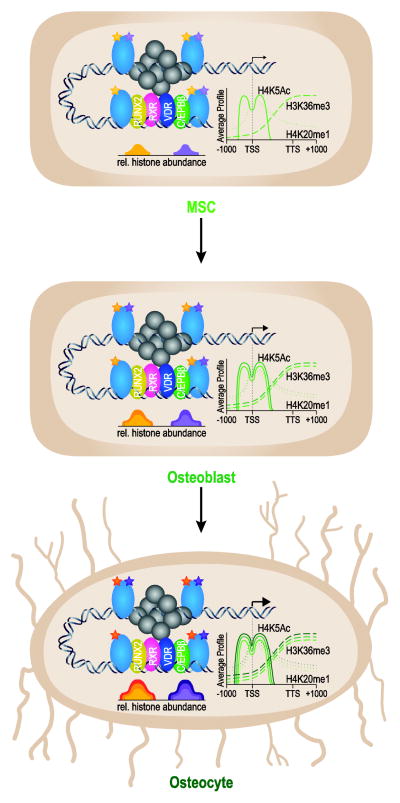
FIGURE 1.
Modulation of transcriptional output during the MSC to osteocyte transition is correlated with epigenetic abundance at enhancers and across the gene body. A model of gene up-regulation is shown schematically with associated changes in several representative histone (light blue) modifications (yellow and purple stars) and their quantitated abundance (bottom peaks) at osteoblast lineage enhancers marked by RUNX2 (yellow), RXR (pink), VDR (blue), and C/EBPβ (green). CEAS (Cis-regulatory element annotation system) analyzed histone modifications (right side) for H3K36me3 (dashed line), H4K20me1 (dotted line), and H4K5ac (solid line) across the gene body are also associated, and increased, with gene expression changes during this transition. The transcriptional start site is depicted by a black arrow (arrow size represents transcriptional output). (See references 15, 17 and 31).
Acknowledgments
We acknowledge the helpful contributions of members of the Pike laboratory in this review and financial support from National Institutes of Health grants from NIDDK (DK-072281, DK-074993) and NIAMS (AR-045173 and AR-062442) to JWP.
Footnotes
Disclosure: The authors declare no conflict of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nat Rev Genet. 112010:476–486. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, Khatun J, Lajoie BR, Landt SG, Lee BK, Pauli F, Rosenbloom KR, Sabo P, Safi A, Sanyal A, Shoresh N, Simon JM, Song L, Trinklein ND, Altshuler RC, Birney E, Brown JB, Cheng C, Djebali S, Dong X, Ernst J, Furey TS, Gerstein M, Giardine B, Greven M, Hardison RC, Harris RS, Herrero J, Hoffman MM, Iyer S, Kelllis M, Kheradpour P, Lassman T, Li Q, Lin X, Marinov GK, Merkel A, Mortazavi A, Parker SC, Reddy TE, Rozowsky J, Schlesinger F, Thurman RE, Wang J, Ward LD, Whitfield TW, Wilder SP, Wu W, Xi HS, Yip KY, Zhuang J, Bernstein BE, Green ED, Gunter C, Snyder M, Pazin MJ, Lowdon RF, Dillon LA, Adams LB, Kelly CJ, Zhang J, Wexler JR, Good PJ, Feingold EA, Crawford GE, Dekker J, Elinitski L, Farnham PJ, Giddings MC, Gingeras TR, Guigó R, Hubbard TJ, Kellis M, Kent WJ, Lieb JD, Margulies EH, Myers RM, Starnatoyannopoulos JA, Tennebaum SA, Weng Z, White KP, Wold B, Yu Y, Wrobel J, Risk BA, Gunawardena HP, Kuiper HC, Maier CW, Xie L, Chen X, Mikkelsen TS, Gillespie S, Goren A, Ram O, Zhang X, Wang L, Issner R, Coyne MJ, Durham T, Ku M, Truong T, Eaton ML, Dobin A, Lassmann T, Tanzer A, Lagarde J, Lin W, Xue C, Williams BA, Zaleski C, Röder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Batut P, Bell I, Bell K, Chakrabortty S, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Li G, Luo OJ, Park E, Preall JB, Presaud K, Ribeca P, Robyr D, Ruan X, Sammeth M, Sandu KS, Schaeffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Hayashizaki Y, Reymond A, Antonarakis SE, Hannon GJ, Ruan Y, Carninci P, Sloan CA, Learned K, Malladi VS, Wong MC, Barber GP, Cline MS, Dreszer TR, Heitner SG, Karolchik D, Kirkup VM, Meyer LR, Long JC, Maddren M, Raney BJ, Grasfeder LL, Giresi PG, Battenhouse A, Sheffield NC, Showers KA, London D, Bhinge AA, Shestak C, Schaner MR, Kim SK, Zhang ZZ, Mieczkowski PA, Mieczkowska JO, Liu Z, McDaniell RM, Ni Y, Rashid NU, Kim MJ, Adar S, Zhang Z, Wang T, Winter D, Keefe D, Iyer VR, Sandhu KS, Zheng M, Wang P, Gertz J, Vielmetter J, Partridge EC, Varley KE, Gasper C, Bansal A, Pepke S, Jain P, Amrhein H, Bowling KM, Anaya M, Cross MK, Muratet MA, Newberry KM, McCue K, Nesmith AS, Fisher-Aylor KI, Pusey B, DeSalvo G, Parker SL, Balasubramanian S, Davis NS, Meadows SK, Eggleston T, Newberry JS, Levy SE, Absher DM, Wong WH, Blow MJ, Visel A, Pennachio LA, Elnitski L, Petrykowska HM, Abyzov A, Aken B, Barrell D, Barson G, Berry A, Bignell A, Boychenko V, Bussotti G, Davidson C, Despacio-Reyes G, Diekhans M, Ezkurdia I, Frankish A, Gilbert J, Gonzalez JM, Griffiths E, Harte R, Hendrix DA, Hunt T, Jungreis I, Kay M, Khurana E, Leng J, Lin MF, Loveland J, Lu Z, Manthravadi D, Mariotti M, Mudge J, Mukherjee G, Notredame C, Pei B, Rodriguez JM, Saunders G, Sboner A, Searle S, Sisu C, Snow C, Steward C, Tapanari E, Tress ML, van Baren MJ, Washieti S, Wilming L, Zadissa A, Zhengdong Z, Brent M, Haussler D, Valencia A, Raymond A, Addleman N, Alexander RP, Auerbach RK, Bettinger K, Bhardwaj N, Boyle AP, Cao AR, Cayting P, Charos A, Cheng Y, Eastman C, Euskirchen G, Fleming JD, Grubert F, Habegger L, Hariharan M, Harmanci A, Iyenger S, Jin VX, Karczewski KJ, Kasowski M, Lacroute P, Lam H, Larnarre-Vincent N, Lian J, Lindahl-Allen M, Min R, Miotto B, Monahan H, Moqtaderi Z, Mu XJ, O’Geen H, Ouyang Z, Patacsil D, Raha D, Ramirez L, Reed B, Shi M, Slifer T, Witt H, Wu L, Xu X, Yan KK, Yang X, Struhl K, Weissman SM, Tenebaum SA, Penalva LO, Karmakar S, Bhanvadia RR, Choudhury A, Domanus M, Ma L, Moran J, Victorsen A, Auer T, Centarin L, Eichenlaub M, Gruhl F, Heerman S, Hoeckendorf B, Inoue D, Kellner T, Kirchmaier S, Mueller C, Reinhardt R, Schertel L, Schneider S, Sinn R, Wittbrodt B, Wittbrodt J, Jain G, Balasundaram G, Bates DL, Byron R, Canfield TK, Diegel MJ, Dunn D, Ebersol AK, Frum T, Garg K, Gist E, Hansen RS, Boatman L, Haugen E, Humbert R, Johnson AK, Johnson EM, Kutyavin TM, Lee K, Lotakis D, Maurano MT, Neph SJ, Neri FV, Nguyen ED, Qu H, Reynolds AP, Roach V, Rynes E, Sanchez ME, Sandstrom RS, Shafer AO, Stergachis AB, Thomas S, Vernot B, Vierstra J, Vong S, Weaver MA, Yan Y, Zhang M, Akey JA, Bender M, Dorschner MO, Groudine M, MacCoss MJ, Navas P, Stamatoyannopoulos G, Stamatoyannopoulos JA, Beal K, Brazma A, Flicek P, Johnson N, Lukk M, Luscombe NM, Sobral D, Vaquerizas JM, Batzoglou S, Sidow A, Hussami N, Kyriazopoulou-Panagiotopoulou S, Libbrecht MW, Schaub MA, Miller W, Bickel PJ, Banfai B, Boley NP, Huang H, Li JJ, Noble WS, Bilmes JA, Buske OJ, Sahu AO, Kharchenko PV, Park PJ, Baker D, Taylor J, Lochovsky L, Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28(8):817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffman MM, Ernst J, Wilder SP, Kundaje A, Harris RS, Libbrecht M, Giardine B, Ellenbogen PM, Bilmes JA, Birney E, Hardison RC, Dunham I, Kellis M, Noble WS. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013;41(2):827–841. doi: 10.1093/nar/gks1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, Min R, Alves P, Abyzov A, Addleman N, Bhardwaj N, Boyle AP, Cayting P, Charos A, Chen DZ, Cheng Y, Clarke D, Eastman C, Euskirchen G, Frietze S, Fu Y, Gertz J, Grubert F, Harmanci A, Jain P, Kasowski M, Lacroute P, Leng J, Lian J, Monahan H, O’Geen H, Ouyang Z, Partridge EC, Patacsil D, Pauli F, Raha D, Ramirez L, Reddy TE, Reed B, Shi M, Slifer T, Wang J, Wu L, Yang X, Yip KY, Zilberman-Schapira G, Batzoglou S, Sidow A, Farnham PJ, Myers RM, Weissman SM, Snyder M. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489(7414):91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12(1):7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 7.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13(5):343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corradin O, Scacheri PC. Enhancer variants: evaluating functions in common disease. Genome Med. 2014;6(10):85. doi: 10.1186/s13073-014-0085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L, Wei D. Bioinformatics tools for discovery and functional analysis of single nucleotide polymorphisms. Adv Exp Med Biol. 2015;827:287–310. doi: 10.1007/978-94-017-9245-5_17. [DOI] [PubMed] [Google Scholar]
- 10.Brookes E, Shi Y. Diverse epigenetic mechanisms of human disease. Annu Rev Genet. 2014;48:237–268. doi: 10.1146/annurev-genet-120213-092518. [DOI] [PubMed] [Google Scholar]
- 11.Conte M, Altucci L. Molecular pathways: the complexity of the epigenome in cancer and recent clinical advances. Clin Cancer Res. 2012;18(20):5526–5534. doi: 10.1158/1078-0432.CCR-12-2037. [DOI] [PubMed] [Google Scholar]
- 12.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yun M, Wu J, Workman JL, Li B. Readers of histone modifications. Cell Res. 2011;21(4):564–578. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linghu C, Zheng H, Zhang L, Zhang J. Discovering common combinatorial histone modification patterns in the human genome. Gene. 2013;518(1):171–178. doi: 10.1016/j.gene.2012.11.038. [DOI] [PubMed] [Google Scholar]
- 15.Meyer MB, Benkusky NA, Pike JW. The RUNX2 cistrome in osteoblasts: characterization, down-regulation following differentiation, and relationship to gene expression. J Biol Chem. 2014;289(23):16016–16031. doi: 10.1074/jbc.M114.552216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu H, Whitfield TW, Gordon JA, Dobson JR, Tai PW, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Genomic occupancy of Runx2 with global expression profiling identifies a novel dimension to control of osteoblastogenesis. Genome Biol. 2014;15(3):R52. doi: 10.1186/gb-2014-15-3-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.St John HC, Bishop KA, Meyer MB, Benkusky NA, Leng N, Kendziorski C, Bonewald LF, Pike JW. The osteoblast to osteocyte transition: epigenetic changes and response to the vitamin d3 hormone. Mol Endocrinol. 2014;28(7):1150–1165. doi: 10.1210/me.2014-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bishop KA, Coy HM, Nerenz RD, Meyer MB, Pike JW. Mouse Rankl expression is regulated in T cells by c-Fos through a cluster of distal regulatory enhancers designated the T cell control region. J Biol Chem. 2011;286(23):20880–20891. doi: 10.1074/jbc.M111.231548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463(7280):474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vigano MA, Ivanek R, Balwierz P, Berninger P, van Nimwegen E, Karjalainen K, Rolink A. An epigenetic profile of early T-cell development from multipotent progenitors to committed T-cell descendants. Eur J Immunol. 2014;44(4):1181–1193. doi: 10.1002/eji.201344022. [DOI] [PubMed] [Google Scholar]
- 21.Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149(2):467–482. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, Ideker T, Glass CK, Murre C. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11(7):635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, Glass CK. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51(3):310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stender JD, Pascual G, Liu W, Kaikkonen MU, Do K, Spann NJ, Boutros M, Perrimon N, Rosenfeld MG, Glass CK. Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20. Mol Cell. 2012;48(1):28–38. doi: 10.1016/j.molcel.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang DX, Glass CK. Towards an understanding of cell-specific functions of signal-dependent transcription factors. J Mol Endocrinol. 2013;51(3):T37–50. doi: 10.1530/JME-13-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol. 2012;13(1):27–38. doi: 10.1038/nrm3254. [DOI] [PubMed] [Google Scholar]
- 27.Karsenty G. Transcriptional control of skeletogenesis. Annu Rev Genomics Hum Genet. 2008;9:183–196. doi: 10.1146/annurev.genom.9.081307.164437. [DOI] [PubMed] [Google Scholar]
- 28.Stein GS, Stein JL, van Wijnen AJ, Lian JB, Zaidi SK, Nickerson JA, Montecino MA, Young DW. An architectural genetic and epigenetic perspective. Integr Biol (Camb) 2011;3(4):297–303. doi: 10.1039/c0ib00103a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dudakovic A, Evans JM, Li Y, Middha S, McGee-Lawrence ME, van Wijnen AJ, Westendorf JJ. Histone deacetylase inhibition promotes osteoblast maturation by altering the histone H4 epigenome and reduces Akt phosphorylation. J Biol Chem. 2013;288(40):28783–28791. doi: 10.1074/jbc.M113.489732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westendorf JJ. Histone deacetylases in control of skeletogenesis. J Cell Biochem. 2007;102(2):332–340. doi: 10.1002/jcb.21486. [DOI] [PubMed] [Google Scholar]
- 31.Meyer MB, Goetsch PD, Pike JW. Genome-wide analysis of the VDR/RXR cistrome in osteoblast cells provides new mechanistic insight into the actions of the vitamin D hormone. J Steroid Biochem Mol Biol. 2010;121(1–2):136–141. doi: 10.1016/j.jsbmb.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer MB, Goetsch PD, Pike JW. VDR/RXR and TCF4/β-catenin cistromes in colonic cells of colorectal tumor origin: impact on c-FOS and c-MYC gene expression. Mol Endocrinol. 2012;26(1):37–51. doi: 10.1210/me.2011-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heikkinen S, Väisänen S, Pehkonen P, Seuter S, Benes V, Carlberg C. Nuclear hormone 1{alpha},25-dihydroxyvitamin D3 elicits a genome-wide shift in the locations of VDR chromatin occupancy. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramagopalan S, Heger A, Berlanga A, Maugeri N, Lincoln M, Burrell A, Handunnetthi L, Handel A, Disanto G, Orton S, Watson C, Morahan J, Giovannoni G, Ponting C, Ebers G, Knight J. A ChIP-seq defined genome-wide map of vitamin D receptor binding: Associations with disease and evolution. Genome Res. 2010 doi: 10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer MB, Benkusky NA, Lee CH, Pike JW. Genomic determinants of gene regulation by 1,25-dihydroxyvitamin D3 during osteoblast-lineage cell differentiation. J Biol Chem. 2014;289(28):19539–19554. doi: 10.1074/jbc.M114.578104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding N, Yu RT, Subramaniam N, Sherman MH, Wilson C, Rao R, Leblanc M, Coulter S, He M, Scott C, Lau SL, Atkins AR, Barish GD, Gunton JE, Liddle C, Downes M, Evans RM. A vitamin D receptor/SMAD genomic circuit gates hepatic fibrotic response. Cell. 2013;153(3):601–613. doi: 10.1016/j.cell.2013.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim S, Shevde N, Pike J. 1,25-Dihydroxyvitamin D3 stimulates cyclic vitamin D receptor/retinoid X receptor DNA-binding, co-activator recruitment, and histone acetylation in intact osteoblasts. J Bone Miner Res. 2005;20(2):305–317. doi: 10.1359/JBMR.041112. [DOI] [PubMed] [Google Scholar]
- 38.Zella LA, Kim S, Shevde NK, Pike JW. Enhancers located within two introns of the vitamin D receptor gene mediate transcriptional autoregulation by 1,25-dihydroxyvitamin D3. Mol Endocrinol. 2006;20(6):1231–1247. doi: 10.1210/me.2006-0015. [DOI] [PubMed] [Google Scholar]
- 39.Kim S, Yamazaki M, Zella LA, Shevde NK, Pike JW. Activation of receptor activator of NF-kappaB ligand gene expression by 1,25-dihydroxyvitamin D3 is mediated through multiple long-range enhancers. Mol Cell Biol. 2006;26(17):6469–6486. doi: 10.1128/MCB.00353-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sutton AL, MacDonald PN. Vitamin D: more than a “bone-a-fide” hormone. Mol Endocrinol. 2003;17(5):777–791. doi: 10.1210/me.2002-0363. [DOI] [PubMed] [Google Scholar]
- 41.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 42.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 43.Pan YF, Wansa KD, Liu MH, Zhao B, Hong SZ, Tan PY, Lim KS, Bourque G, Liu ET, Cheung E. Regulation of estrogen receptor-mediated long range transcription via evolutionarily conserved distal response elements. J Biol Chem. 2008;283(47):32977–32988. doi: 10.1074/jbc.M802024200. [DOI] [PubMed] [Google Scholar]
- 44.Meyer MB, Goetsch PD, Pike JW. A downstream intergenic cluster of regulatory enhancers contributes to the induction of CYP24A1 expression by 1alpha,25-dihydroxyvitamin D3. J Biol Chem. 2010;285(20):15599–15610. doi: 10.1074/jbc.M110.119958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25(1):15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 46.Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49(5):825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gordon JA, Montecino MA, Aqeilan RI, Stein JL, Stein GS, Lian JB. Epigenetic pathways regulating bone homeostasis: potential targeting for intervention of skeletal disorders. Curr Osteoporos Rep. 2014;12(4):496–506. doi: 10.1007/s11914-014-0240-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirata M, Kugimiya F, Fukai A, Ohba S, Kawamura N, Ogasawara T, Kawasaki Y, Saito T, Yano F, Ikeda T, Nakamura K, Chung UI, Kawaguchi H. C/EBPbeta Promotes transition from proliferation to hypertrophic differentiation of chondrocytes through transactivation of p57. PLoS One. 2009;4(2):e4543. doi: 10.1371/journal.pone.0004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tominaga H, Maeda S, Hayashi M, Takeda S, Akira S, Komiya S, Nakamura T, Akiyama H, Imamura T. CCAAT/enhancer-binding protein beta promotes osteoblast differentiation by enhancing Runx2 activity with ATF4. Mol Biol Cell. 2008;19(12):5373–5386. doi: 10.1091/mbc.E08-03-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bikle DD. Vitamin D and bone. Curr Osteoporos Rep. 2012;10(2):151–159. doi: 10.1007/s11914-012-0098-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eisman JA, Bouillon R. Vitamin D: direct effects of vitamin D metabolites on bone: lessons from genetically modified mice. Bonekey Rep. 2014;3:499. doi: 10.1038/bonekey.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lieben L, Masuyama R, Torrekens S, Van Looveren R, Schrooten J, Baatsen P, Lafage-Proust MH, Dresselaers T, Feng JQ, Bonewald LF, Meyer MB, Pike JW, Bouillon R, Carmeliet G. Normocalcemia is maintained in mice under conditions of calcium malabsorption by vitamin D-induced inhibition of bone mineralization. J Clin Invest. 2012;122(5):1803–1815. doi: 10.1172/JCI45890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leibbrandt A, Penninger JM. RANK/RANKL: regulators of immune responses and bone physiology. Ann N Y Acad Sci. 2008;1143:123–150. doi: 10.1196/annals.1443.016. [DOI] [PubMed] [Google Scholar]
- 54.St John HC, Meyer MB, Benkusky NA, Carlson AH, Prideaux M, Bonewald LF, Wesley Pike J. The parathyroid hormone-regulated transcriptome in osteocytes: Parallel actions with 1,25-dihydroxyvitamin D3 to oppose gene expression changes during differentiation and to promote mature cell function. Bone. 2014;72C:81–91. doi: 10.1016/j.bone.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bellido T, Saini V, Pajevic PD. Effects of PTH on osteocyte function. Bone. 2013;54(2):250–257. doi: 10.1016/j.bone.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]