Abstract
Neurogenesis in the brain of Xenopus laevis continues throughout larval stages of development. We developed a 2-tier screen to identify candidate genes controlling neurogenesis in Xenopus optic tectum in vivo. First, microarray and NanoString analyses were used to identify candidate genes that were differentially expressed in Sox2-expressing neural progenitor cells or their neuronal progeny. Then an in vivo, time-lapse imaging-based screen was used to test whether morpholinos against 34 candidate genes altered neural progenitor cell proliferation or neuronal differentiation over 3 days in the optic tectum of intact Xenopus tadpoles. We co-electroporated antisense morpholino oligonucleotides against each of the candidate genes with a plasmid that drives GFP expression in Sox2-expressing neural progenitor cells and quantified the effects of morpholinos on neurogenesis. Of the 34 morpholinos tested, 24 altered neural progenitor cell proliferation or neuronal differentiation. The candidates which were tagged as differentially expressed and validated by the in vivo imaging screen include: actn1, arl9, eif3a, elk4, ephb1, fmr1-a, fxr1-1, fbxw7, fgf2, gstp1, hat1, hspa5, lsm6, mecp2, mmp9, and prkaca. Several of these candidates, including fgf2 and elk4, have known or proposed neurogenic functions, thereby validating our strategy to identify candidates. Genes with no previously demonstrated neurogenic functions, gstp1, hspa5 and lsm6, were identified from the morpholino experiments, suggesting that our screen successfully revealed unknown candidates. Genes that are associated with human disease, such as such as mecp2 and fmr1-a, were identified by our screen, providing the groundwork for using Xenopus as an experimental system to probe conserved disease mechanisms. Together the data identify candidate neurogenic regulatory genes and demonstrate that Xenopus is an effective experimental animal to identify and characterize genes that regulate neural progenitor cell proliferation and differentiation in vivo.
Keywords: Neurogenesis, Neural progenitor cell, Radial glia, Microarray, Morpholino, Candidate gene, In vivo imaging, Proliferation, Differentiation
Introduction
The control of cell proliferation and differentiation is essential for proper development of the central nervous system (CNS). At early stages of CNS development, neural stem cells divide symmetrically to expand the neural stem cell pool (Götz and Huttner, 2005; Hardwick and Philpott, 2014). Neural stem cells change fate and undergo asymmetric regenerative divisions to generate both neural stem cells and neurons, which then organize into nascent circuits. Further cell fate changes occur when neural stem cells become quiescent or exit the cell cycle and differentiate into either neurons or astrocytes (Encinas et al., 2006). These cell fate decisions are essential events that control the patterning of the developing brain and ultimately affect brain function (Geschwind and Rakic, 2013; Kriegstein et al., 2006). Recent work has demonstrated that neurogenic cell fate decisions are influenced by the local environment and neural circuit activity (Alvarez-Buylla et al., 2008; Best-man et al., 2012; Conover and Notti, 2008; Encinas et al., 2006; Giachino and Taylor, 2009; Holmes, 2009; Sharma and Cline, 2010; Vergano-Vera et al., 2009), suggesting that an in vivo screen may reveal novel candidate neurogenic regulators.
The Xenopus laevis tadpole is ideally suited to screen for candidate neurogenic genes. Cell proliferation continues throughout the development of the nervous system in Xenopus. In the visual system, for example, new neurons are generated in the optic tectum throughout larval development and integrate into the developing retinotectal circuit. Because the tadpole is transparent at early stages of development, in vivo time-lapse confocal imaging of GFP-expressing progenitor cells in the Xenopus brain allows direct observations of the fates of the proliferating cell population (Bestman et al., 2012). We developed an in vivo screen to identify candidate genes affecting cell proliferation or differentiation in Xenopus tectum. First, we used cDNA microarrays and NanoString analysis to identify transcripts that are differentially expressed between neural progenitor cells (NPCs) and their progeny. Next, a subset of gene candidates was evaluated in a secondary screen: after morpholinos were electroporated to knockdown candidates, differences in proliferation or differentiation were determined by in vivo time-lapse imaging of NPCs and their neuronal progeny. These analyses identified a diverse range of candidate neurogenic genes that modulate proliferation and neuronal differentiation in the brain, thus implicating a variety of regulatory pathways affecting neurogenesis. Mechanisms controlling cell proliferation and differentiation are highly conserved across evolution (Chapouton et al., 2007; Cheung et al., 2007; Kriegstein et al., 2006; Molnar, 2011; Pevny and Nicolis, 2010; Pierfelice et al., 2011) and are fundamental for the evolution of brain structures (Charvet and Striedter, 2011; Finlay et al., 1998). Therefore, identification of regulatory mechanisms affecting neurogenesis in the Xenopus CNS will likely provide insights into neural stem cell fate decisions during the development of the CNS and during adult neurogenesis. Furthermore, a deeper understanding of the underlying mechanisms controlling the balance between cell proliferation and differentiation may also direct the discovery of potential therapeutics for brain injury, developmental disorders, and interventions to replace cells lost by injury and neurodegenerative diseases.
Results
A screen for differentially expressed transcripts from neural progenitor cells and differentiated neurons
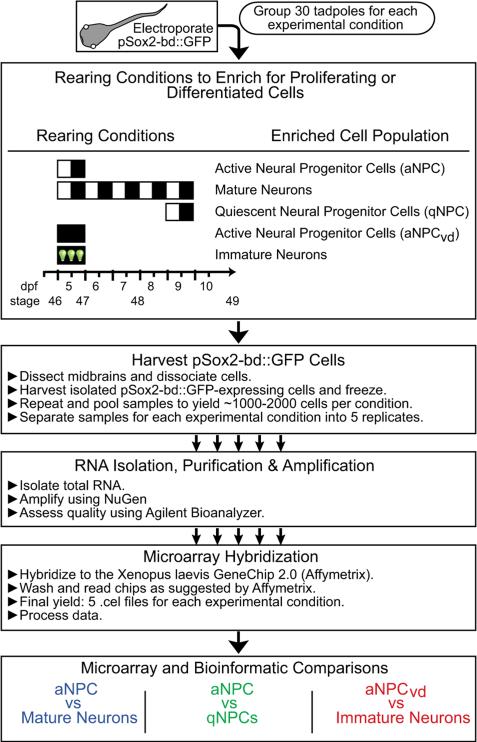
The goal of our study was to identify and evaluate candidate neurogenic genes based on a 2-tiered screen in which microarray and NanoString analyses were used to identify transcripts that might regulate cell proliferation and differentiation in the brain, followed by an in vivo, time-lapse imaging-based screen to test selected candidate genes. We focused our attention on the tadpole optic tectum, where we had established experimental strategies to enrich for actively dividing NPCs, differentiated neurons or quiescent progenitors based on the normal time course of optic tectal cell development and the effects of visual experience (Bestman et al., 2012; Sharma and Cline, 2010). We labeled NPCs and their progeny with a construct that drives GFP reporter expression in Sox2-expressing cells, called pSox2-bd::GFP (Bestman et al., 2012) and isolated GFP-labeled cells that are enriched for active or quiescent NPCs or differentiated neurons (Fig. 1). Our previous work showed that 1 day after transfecting the optic tectum of stage 46 animals with pSox2-bd::GFP, the majority of the GFP-expressing cells are mitotically active NPCs and by three days after transfection, most GFP-expressing cells have differentiated into neurons (Bestman et al., 2012). Pulse-chase labeling tectal progenitors with CldU also demonstrated that the majority of NPCs differentiate into neurons over a two-day period (Sharma and Cline, 2010). Furthermore, rates of cell proliferation in the optic tectum decrease significantly over the five day period between stages 46 and 48 (Sharma and Cline, 2010), suggesting that Sox2-expressing progenitors are relatively quiescent at stage 48/49. We therefore collected GFP-expressing cells at different times during normal rearing to enrich the following cell populations: active NPCs (aNPCs) isolated from animals one day after electroporation at stage 46; Mature Neurons isolated from tadpoles 5 days after electroporation at stage 46; and quiescent progenitors (qNPCs) isolated from the stage 49 tadpoles 1 day after electroporation. Our previous work also showed that rearing stage 46 tadpoles in the dark for 24 h increased the proportion of actively dividing progenitor cells whereas enhanced visual experience drove cells toward neuronal differentiation (Bestman et al., 2012; Sharma and Cline, 2010). Therefore, we manipulated visual experience to produce 2 cell groups enriched for Immature Neurons (isolated from animals that were electroporated at stage 46 and exposed to visual stimulation for the next 24 h) and actively dividing NPCs (isolated from tadpoles that were electroporated at stage 46 and visually deprived for the following 24 h), called aNPCvd (Fig. 1).
Fig. 1.
Flow diagram of the protocols for animal rearing, cell isolation, RNA preparation and microarray hybridization. At stage 46 or 48, tadpoles were electro-porated with a GFP-expression construct and placed in one of three visual experience conditions: normal 12 h light:12 h dark conditions; visual deprivation (vd), or enhanced visual experience. These rearing conditions produced 5 cell groups: active NPCs (aNPCs), Mature Neurons, Immature Neurons, Active NPCs isolated from visually-deprived tadpoles (aNPCvd), and quiescent NPCs (qNPCs). See text for details. GFP+ cells were harvested from dissociated midbrains and RNA was isolated and prepared for microarrays. The bottom panel shows which samples were compared by microarray analysis to identify differentially expressed genes that might be involved in cell proliferation and neurogenesis.
We used multiple microarray analyses to identify transcripts that were differentially expressed in cell populations enriched for aNPCs relative to neurons or qNPCs. To do this, the expression values of the transcripts for five replicates of each of the five experimental conditions were normalized, outliers representing hybridization artifacts were removed and the averages across the probe replicates were calculated. We then made three comparisons of the transcript expression data from the cDNA microarrays between the different experimental conditions described above (Fig. 1). In one comparison, microarray data from aNPCs and Mature Neurons were compared. The second comparison identified transcripts that are differentially expressed in aNPCs compared to the qNPCs. Third, aNPCvd cells were compared to Immature Neurons. Transcripts with p-values <0.05 were considered significant and are provided in Supplementary Table 1. Each of the three comparisons contains an aNPC group that is compared to either qNPCs, Immature Neurons or Mature Neurons. Therefore, we also explore which transcripts in aNPCs are shared across the comparisons to reveal genes that regulate continued cell proliferation in the brain.
Relationships between the multiple microarray comparisons
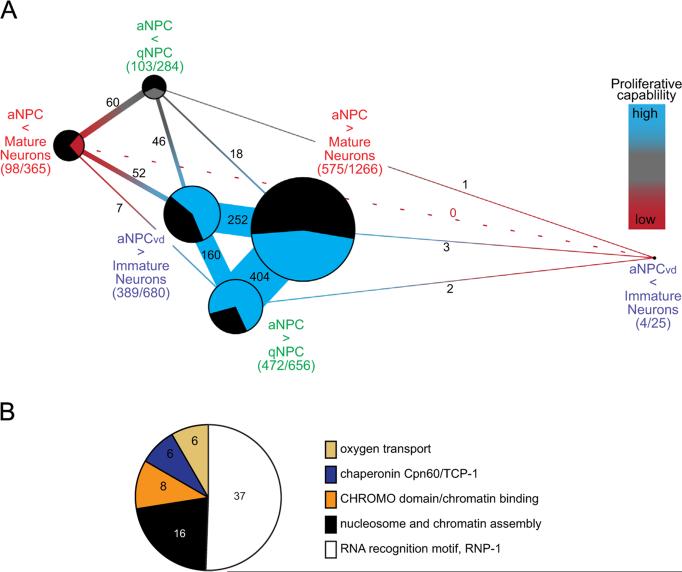
To depict the differentially expressed transcripts shared between aNPCs from the different microarray comparisons, we generated a Venn diagram from the sets of differentially expressed transcripts from the three datasets (Fig. 2A). The areas of the ovals in the diagram represent the relative number of differentially expressed transcripts for each microarray comparison and the total number of transcripts in each set is indicated. The size of the overlap between groups represents the proportion of transcripts shared by the different microarray comparisons. The analysis showed that 309 differentially expressed transcripts were shared between the aNPC vs Mature Neuron comparison and the aNPCvd vs Immature Neuron comparison, whereas 477 differentially expressed transcripts were shared between the aNPC vs qNPC comparison and the aNPCvd vs Immature Neuron comparison. Finally, 210 differentially expressed transcripts were shared between the aNPCvd vs Immature Neuron comparison and the aNPC vs qNPC comparison. 124 (3.8%) differentially expressed transcripts were part of all three microarray comparisons (Fig. 2A, black; Supplementary Table 1). The aNPC vs Mature Neuron set contained the greatest number of unique transcripts (934/1606; 58.2%) that were not shared between the 3 groups, compared to 43.7% (307/702) unique transcripts in the aNPCvd vs Immature Neuron dataset and 38.6% (359/932) unique transcripts in the aNPC vs qNPC dataset.
Fig. 2.
Relationships between the multiple microarray comparisons. (A) Venn diagram of the overlap of the transcripts with differential expression (p<0.05) between the 3 microarray comparisons. About 50% of differentially expressed transcripts were shared between the 3 datasets. Sizes of the ovals represent the number of transcripts showing significant differential expression in each micro-array comparison. The overlap represents the proportion of transcripts shared by multiple microarray comparisons. The aNPC vs Mature Neuron set contains 1606 transcripts, 672 of which were shared with at least one other group. The aNPC vs qNPC comparison has 932 differentially expressed transcripts, and shared 573 transcripts with other comparisons. The aNPCvd vs Immature Neuron comparison had 702 genes with significant differential expression, with 395 shared between the different comparisons. In total, 124 genes were shared between all 3 gene groups. (B) DAVID analyses reveal gene ontology traits from the microarray comparisons. Differentially expressed transcripts with p<0.05 from the aNPC vs qNPC, aNPC vs Immature Neuron, and aNPC vs Mature Neuron microarray comparisons were clustered using the DAVID Functional Annotation Clustering tool. The enriched gene clusters are shown in pie charts for each comparison. The number of transcripts identified in each cluster is indicated in the diagram or in the legend. Transcripts in the nucleosome and chromatin assembly pathways (black) were common to all three microarray comparisons. The RNA recognition RNP-1 family (white) was abundant in both the aNPCvd vs Immature Neuron and the aNPC vs Mature Neurons microarray comparisons. These data are provided in Supplementary Table 2.
The differential expression analysis from the microarrays revealed that well known markers of aNPCs, qNPCs or neurons were significantly expressed in the different cell groups (all genes with significant differential expression are listed in Supplementary Table 1). For example, the intermediate filament protein and marker of radial glial progenitor cells, vimentin, had significant differential expression in each microarray comparison. Other genes associated with proliferating NPCs, the astrocyte specific l-glutamate/l-aspartate transporter, slc1a3/GLAST, and the gluta-mate–ammonia ligase, glul, were differentially expressed in the aNPCvd vs Immature Neuron and aNPC vs qNPC comparisons. The analysis also revealed that empty spiracles homeobox 2 (emx2), a regulator of NPCs (Falcone et al., 2015), showed significant differential expression in all 3 comparisons. Similarly, members of the sox gene family (sox 2, 3, 5, 9, and 11), and the POU domain transcription factor family (pou3f2, pou3f4 and pou4f1), transcription factors which regulate in the maintenance of the progenitor pool and differentiation of neurons (Tantin, 2013; Uy et al., 2015), were differentially expressed in multiple microarray comparisons. Genes that direct neurogenesis or are expressed in post-mitotic neurons, such as members 1, 4 and 6 of the neurogenic differentiation (NeuroD) gene family, members of the α and β-tubulin gene family, and the early B cell factors 1 and 2, for example, were also differentially expressed across the microarray comparisons (Supplementary Table 1). Overall, the significant differential expression of these and other transcripts indicates that our methods for harvesting cells and performing the microarray analyses successfully identified genes known to be differentially expressed in our target cell populations.
Bioinformatic analyses of the differentially expressed transcripts
Analyses of gene families and signaling pathways provide a broader view of transcriptionally regulated cellular processes during neurogenesis than an analysis of single transcripts. We conducted a bioinformatic analysis using DAVID (the Database for Annotation, Visualization and Integrated Discovery (Huang et al., 2009)) to investigate whether gene families or pathways were expressed in the microarray datasets. DAVID identifies biological processes, protein domains and other annotation terms in a gene list that are significantly enriched compared to their corresponding frequencies in a background gene list. We used the transcripts from the Affymetrix X. laevis 2.0 microarray chip as the background gene set. Using the lists of transcripts with significant differential expression from the microarray comparisons, Fig. 2B shows the gene clusters that the DAVID Functional Annotation Clustering algorithm identified as enriched relative to the expected numbers from the microarray background. The DAVID algorithm detected 2 gene clusters in the transcript list from the aNPC–qNPCs comparison, 3 gene clusters in the list from the aNPCvd–Immature Neurons comparison, and 9 gene clusters in the list from the aNPC–Mature Neurons comparison, shown in the pie charts in Fig. 2B. The DAVID-generated gene lists, p-values and Benjamini false discovery rates are provided in Supplementary Table 2.
In all three microarray comparisons, DAVID identified a cluster of “nucleosome and chromatin assembly” genes (black, Fig. 2B), a family known to regulate the balance between cell proliferation and differentiation in the brain (Lilja et al., 2013; Fig. 2B and Supplementary Table 2). The “RNA recognition motif, RNP-1” gene cluster (white, Fig. 2B), which includes the proliferation regulator Musashi (Okano et al., 2005), was identified in both the aNPC vs qNPC and the aNPC vs Mature Neuron microarray comparisons. Two additional gene clusters involved in RNA regulation were enriched in the aNPC vs Mature Neuron microarray comparison: the “Sm-like ribonucleoprotein, core” protein family (yellow) composed mainly of heterogeneous nuclear ribonucleoproteins and RNA splicing factors, and the functionally diverse genes represented in the “nucleoside binding/tRNA aminoacylation” cluster (gray). The aNPC vs Mature Neuron microarray comparison also contained two clusters associated with mitochondrial function, the “purine nucleotide metabolic processes/ATP synthesis coupled proton transport” cluster (magenta) and the “cytochrome c oxidase activity” cluster (light blue), which may reflect the increase in mitochondrial abundance that accompanies cell differentiation (Wanet et al., 2012). In addition, the “proteasome core complex” (purple) and “actin cytoskeleton regulation” (red) clusters were also enriched among the differentially expressed genes from the aNPC vs Mature Neuron microarray comparison. Two annotation clusters were only associated in the aNPCvd vs Immature Neuron comparison: “iron storage” (blue) and “oxygen transport” (tan). Genes in the “iron storage” cluster, such ferritin light and heavy chain proteins, and in the “oxygen transport” cluster, such as hemoglobin genes, have been associated with oxygen uptake in neural tissue (Schelshorn et al., 2009). Therefore the DAVID analysis identified gene families connected to the processes that govern cell proliferation or neuronal function.
Fig. 2A demonstrated that ~50% of all the differentially expressed genes are shared between at least two of the microarray comparisons. To explore this further, we were interested in whether the differentially expressed genes were similarly regulated in each group, that is, whether they were up- or down-regulated only in cells with high proliferative capacity or only the cells with low proliferative capacity. In all three microarray comparisons, positive differential expression values represent genes that are more highly expressed under conditions that favor active cell proliferation (aNPC or aNPCvd populations) compared to conditions of low proliferation (qNPC, Immature Neuron, or Mature Neuron populations). We expected that genes shared across groups would be similarly regulated (e.g. positive differential expression value in multiple aNPC groups). To determine whether the transcripts shared between groups were similarly regulated, we determined the ratio of upregulated/total differentially expressed transcripts and mapped the network of shared transcripts using the Gephi clustering algorithm (Bastian et al., 2009; Fig. 3A). The gene network generated by Gephi plots the transcripts that were upregulated in aNPCs compared to the other cell groups (blue circles), transcripts that were upregulated in the two neuron groups compared to aNPCs (red), and transcripts that were upregulated in the qNPC group (gray) compared to aNPCs (Fig. 3A). The sizes of the circles, or nodes, represent the relative number of differentially expressed transcripts in each paired comparison. The blue portions of the circles at each node of the network represent the proportion of genes that are upregulated and the black portions show the proportion of genes that are unique to the group and unshared. The ratio of upregulated/total differentially expressed transcripts is shown next to each node. The aNPCvd vs Immature Neuron microarray comparison contained 389/680 transcripts with higher expression in the aNPCs (blue) and 4/25 that were more highly expressed in immature neurons (red; Fig. 3A). Similarly, the 575/1266 transcripts in the aNPC vs Mature Neuron comparison (blue) were upregulated in the NPCs compared to 98/365 transcripts that were more highly expressed in Mature Neurons (red). Finally, in the aNPC vs qNPC comparison, 472/656 transcripts were upregulated in the aNPCs compared to 103/284 upregulated transcripts in qNPCs (gray). This analysis also demonstrates a high degree of overlap in the upregulated transcripts across different aNPC groups, indicated as the connecting blue bars between the blue and black nodes. The aNPCvd group shares 252 upregulated transcripts with the aNPCs in the aNPC vs Mature Neuron comparison and 160 upregulated transcripts with the aNPCs of the aNPC vs qNPC group. Furthermore, the aNPC groups shared 404 transcripts between the Mature Neurons and qNPCs comparisons.
Fig. 3.
aNPCs share networks of differentially expressed transcripts. (A) Networks of differentially-expressed transcripts that are shared between NPCs. Transcripts with positive expression values (more highly expressed in NPCs) or negative expression values (more highly expressed in neurons or quiescent NPCs) were analyzed separately. Each node of the network indicates the groups of positively (blue) or negatively (red)-expressed genes for each microarray comparison, nodes separated by short distances and thicker connections indicate that the groups share many transcripts. The size of the pie diagram at each node represents the total number of differentially expressed genes and the number of unique genes unshared (black) and shared between groups (blue, gray or red). Blue represents genes enriched in aNPCs with high proliferative capability, gray represents enrichment in qNPCs and red indicates enrichment in neurons with the lowest proliferative capability. Numbers on the connecting segments are the numbers of transcripts in common between the groups. (B) DAVID Functional Annotation Clustering tool identified 5 gene families that are enriched in multiple aNPC datasets: nucleosome and chromatin assembly genes, RNA recognition motif RNP-1 family, CHROMO domain containing chromatin binding genes, the TCP-1 chaperonin family and genes associated with oxygen transport.
In contrast, Gephi identified relatively few transcripts that are upregulated in the cell groups with low proliferative capabilities (the aNPC<Mature Neurons (red), aNPC<qNPC (gray) and aNPCvd<Immature Neuron groups), and fewer transcripts were shared between these conditions. For example, only 4 of the 25 differentially expressed transcripts were upregulated in the Immature Neurons compared to aNPCvd group, and none were shared with the upregulated transcripts in the Mature Neuron group. Of the 98/365 upregulated transcripts in the Mature Neuron group, 60 were similarly upregulated in the qNPC set and 52 were shared with the aNPCvd group. Similarly, the transcripts that were upregulated in qNPCs compared to aNPCs were roughly equally shared with cell groups with low proliferative capability (60 transcripts in common with upregulated transcripts in Mature Neurons) and with cell groups with higher proliferative activity (18 with the aNPC>Mature Neurons and 46 with the aNPCvd>Immature Neuron). Together these analyses indicate that upregulated transcripts are a larger proportion of the differentially expressed transcripts in aNPCs and that more upregulated transcripts are shared between aNPCs under different conditions, but fewer upregulated transcripts are shared between the cell groups with lower proliferative activity.
We were interested in whether applying the DAVID Functional Clustering tools to the list of genes that were either shared between two or more of groups with high proliferative capability or low proliferative capability would reveal enriched biological pathways or processes. Of the 61 genes shared between the groups with low proliferative capabilities, there were 44 unique genes, and the DAVID analysis did not reveal any enriched biological processes. The 815 genes shared between at least 2 different aNPC groups (blue connecting lines in Fig. 3A) included 615 unique transcripts. We reasoned that a bioinformatic analysis of these transcripts might identify genes that were particularly important for maintaining cell proliferation in the brain. DAVID Functional Annotation Clustering tools identified 5 biological pathways or processes that were abundant in this transcript group (Fig. 3B and Supplementary Table 2). As expected, we found that some annotation pathways were shared with the gene sets in Fig. 2B, including the “RNA recognition motif, RNP-1” family, “nucleosome and chromatin assembly” transcripts, and transcripts involved in “oxygen transport”. Two unique gene groups were also identified: “CHROMO domain/chromatin binding” transcripts, which encode heterochromatin proteins involved in the regulation of transcription, and transcripts in the “chaperonin Cpn60/TCP-1” family, known regulators of the cytoskeleton and cell cycle (Brackley and Grantham, 2009; Fig. 3B).
Identification of candidate human genes and disease pathways
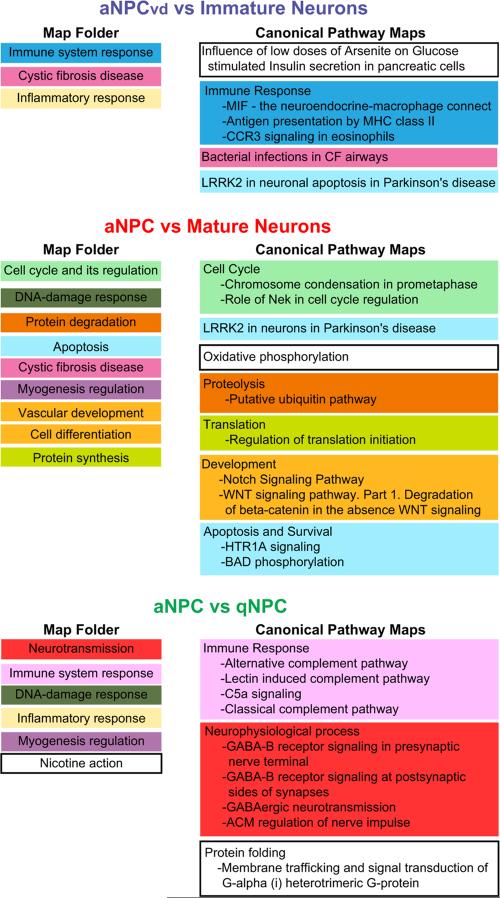
A second analysis of the differentially expressed transcripts from the three microarray comparisons was conducted using MetaCore, which identifies enriched canonical pathways compared to a background gene list using a proprietary literature database of human genes. Consequently, analysis using MetaCore is intended to identify conserved pathways and to highlight candidate neurogenic pathways that may be active in the human brain. Starting with transcripts that are differentially expressed in the Xenopus micro-arrays, we identified their human homologs and used them to search MetaCore. MetaCore analysis of the differentially expressed genes from both the aNPCvd vs Immature Neuron and the aNPC vs Mature Neuron microarray comparisons identified pathways in the immune, inflammatory and stress response pathways (Fig. 4, Supplementary Table 3). It is noteworthy that many genes that represent immune and stress responses in MetaCore have also been shown to associate with cell cycle processes. For example, thioredoxin was identified in the aNPCvd vs Immature Neuron comparison. It is a key component of redox regulation, and is part of the stress response pathway, however, thioredoxin also regulates the G1 phase of the cell cycle by controlling cyclin D1 transcription and the ERK/AP-1 signaling pathways (Mochizuki et al., 2009). The genes AP-1, c-Jun, Rac2 and the C3 and C5a complement signaling components are part of the immune response pathways identified in the aNPC vs qNPC comparison, although each of these genes is also involved in cell cycle control (Daveau et al., 2004; Gu et al., 2003; Schonthaler et al., 2011; Shaulian and Karin, 2001). In addition, analysis of the differentially expressed transcripts from the aNPC vs Mature Neuron group revealed that ubiquitin proteolytic pathways, as well as WNT and Notch signaling pathways, which are known regulators of cell fate and cell proliferation (Gaiano et al., 2000), were up-regulated in aNPCs.
Fig. 4.
MetaCore analysis of differentially expressed transcripts in NPCs and Neurons. Map Folders (left column), which identify broad functional categories, and Canonical Pathways Maps (right column), which identify more specific candidate interaction pathways, are listed in the order of significance, from top to bottom of the lists. Pathway Maps that are within Map Folders are color coded. The top 10 significant pathways from MetaCore (p<0.05 and False Discovery Rate <0.05) are presented here. Specific components of the Pathway Maps that were identified in the microarray comparisons are shown in Supplementary Table 3.
NanoString analysis of NPC and neuronal transcripts
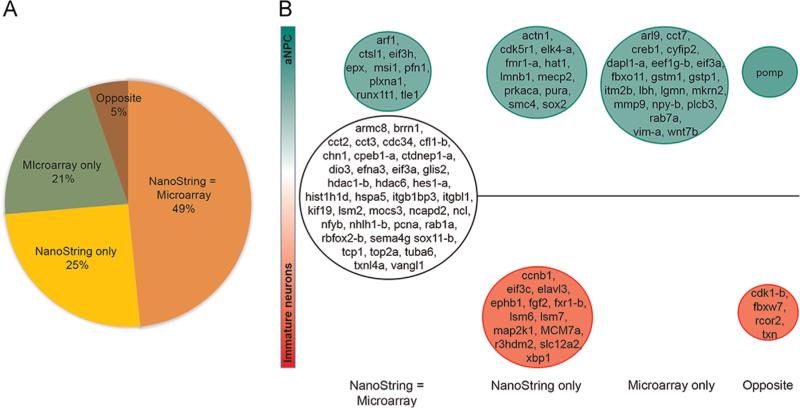
We used NanoString as an independent means to compare expression of a subset of transcripts from independent samples of aNPCvds and Immature Neurons. We selected 95 transcripts that included a subset of differentially-expressed transcripts identified by the microarray analysis and other transcripts in signaling pathways flagged by the differentially-expressed candidates. Of these, 46 transcripts shared the same expression profile in both the microarray and NanoString assays, 20 transcripts were differentially expressed in the microarray assays but not in the Nano-String, and 24 transcripts were differentially expressed in the NanoString assay but not the microarrays. Only 5 transcripts showed differential expression in opposite directions in the aNPCvd and Immature Neuron comparison in the NanoString and microarray analyses (Fig. 5, Supplementary Table 4). As further validation of our strategy to isolate populations of cells enriched for neurons or NPCs, elavl3, a neuron-specific RNA-binding protein, was more highly expressed in the Immature Neurons than aNPCvds. Furthermore, aNPCvd samples had higher expression of NPC transcripts sox2 and musashi1, which are known to be more highly expressed in Xenopus NPCs than neurons (Sharma and Cline, 2010). These data provide strong, independent corroboration of the differential expression detected by microarray comparisons and specifically demonstrate concordance between NanoString and microarray data for 49% of tested transcripts.
Fig. 5.
Concordance of differentially-expressed transcripts detected by NanoString and microarrays. (A) Pie chart illustrating the degree of concordance of 95 transcripts analyzed by NanoString and microarrays. 49% of transcripts tested by NanoString share the same expression profile as microarray; 25% of transcripts were detected as differentially expressed by only NanoString analysis and 21% were detected as differentially expressed only by microarray analysis. Only 5% of the transcripts that were differentially expressed in the NanoString analysis exhibited differential expression in opposite directions in the microarray analysis. (B) Differential expression of transcripts analyzed by NanoString and microarrays for aNPCvd and Immature Neurons. Transcripts that are more highly expressed in aNPCvd than Immature Neurons (green), more highly expressed in Immature Neurons (red) or not differentially expressed (white) are shown for concordant transcripts (NanoString = Microarray) or those that were detected as differentially expressed only by NanoString or microarray. Transcripts to the far right were differentially expressed but in opposite directions between NanoString and microarray analyses.
In vivo screen identifies candidate gene mechanisms controlling neurogenesis
We selected 34 candidate genes for analysis of their roles in cell proliferation and neuronal differentiation based on the microarray and NanoString analyses (Table 1). We designed an in vivo imaging strategy to test whether antisense morpholinos directed against the candidate genes affected cell proliferation and differentiation (Fig. 6). To conduct the morpholino knockdown screen, cells in the tectum were electroporated with pSox2-bd::FP reporter alone, or mixed with either a fluorescently-tagged control morpholino or a specific morpholino oligonucleotide designed to block translation of a target gene (Bestman and Cline, 2014; Eisen and Smith, 2008). Coelectroporation of lissamine-tagged morpholinos and pSox2-bd::FP shows that morpholinos distribute widely through the optic tectum and are not limited to the cells expressing the pSox2-bd::FP reporter (Bestman and Cline, 2014), as previously indicated (Falk et al., 2007). After one day, we screened tadpoles for those with sparsely labeled tectal lobes (approximately 20 pSox2-bd::FP-labeled cells) for time-lapse imaging and analysis. This initial density of labeled cells allowed accurate quantification of cell proliferation and differentiation within the 3D confocal stacks of the optic tectal lobes (Bestman et al., 2012). Over the next 2 days, we collected complete confocal z-series through 6–13 tectal lobes per group. We analyzed the time-lapse images using Volocity software to determine the average cell proliferation rate of FP-labeled cells over the 48-h period, and determined the proportion of FP-labeled neurons and NPCs at each time point for animals electroporated with morpholinos. The effects of morpholino treatments were compared to results from animals electroporated with control morpholinos.
Table 1.
Morpholino sequences and their target genes.
| MO sequence | X. laevis symbol | Full name | Ref. seq | Unigene ID |
|---|---|---|---|---|
| GACCCAGGTTTCGCAGACCAGACAT | arl9 | ADP-ribosylation factor-like 9 | NM_001096919 | Xl.66117 |
| GAGGCTCCAACACGCACGCCATCTT | armc8 | Armadillo repeat containing 8 | NM_001096774 | Xl.19526 |
| CGTTGTCGAATAAAGTGAGAGCCAT | chn1 | Chimerin (chimaerin) 1 | NP_001080335 | Xl.56738 |
| GCCAAATTGAATTTCAATGCAATCG | cpeb1-a | Cytoplasmic polyadenylation element binding protein 1 | NM_001090603 | Xl.984 |
| AGGAGCCCCGGAGTCCGCATCATCC | ctdnep1-a | CTD nuclear envelope phosphatase 1 (dullard) | NM_001096787 | Xl.76057 |
| GCAGTGCAACATGGTGCTCAGCCCT | dio3 | Deiodinase, iodothyronine, type 3 | NM_001087863 | Xl.862 |
| AAAGAAGCACAAACACCACAGCCAT | efna3 | Ephrin-A3 | NM_001087027 | Xl.47030 |
| AAGTAGACCGGCATTGCGGCAGATA | eif3a | Eukaryotic translation initiation factor 3 subunit A | NM_001091816 | Xl.3189 |
| GCCACAAAGTGATAGCACTATCCAT | elk4 | ELK4, ETS-domain protein (SRF accessory protein 1) | NM_001085854 | Xl.782 |
| GTAGCAGCAGCACATTTAGTTCCAT | ephb1 | EPH receptor B1 | NM_001090601 | Xl.1028 |
| CAGGAGCCATTTTCTGTAGCACAAA | epx | Eosinophil peroxidase | NM_001088379 | Xl.424 |
| CCACAGACAGCAGTTCCTGATTCAT | fbxw7 | F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase | NM_001095717 | Xl.32837 |
| GAGTTGTGATGCTCCCTGCCGCCAT | fgf2 | Fibroblast growth factor 2 (basic) | NM_001099871 | Xl.76214 |
| AGCTCCTCCATGTTGCGTCCGCACA | fmr1-a | Fragile X mental retardation 1 | NM_001085687 | Xl.3332 |
| GCACTTCCACCGTCATGTCCTCCAT | fxr1-a | Fragile X mental retardation, autosomal homolog 1 | NM_001088317 | Xl.331 |
| TCTTCTTTGGTCTTTCCAAAATGCC | glis2-a | GLIS family zinc finger 2 | NM_001088623 | Xl.30097 |
| AATAGGTGAGGACGTAGCCAGGCAT | gstp1 | Glutathione S-transferase pi 1 | NM_001088783 | Xl.54920 |
| ACTCAGCCAATTTCTTTTCCATAGC | hat1 | Histone acetyltransferase 1 | NM_001094404 | Xl.43663 |
| GGGCACTAGACACAAACAAGTGCAC | hdac6 | Histone deacetylase 6 | NM_001087017 | Xl.8310 |
| AGGCAAACAGCTTCATGGTCACCAT | hspa5 | Heat shock 70 kDa protein 5 (glucose-regulated protein, 78 kDa) | NM_001086595 | Xl.21814 |
| GCGCAGGCTCATCCTAGTTTCCTTT | lsm6 | LSM6 homolog, U6 small nuclear RNA associated | NM_001093849 | Xl.48776 |
| CGCTCGGCGCAGCGGCCATTTTCTC | mecp2 | Methyl CpG binding protein 2 (Rett syndrome) | NM_001088385 | Xl.439 |
| ACGTGTTTGATGCTCATTGCCGCTC | mkrn2 | Makorin ring finger protein 2 | NM_001096637 | Xl.84320 |
| ATTGTGCTCCTCATAATGATCCATC | actn1/MGC81191 | Uncharacterized protein MGC81191; homolog to ACTN1, actinin, alpha 1 | NM_001091370 | |
| AGACTAAAACTCCCACCCTACCCAT | mmp9 | Matrix metallopeptidase 9 (92 kDa gelatinase/type IV collagenase) | NM_001086503 | Xl.526 |
| ATCTGTGAATCCGTTTCATCCATTC | mocs3 | Molybdenum cofactor synthesis 3 | NM_001095850 | Xl.52919 |
| TCTTTGTGGTAGCCGCGTTGCCCAT | prkaca | Protein kinase, cAMP-dependent, catalytic, alpha | NM_001099869 | Xl.83942 |
| CGCTTCCACTGTCTCTATCCGCCAT | pura | Purine-rich element binding protein A | NM_001093440 | Xl.3084 |
| CATGGGACATCTTCAGCATAATACA | r3hdm2 | R3H domain containing 2 | NM_001095964 | Xl.15158 |
| CATCAGACATTACAGCATCTGCCAT | rbfox2-b | RNA binding protein, fox-1 homolog 2 | NM_001091634 | Xl.14636 |
| AGCCCGGTTTTCCTGCTTGCTCCAT | slc12a2 | Solute carrier family 12 (Na+/K+/Cl- transporters), member 2 (NKCC1) | NM_001122599 | Xl.84328 |
| TCGGAGGTCTGTTTTGAGGGAACAT | tle1 | Transducin-like enhancer of split 1 (E(sp1) homolog) | NM_001096979 | Xl.57178 |
| AGTGGTTGGATTCCGTGTCCATATC | vangl1 | VANGL planar cell polarity protein 1 | NM_001096375 | Xl.2505 |
| TCCAGTTGCGGAAGTGTCTGTGCAT | wnt7b | Wingless-type MMTV integration site family, member 7B | NM_001090733 | Xl.229 |
| GCGAAATTCAATTTGAATCCAATGG | Control | - | - | - |
Fig. 6.

In vivo time-lapse imaging protocol. We electroporated optic tecta of stage 46 tadpoles with pSox2-bd::tGFP and control morpholinos (MO) or MOs targeted against genes of interest. After 24 h, all tGFP-labeled cells in each tectal lobe were imaged at daily intervals over 3 days.Cell proliferation over 2 days and the proportions of labeled NPCs and neurons were determined for each timepoint.
Images collected the first day after electroporation of stage 46 tadpoles with pSox2-bd::tGFP under control conditions revealed that approximately half of the transfected cells are NPCs (Bestman et al., 2012). Animals electroporated with pSox2-bd::FP alone, or combined with control morpholinos, had comparable proliferation rates (Tables 2–5). An example of a time-lapse series acquired from the optic tectal lobe of an animal that was electroporated with the control morpholino is shown in Fig. 7A1–3. The NPCs are distinguished from neurons by their radial glial morphology (Bestman et al., 2012; Morest and Silver, 2003; Tremblay et al., 2009). NPCs have triangular or elongated cell bodies at the ventricular surface of the tectum and extend a slender radial process that spans from the cell body to the endfeet on the pial surface, examples of which are shown in Fig. 7 where the arrows point to the distal pial endfoot of NPCs. By contrast, neurons have round or pear-shaped somata and tend to extend a single process from which both the axon and the dendritic arbor elaborate (the asterisk is next to neuronal somata in Fig. 7). The quantification of the changing proportions of NPCs versus neurons that occurs over the 3 day window is summarized for all control animals in Fig. 7A4 and A5 and Tables 2–4. About 10% of FP-labeled cells could not be classified as neurons or NPCs based on morphology and are quantified as ‘unclassified’ (Tables 2–4). These data show that while NPC numbers decreased over the course of the experiment, the proportion of neurons increased as the proliferating cells generated neurons and terminally differentiated. From the 23 control animals imaged, we found that 49.5±2.7% of cells on the first day of the experiment were NPCs, dropping significantly to 24.9±2.9% on day 3 (Fig. 7A4 and Tables 2–4). During this same window, the proportion of differentiated neurons increased from 37.8±2.5% on day 1 to 59.2±3.7% on day 3 (Fig. 7A5 and Tables 2–4).
Table 2.
Average proportions of cell types on day 1 with comparisons to control values.
| Radial glial progenitors |
Neurons |
Unclassified |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Morpholino (N) | %± SEM | Mann-Whitney unpaired p- values |
% Of control levels ± SEM |
%± SEM | Mann-Whitney unpaired p- values |
% Of control levels ± SEM |
%± SEM | Mann-Whitney unpaired p- values |
% Of control levels ± SEM |
| conMO Group 1 (11) | 43.1 ± 3.4 | - | 99.9 ± 8 | 39.8 ± 3.4 | - | 100 ± 8.6 | 17 ± 4.1 | - | 100 ± 24 |
| arl9 (8) | 51.7 ± 3.4 | 0.5889 | 93.5 ± 6.2 | 41 ± 4.4 | 0.3347 | 114 ± 12.4 | 7.1 ± 3.4 | 0.7803 | 82.5 ± 39.9 |
| armc8 (8) | 54.8 ± 6.6 | 0.6158 | 99.1 ± 11.9 | 29.8 ± 9 | 0.2468 | 82.8 ± 25 | 15.3 ± 4.5 | 0.2407 | 177 ± 52.9 |
| chn1 (7) | 59.3 ± 5.6 | 0.5538 | 107.2 ± 10.1 | 19.9 ± 2.2 | 0.0052 | 55.3 ± 6.1 | 20.7 ± 4.6 | 0.0218 | 239.5 ± 53.9 |
| ctdnep1-a (8) | 70.1 ± 5.5 | 0.0038 | 126.6 ± 9.9 | 22.5 ± 6.5 | 0.0474 | 62.7 ± 18.1 | 7.3 ± 2.3 | 0.1157 | 84.5 ± 27.6 |
| eif3a (8) | 65.8 ± 8.4 | 0.1322 | 119 ± 15.2 | 20.5 ± 6.1 | 0.0252 | 57.1 ± 17.1 | 13.5 ± 3.4 | 0.2763 | 156.2 ± 40.2 |
| fbxw7 (8) | 54.7 ± 2.2 | 0.9385 | 98.8 ± 3.9 | 30.6 ± 2.6 | 0.5627 | 84.9 ± 7.2 | 14.7 ± 2.9 | 0.1519 | 169.9 ± 34 |
| fgf2 (9) | 50.4 ± 4.4 | 0.4136 | 91.2 ± 7.9 | 37.1 ± 4.4 | 1 | 103.1 ± 12.3 | 12.3 ± 2.5 | 0.2106 | 143.1 ± 29.3 |
| glis2 (8) | 59.5 ± 6.8 | 0.6159 | 107.6 ± 12.3 | 30.4 ± 3.9 | 0.5883 | 84.5 ± 10.9 | 10 ± 3.4 | 0.7544 | 115.7 ± 40 |
| hat1 (8) | 52.7 ± 3.7 | 0.7574 | 95.2 ± 6.6 | 27.8 ± 4.6 | 0.4177 | 77.3 ± 12.9 | 19.4 ± 2.8 | 0.0247 | 224.7 ± 32.5 |
| lsm6 (8) | 58.5 ± 3.6 | 0.9692 | 105.8 ± 6.6 | 25.2 ± 2.9 | 0.0825 | 70.1 ± 8.2 | 16.1 ± 1.7 | 0.0223 | 186.8 ± 20.2 |
| mecp2 (8) | 38.8 ± 4.5 | 0.015 | 70.1 ± 8.1 | 48.3 ± 3.8 | 0.0371 | 134.3 ± 10.5 | 12.8 ± 2.2 | 0.1752 | 148.1 ± 25.6 |
| MGC81190/actn1 (9) | 59.7 ± 3.5 | 0.3552 | 107.9 ± 6.5 | 26.9 ± 2.7 | 0.145 | 74.9 ± 7.7 | 13.2 ± 2.4 | 0.1524 | 153.3 ± 28.2 |
| mkrn2 (8) | 50.2 ± 7.3 | 0.3742 | 90.7 ± 13.2 | 40.2 ± 6.1 | 0.4174 | 111.8 ± 16.9 | 9.5 ± 3.5 | 0.9377 | 109.7 ± 40.9 |
| mocs3 (9) | 62 ± 6.6 | 0.5218 | 112.1 ± 12 | 27.1 ± 4.8 | 0.3369 | 75.2 ± 13.4 | 10.8 ± 3.5 | 0.6121 | 125.3 ± 41.1 |
| pura (8) | 57 ± 7.4 | 1 | 103.1 ± 13.4 | 36.5 ± 6.3 | 0.7871 | 101.5 ± 17.7 | 6.3 ± 2.7 | 0.55 | 73.5 ± 31.5 |
| r3hdm2 (8) | 62 ± 5.1 | 0.2027 | 112 ± 9.2 | 28.1 ± 4.8 | 0.2797 | 78.2 ± 13.4 | 9.8 ± 1.9 | 0.5599 | 113.4 ± 22.4 |
| rbfox2b (9) | 42.6 ± 5.2 | 0.0593 | 76.9 ± 9.5 | 47.7 ± 5.7 | 0.081 | 132.6 ± 15.9 | 9.6 ± 1.8 | 0.5914 | 111.3 ± 21.3 |
| slc12a2 (9) | 54.8 ± 6.7 | 0.6958 | 99 ± 12.2 | 27.8 ± 4.6 | 0.3026 | 77.5 ± 13 | 17.2 ± 4.2 | 0.0998 | 199.5 ± 48.5 |
| tle1 (8) | 46 ± 3.6 | 0.0825 | 83.1 ± 6.5 | 43.9 ± 3.1 | 0.1135 | 121.9 ± 8.8 | 10 ± 0.9 | 0.4385 | 116.1 ± 10.6 |
| vangl1 (7) | 51.7 ± 3.9 | 0.4465 | 93.5 ± 7.1 | 32 ± 4.3 | 0.7036 | 89 ± 12.1 | 16.1 ± 2.5 | 0.0745 | 186.6 ± 29.2 |
| conMO Group 2 (12) | 55.3 ± 3.3 | - | 100 ± 6.1 | 35.9 ± 3.7 | - | 100 ± 10.4 | 8.6 ± 2.6 | - | 99.9 ± 31.1 |
| cpeb1-a (10) | 32.2 ± 4.4 | 0.0317 | 74.7 ± 10.3 | 54.7 ± 3.5 | 0.0112 | 137.5 ± 8.9 | 13 ± 2.2 | 1 | 76.3 ± 13.4 |
| dio3 (9) | 42.6 ± 8.2 | 0.9092 | 98.8 ± 19.1 | 42.5 ± 6.7 | 0.8197 | 106.9 ± 16.8 | 14.7 ± 2.7 | 0.7034 | 86.7 ± 16.3 |
| efna3 (6) | 29.7 ± 7.7 | 0.1445 | 68.8 ± 18 | 57.2 ± 7.4 | 0.0786 | 143.7 ± 18.6 | 13 ± 4.6 | 0.8798 | 76.7 ± 27.4 |
| elk4 (11) | 27.9 ± 4.8 | 0.0126 | 64.8 ± 11.3 | 47.4 ± 4.8 | 0.2638 | 119.1 ± 12 | 24.5 ± 3.9 | 0.0817 | 144.4 ± 23.1 |
| ephb1 (5) | 56.3 ± 11.5 | 0.3349 | 130.4 ± 26.8 | 16 ± 6.3 | 0.0127 | 40.2 ± 16 | 27.6 ± 8.9 | 0.2811 | 162.3 ± 52.6 |
| epx (14) | 36.8 ± 3.5 | 0.1887 | 85.3 ± 8.2 | 40.4 ± 4.5 | 0.8911 | 101.5 ± 11.4 | 22.7 ± 3.4 | 0.171 | 133.4 ± 20.5 |
| fmr1-a (8) | 50.5 ± 11.5 | 0.9341 | 117 ± 26.6 | 25.8 ± 7.1 | 0.1264 | 65 ± 18 | 23.5 ± 7.4 | 0.6494 | 138.4 ± 43.8 |
| fxr1-a (10) | 36 ± 7.9 | 0.0778 | 83.5 ± 18.3 | 39.8 ± 8.2 | 0.9438 | 100.1 ± 20.6 | 24 ± 7.6 | 0.7245 | 141.4 ± 44.7 |
| gstp1 (7) | 12.4 ± 4.2 | 0.0011 | 28.8 ± 9.8 | 74.1 ± 4.4 | 0.0006 | 186.3 ± 11 | 13.3 ± 3.6 | 0.4685 | 78.5 ± 21.6 |
| hdac6 (6) | 15.7 ± 6.1 | 0.0103 | 36.5 ± 14.2 | 61.7 ± 6.8 | 0.0307 | 155.1 ± 17.2 | 22.4 ± 7.4 | 0.2079 | 132 ± 43.8 |
| hspa5 (7) | 46.2 ± 8 | 0.8207 | 107 ± 18.7 | 45.3 ± 6.4 | 0.6505 | 113.8 ± 16.1 | 8.5 ± 2.7 | 0.2364 | 49.8 ± 15.9 |
| mmp9 (6) | 25.3 ± 7.9 | 0.1188 | 58.5 ± 18.4 | 56.6 ± 7.2 | 0.0873 | 142.3 ± 18.2 | 18 ± 4 | 0.546 | 106 ± 23.5 |
| prkaca (6) | 22 ± 8.3 | 0.0267 | 51 ± 19.4 | 61.2 ± 8.7 | 0.0347 | 153.9 ± 21.8 | 16.7 ± 3.2 | 0.6507 | 98.1 ± 18.8 |
| Wnt7b (8) | 46.8 ± 4.6 | 0.5913 | 108.6 ± 10.8 | 37.6 ± 5.6 | 0.4818 | 94.6 ± 14.1 | 15.4 ± 3.2 | 0.7097 | 90.5 ± 19.2 |
| noMO Control (13) | 35.3 ± 4.4 | 0.2022 | 81.8 ± 10.3 | 45.6 ± 3.5 | 0.4512 | 114.7 ± 8.8 | 19 ± 4.3 | 0.9077 | 111.6 ± 25.7 |
| conMO Combined (23) | 49.5 ± 2.7 | - | - | 37.8 ± 2.5 | - | - | 12.67 ± 2.52 | - | - |
Each control morpholino group (conMO group 1 and group 2) is compared to the experimental MO data acquired over the same period (experimental results grouped below each control group). All pSox2-bd::FP labeled cells in a tectal lobe were categorized as neurons, radial glial progenitors or unclassifiable/undifferentiated cells and the mean percent of each cell type and standard error of the mean (SEM) are given. The % of control values ± SEM is calculated by dividing the cell percentage of the experimental group by the control value. Mann-Whitney Unpaired tests were conducted for differences in the percent of each cell type compared to control values and the p-values are given. Significant differences from control values are in bold. N=number of electroporated tecta examined. The “conMO Combined” data averages all control MO values and is displayed in Figs. 7 and 8; it is not used for statistical comparisons.
Table 5.
Average proliferation rates between day 1 and day 3 with comparisons to control values.
| Morpholino | Tectal lobe# | Total cell # | Cell proliferation rate between day 1 and day 3 mean ± SEM | Mann-Whitney Unpaired p-values | % of control cell proliferation rate mean ± SEM |
|---|---|---|---|---|---|
| conMO Group 1 | 12 | 247 | 23.5 ± 5.5 | 0.8052 | 100 ± 23.5 |
| arl9 | 8 | 304 | 73.8 ± 15.4 | 0.005 | 314.8 ± 65.6 |
| armc8 | 8 | 277 | 179.1 ± 49.8 | 0.003 | 763.9 ± 212.7 |
| chn1 | 7 | 371 | 96.4 ± 23.1 | 0.002 | 411.1 ± 98.5 |
| ctdnep1 | 8 | 215 | 97.4 ± 26.9 | 0.006 | 415.5 ± 114.5 |
| eif3a | 8 | 300 | 40.9 ± 8.1 | 0.097 | 174.4 ± 34.5 |
| fbxw7 | 8 | 423 | 50.8 ± 18.4 | 0.202 | 216.4 ± 78.7 |
| fgf2 | 9 | 407 | 64.7 ± 9.4 | 0.002 | 275.9 ± 40.3 |
| glis2 | 8 | 300 | 63.8 ± 15.4 | 0.012 | 271.6 ± 65.7 |
| hat1 | 8 | 417 | 46.9 ± 5.4 | 0.018 | 200 ± 23.1 |
| lsm6 | 8 | 438 | 78.8 ± 16.3 | 0.005 | 336.4 ± 69.9 |
| mecp2 | 8 | 288 | 44 ± 7.3 | 0.058 | 187.8 ± 31.3 |
| MGC8U91/actn1 | 9 | 367 | 69.3 ± 15.1 | 0.006 | 295.7 ± 64.3 |
| mkrn2 | 8 | 221 | 9.3 ± 9.5 | 0.177 | 39.5 ± 40.5 |
| mocs3 | 8 | 238 | 124.5 ± 31.1 | 0.002 | 531.3 ± 132.5 |
| pura | 8 | 232 | 48.1 ± 11.3 | 0.089 | 205 ± 48.2 |
| r3hdm2 | 8 | 278 | 37.9 ± 8.2 | 0.203 | 161.6 ± 34.9 |
| rbfox2b | 9 | 394 | 44.4 ± 21.1 | 0.722 | 189.3 ± 90 |
| slc12a2 | 8 | 309 | 31 ± 6.4 | 0.396 | 132.1 ± 27.1 |
| tle1 | 8 | 300 | 20.1 ± 4.5 | 0.877 | 85.8 ± 19.3 |
| vangl1 | 7 | 237 | 34.4 ± 21.8 | 0.704 | 146.5 ± 92.9 |
| conMO Group 2 | 11 | 296 | 20.8 ± 4.1 | 0.8052 | 100 ± 19.8 |
| cpeb1-a | 10 | 157 | 6.2 ± 3.8 | 0.029 | 29.5 ± 18.7 |
| dio3 | 9 | 104 | 34.8 ± 14.4 | 0.676 | 167.3 ± 69.3 |
| efna3 | 6 | 66 | 12.9 ± 5.1 | 0.159 | 62.1 ± 24.7 |
| elk4 | 11 | 322 | 37.7 ± 8.04 | 0.048 | 181.7 ± 38.7 |
| ephB1 | 5 | 91 | 115.6 ± 30.2 | 0.002 | 556.7 ± 145.5 |
| epx | 14 | 385 | 15.1 ± 4.2 | 0.338 | 73.1 ± 20.4 |
| fmr1-a | 9 | 185 | –10.5 ± 11.2 | 0.033 | –47.8 ± 61.1 |
| fxr1-a | 10 | 101 | –3.2 ± 13.9 | 0.024 | –15.4 ± 66.8 |
| gstp1 | 7 | 163 | 1.5 ± 6.3 | 0.013 | 7.4 ± 30.7 |
| hdac6 | 6 | 172 | 14.4 ± 13.5 | 0.291 | 68.8 ± 65.1 |
| hspa5 | 7 | 121 | –2.6 ± 14.6 | 0.024 | –12.2 ± 70.2 |
| mmp9 | 6 | 117 | –1.8 ± 8.3 | 0.027 | –8.7 ± 39.8 |
| prkaca | 6 | 98 | –10.7 ± 6.4 | 0.003 | –51.5 ± 30.7 |
| wnt7b | 8 | 119 | 11.2 ± 11.6 | 0.231 | 53.8 ± 56.1 |
| noMO Control | 13 | 208 | 52.7 ± 14 | 0.118 | 253.7 ± 67.3 |
| conMO Combined | 23 | 543 | - | 100 ± 15.15 |
Each control morpholino group (conMO group 1 and group 2) is compared to the experimental MO data acquired over the same period (experimental results grouped below each control group). All pSox2-bd::FP labeled cells in a tectal lobe were counted on days 1 and 3 and the mean differences and standard error of the mean (SEM) are given. The % of control levels ± SEM is calculated by dividing the proliferation rate of the experimental group by the mean control proliferation rate. Mann-Whitney Unpaired tests were conducted for differences in the proliferation rates to the control values and the p-values are given. Significant differences from control values are in bold. The “conMO Combined” value is the average averages of all MO control values and is displayed in Figs. 7 and 8, but it is not used for statistical comparisons.
Fig. 7.
Morpholinos against candidate neurogenic genes alter cell proliferation and differentiation. A1–D3 Projections of confocal stacks of the right tectal lobe imaged 1 day after co-electroporation with pSox2-bd::tGFP and control morpholinos (A1–A3), or morpholinos against glutathione S-transferase pi 1 (gstp1; B1–B3), armadillo repeat containing 8 (armc8; C1–C3), or heat shock protein 5 (hspa5; D1–D3), GFP-labeled cells are relatively sparse on day 1 (A1, B1, C1, D1). Arrows point to the distal pial endfoot of example neural progenitor cells and asterisks indicate neurons. Under control conditions, the number of NPCs decreases over the subsequent two days (A2 and A3). The tectal lobes with targeted gene knockdown show decreases (gstp1 and hspa5) and increases (armc8) in cell proliferation, as well as higher proportions of NPCs (hspa5) or neurons (gstp1 and armc8) on the third day of imaging. A4–D4, A5–D5 Summary graphs of changes in the proportion of cells in the tectum of the control (A) and morpholino-treated (B–D) animals that are NPCs (A4–D4) or neurons (A5–D5). Each line represents data from a separate animal. An asterisk over day 1 or day 3 indicates a significant difference from the mean control values (Mann–Whitney U test, p<0.05) and an asterisk over the center bracket indicates that there was a significant change between day 1 and day 3 levels (Wilcoxon Signed Rank test, p<0.05). Summary graphs for all control and morpholino results are provided in Supplementary Fig. 1. Data are shown in Tables 2–5.
Table 4.
Pairwise comparisons of changing percentages of cell types between day 1 and day 3.
| % Neural progenitor cells |
% Neurons |
% Unclassified |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Morpholino | Day 1 | Day 3 | Wilcoxon p-value | Day 1 | Day 3 | Wilcoxon p-value | Day 1 | Day 3 | Wilcoxon p-value |
| conMO group 1 | 55.4 ± 3.4 | 27.9 ± 4.7 | 0.0005 | 36 ± 3.8 | 61.7 ± 5 | 0.0024 | 8.7 ± 2.7 | 10.3 ± 2.7 | 0.5693 |
| arl9 | 51.8 ± 3.5 | 18.8 ± 3.5 | 0.0078 | 41 ± 4.5 | 62.3 ± 4.6 | 0.0078 | 7.1 ± 3.5 | 18.9 ± 3.2 | 0.0078 |
| armc8 | 54.9 ± 6.6 | 10.1 ± 2.4 | 0.0078 | 29.8 ± 9 | 74.7 ± 4.5 | 0.0078 | 15.3 ± 4.6 | 15.2 ± 2.9 | 0.9453 |
| chn1 | 59.4 ± 5.6 | 8.3 ± 1.9 | 0.0156 | 19.9 ± 2.2 | 67.5 ± 3.3 | 0.0156 | 20.7 ± 4.7 | 24.3 ± 2.9 | 0.4688 |
| ctdnep1-a | 70.1 ± 5.5 | 21.6 ± 4.2 | 0.0078 | 22.6 ± 6.5 | 64.7 ± 3.9 | 0.0078 | 7.3 ± 2.4 | 13.7 ± 2.5 | 0.1953 |
| fbxw7 | 54.7 ± 2.2 | 22.2 ± 3 | 0.0078 | 30.6 ± 2.6 | 58.2 ± 3.8 | 0.0078 | 14.7 ± 2.9 | 19.6 ± 1.7 | 0.1094 |
| fgf2 | 50.5 ± 4.4 | 23.9 ± 4.8 | 0.0039 | 37.1 ± 4.5 | 62.4 ± 5.1 | 0.0078 | 12.4 ± 2.5 | 13.7 ± 1.7 | 0.8209 |
| glis2 | 59.6 ± 6.9 | 13.7 ± 2.9 | 0.0078 | 30.4 ± 4 | 75.9 ± 3.2 | 0.0078 | 10 ± 3.5 | 10.4 ± 2.8 | 0.2969 |
| hat1 | 52.7 ± 3.7 | 25.2 ± 2.2 | 0.0078 | 27.8 ± 4.7 | 56.1 ± 1.9 | 0.0078 | 19.4 ± 2.8 | 18.7 ± 0.9 | 0.7422 |
| lsm6 | 58.6 ± 3.7 | 18.6 ± 2.3 | 0.0078 | 25.3 ± 3 | 62.5 ± 3.5 | 0.0078 | 16.2 ± 1.7 | 19 ± 2.3 | 0.25 |
| mecp2 | 38.8 ± 4.5 | 17.3 ± 3.3 | 0.0078 | 48.4 ± 3.8 | 68.2 ± 3.4 | 0.0234 | 12.8 ± 2.2 | 14.5 ± 3.1 | 0.8438 |
| mkrn2 | 50.2 ± 7.3 | 20.5 ± 4 | 0.0078 | 40.3 ± 6.1 | 71.1 ± 4.4 | 0.0078 | 9.5 ± 3.5 | 8.4 ± 2.2 | 0.8438 |
| MGC81191/actn1 | 59.8 ± 3.6 | 17.3 ± 3 | 0.0039 | 27 ± 2.8 | 69.7 ± 3.5 | 0.0039 | 13.3 ± 2.4 | 13 ± 2.7 | 0.9102 |
| mocs3 | 62.1 ± 6.7 | 17.9 ± 4 | 0.0039 | 27.1 ± 4.9 | 73 ± 3.6 | 0.0039 | 10.8 ± 3.6 | 9.1 ± 1.5 | 0.6523 |
| pura | 57.1 ± 7.4 | 23.1 ± 3.1 | 0.0156 | 36.6 ± 6.4 | 68.2 ± 3.2 | 0.0156 | 6.4 ± 2.7 | 8.7 ± 2.3 | 0.4375 |
| r3hdm2 | 62 ± 5.1 | 27.4 ± 5.1 | 0.0078 | 28.2 ± 4.8 | 55.7 ± 4.3 | 0.0078 | 9.8 ± 1.9 | 16.9 ± 2 | 0.0156 |
| rbfox2b | 42.6 ± 5.3 | 22.7 ± 3.5 | 0.0391 | 47.8 ± 5.7 | 66 ± 4.8 | 0.0195 | 9.6 ± 1.8 | 11.3 ± 3.1 | 0.7344 |
| slc12a2 | 54.8 ± 6.8 | 22.9 ± 4.6 | 0.0039 | 27.9 ± 4.7 | 62.9 ± 5.2 | 0.0039 | 17.3 ± 4.2 | 14.2 ± 2.4 | 0.3594 |
| tle1 | 46 ± 3.6 | 20.5 ± 2.7 | 0.0078 | 43.9 ± 3.2 | 70.8 ± 3 | 0.0078 | 10.1 ± 0.9 | 8.7 ± 1.8 | 0.3125 |
| vangl1 | 51.8 ± 3.9 | 16.3 ± 3.1 | 0.0156 | 32.1 ± 4.4 | 74 ± 3.6 | 0.0156 | 16.1 ± 2.5 | 9.7 ± 1.1 | 0.1094 |
| conMO group 2 | 43.2 ± 3.5 | 21.6 ± 3.3 | < 0.0001 | 39.8 ± 3.4 | 56.5 ± 5.6 | < 0.0001 | 17 ± 4.1 | 21.9 ± 4.4 | 0.1973 |
| cpeb1-a | 32.3 ± 4.5 | 17.5 ± 4.4 | 0.0195 | 54.7 ± 3.6 | 70.7 ± 4.9 | 0.0078 | 13 ± 2.3 | 11.8 ± 2.1 | 1 |
| dio3 | 42.7 ± 8.3 | 28.1 ± 7.4 | 0.0313 | 42.6 ± 6.7 | 51.9 ± 5.8 | 0.1094 | 14.8 ± 2.8 | 20 ± 3.1 | 0.1094 |
| efna3 | 29.7 ± 7.8 | 27 ± 3.9 | 0.6875 | 57.2 ± 7.4 | 57.4 ± 4 | 1 | 13.1 ± 4.7 | 15.6 ± 1.7 | 0.6875 |
| eif3a | 65.9 ± 8.5 | 12.6 ± 2.5 | 0.0078 | 20.6 ± 6.2 | 71.9 ± 3.1 | 0.0078 | 13.5 ± 3.5 | 15.6 ± 2.1 | 0.5469 |
| elk4 | 28 ± 4.9 | 14.9 ± 2 | 0.0029 | 47.4 ± 4.8 | 60.8 ± 4.8 | 0.0273 | 24.6 ± 3.9 | 24.3 ± 3.7 | 0.7646 |
| ephB1 | 56.3 ± 11.6 | 26.1 ± 4.6 | 0.0625 | 16 ± 6.4 | 50.9 ± 5 | 0.0625 | 27.6 ± 9 | 23 ± 4 | 0.8125 |
| epx | 36.9 ± 3.6 | 18.6 ± 3 | 0.0004 | 40.4 ± 4.6 | 58.1 ± 3.5 | 0.0023 | 22.7 ± 3.5 | 23.4 ± 3.3 | 0.9515 |
| fmr1a | 50.5 ± 11.5 | 20.3 ± 5.5 | 0.0078 | 25.9 ± 7.2 | 60.6 ± 6.9 | 0.0078 | 23.6 ± 7.5 | 19 ± 4.7 | 0.8125 |
| fxr1a | 36.1 ± 7.9 | 21.4 ± 6.1 | 0.0117 | 39.9 ± 8.2 | 58 ± 6.4 | 0.0039 | 24.1 ± 7.6 | 20.6 ± 6.4 | 0.4375 |
| gstp1 | 12.5 ± 4.3 | 5.7 ± 1.9 | 0.1563 | 74.2 ± 4.4 | 85.5 ± 2.3 | 0.0781 | 13.4 ± 3.7 | 8.8 ± 1.6 | 0.2188 |
| hdac6 | 15.8 ± 6.2 | 8.7 ± 4.9 | 0.1563 | 61.7 ± 6.9 | 71.5 ± 9.2 | 0.1563 | 22.5 ± 7.5 | 19.8 ± 6.3 | 0.8438 |
| hspa5 | 46.2 ± 8.1 | 50.6 ± 6.7 | 0.4688 | 45.3 ± 6.4 | 36.9 ± 4.1 | 0.2969 | 8.5 ± 2.7 | 12.5 ± 4.6 | 0.2188 |
| mmp9 | 25.3 ± 7.9 | 10.6 ± 5.5 | 0.0313 | 56.7 ± 7.3 | 73.2 ± 8.3 | 0.1563 | 18.1 ± 4 | 16.2 ± 5.5 | 1 |
| prkaca | 22 ± 8.4 | 11.3 ± 5.2 | 0.125 | 61.3 ± 8.7 | 73.1 ± 7 | 0.1563 | 16.7 ± 3.2 | 15.6 ± 3.4 | 1 |
| wnt7b | 46.9 ± 4.7 | 34.8 ± 7.9 | 0.3125 | 37.7 ± 5.6 | 53.6 ± 6.1 | 0.0625 | 15.4 ± 3.3 | 11.5 ± 5.3 | 0.2969 |
| No morpholino | 35.3 ± 4.5 | 14.2 ± 3.4 | 0.0017 | 45.7 ± 3.5 | 63 ± 3.8 | 0.0017 | 19 ± 4.4 | 22.9 ± 4.3 | 0.2163 |
| conMO Combined | 49.5 ± 2.7 | 24.9 ± 2.9 | - | 37.8 ± 2.5 | 59.2 ± 3.7 | - | 12.7 ± 2.5 | 15.9 ± 2.8 | - |
Tecta were electroporated with pSox2-bd::FP (either Kaede or tGFP) alone or combined with the control morpholino (conMO) or one of the 34 experimental morpholinos. The proportions of each cell type were calculated on days 1 and 3 and the means, standard error of the mean (SEM) are given. Wilcoxon Signed Rank tests were used to determine whether the proportions of the neural progenitor cells, neurons or unclassifiable cells changed significantly between day 1 and day 3 p-values. Significant differences from control values are in bold.
Proliferation rates in the tectum can be affected by multiple factors: cell survival, cell cycle length, whether the NPCs divide symmetrically to expand the pool of proliferating cells, whether the NPCs divide asymmetrically to maintain the progenitor pool, and whether the NPCs terminally differentiate and thereby shrink the progenitor pool. We found that time-lapse imaging data was critical for the interpretation of our results, and the examples we describe below reveal that there were mixed relationships between increases or decreases in proliferation rates and whether there were greater or fewer progenitor cells. Fig. 7B–D shows projections of representative time lapse images of optic tectal lobes co-electroporated with the pSox2-bd::FP reporter and morpholinos designed to block translation of genes from our list of candidate neurogenic regulators: glutathione S-transferase pi 1 (gstp1; Fig. 7B1–3), heat shock protein 5 (hspa5; Fig. 7C1–3) and armadillo repeat containing protein 8 (armc8; Fig. 7D1–3). These three examples show the range of cell proliferation and cell differentiation outcomes generated by the different morpholinos we tested.
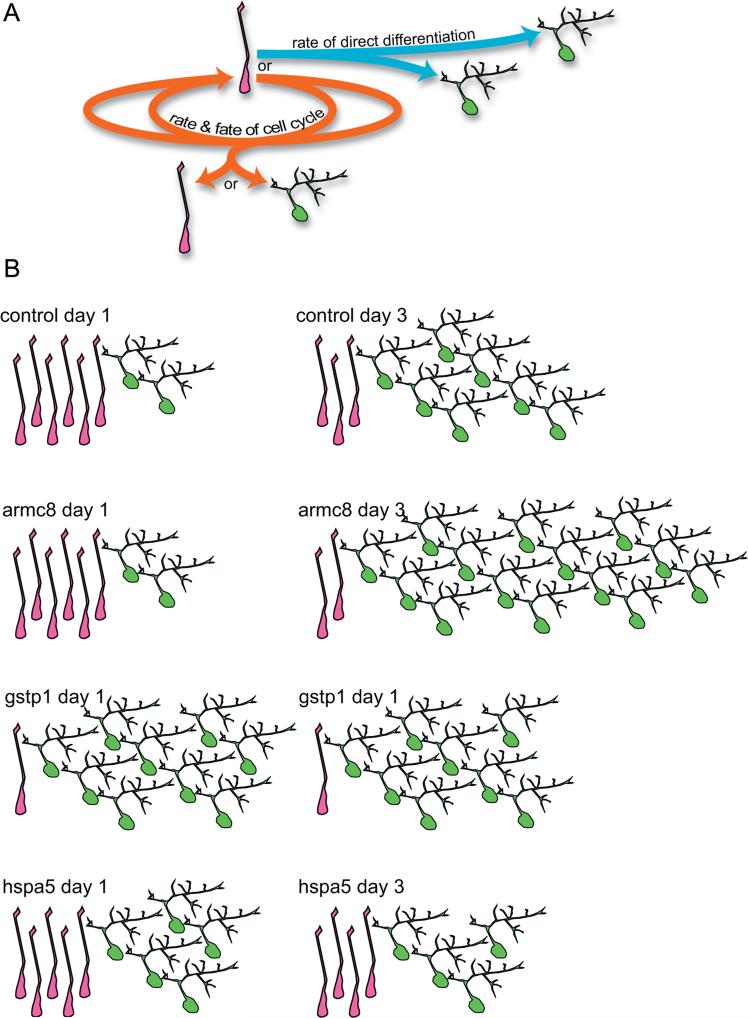
Animals treated with morpholinos directed against gstp1 represent a category of candidate genes that affected the proportion of NPCs within 24 h after electroporation. Fig. 7B1–3 shows a time lapse series from the right optic tectal lobe of a gstp1-MO treated animal with a high number of differentiated neurons on day 1 (asterisk in Figs. 7B1–3 indicates a neuron with a mature dendritic arbor that is visible at each time point). Figs. 7B4 and B5 show the results from the 7 animals that we imaged, indicating that by the time the first image was acquired, electroporating optic tecta treated with the gstp1 morpholino had reduced the NPC population to 12.5±4.2% of all cells in the optic tectum (p=0.001) or 28.8±9.9% of control levels. gstp1 MO treatment concurrently increased the neurons to 72.2±4.4% of the cell population (p<0.01) or 186.3±11.1% of control levels (Fig. 7B4–5 and Table 2). By day 3, the NPCs made up only 5.6±4.9% of the total cell population, just 26.2±8.5% of control levels (p=0.005), and at 85.4±2.3% of the cell population, neurons were 151.2±4.1% of control levels (p=0.001). Compared to control conditions, treatment with gstp1-MO also inhibited the rate at which NPCs differentiated, yielding fewer cells by day 3 of the time lapse series (Fig. 7B3 and Table 4). Consequently, we did not detect changes in the proportion of the different cell types over the 3 day experiment compared to controls (Table 3).
Table 3.
Proportions of cell types generated by day 3 with comparisons to control values.
| Proportion of cell types, day 3 |
% of control levels ± SEM |
Pearson's Chi-square | Mann Whitney Unpaired p-values |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Morpholino | % Progenitors | % Neurons | % Unclassified | Progenitors | Neurons | Unclassified | Progenitors | Neurons | Unclassified | |
| conMO Group 1 | 21.5 ± 10.8 | 56.5 ± 5.5 | 21.9 ± 4.4 | 100 ± 15.1 | 100 ± 9.8 | 100 ± 20.3 | - | - | - | - |
| arl9 | 18.8 ± 9.8 | 62.2 ± 4.5 | 18.8 ± 3.1 | 67.4 ± 12.4 | 100.8 ± 7.4 | 182.8 ± 30.5 | 0.003 | 0.2311 | 0.589 | 0.059 |
| armc8 | 10.1 ± 6.8 | 74.7 ± 4.5 | 15.1 ± 2.9 | 36.2 ± 8.6 | 121 ± 7.3 | 146.8 ± 28.4 | < 0.0001 | 0.0485 | 0.037 | 0.203 |
| chn1 | 8.2 ± 4.8 | 67.4 ± 3.3 | 24.2 ± 2.9 | 29.6 ± 6.6 | 109.2 ± 5.4 | 235 ± 28.4 | < 0.0001 | 0.0222 | 0.22 | 0.008 |
| ctdnep1-a | 21.6 ± 11.9 | 64.6 ± 3.9 | 13.7 ± 2.5 | 77.2 ± 15.1 | 104.7 ± 6.3 | 132.8 ± 24.5 | 0.081 | 0.2969 | 0.487 | 0.216 |
| eif3a | 12.5 ± 6.9 | 71.8 ± 3.1 | 15.5 ± 2 | 44.9 ± 8.8 | 116.4 ± 5 | 150.9 ± 20.1 | 0.001 | 0.0583 | 0.083 | 0.105 |
| fbxw7 | 22.2 ± 8.3 | 58.2 ± 3.8 | 19.6 ± 1.7 | 79.4 ± 10.6 | 94.2 ± 6.1 | 190 ± 16.5 | 0.012 | 0.2463 | 0.908 | 0.015 |
| fgf2 | 23.9 ± 14.5 | 62.3 ± 5.1 | 13.6 ± 1.6 | 85.6 ± 17.3 | 101 ± 8.2 | 132.5 ± 16.2 | 0.046 | 0.569 | 0.57 | 0.145 |
| glis2 | 13.7 ± 8.2 | 75.8 ± 3.1 | 10.4 ± 2.7 | 49 ± 10.4 | 122.8 ± 5.1 | 101.2 ± 26.7 | < 0.0001 | 0.0693 | 0.083 | 0.816 |
| hat1 | 25.2 ± 6.3 | 56 ± 1.9 | 18.7 ± 0.9 | 90.2 ± 8 | 90.8 ± 3.1 | 181.4 ± 8.8 | 0.024 | 0.3341 | 0.7 | 0.019 |
| lsm6 | 18.6 ± 6.5 | 62.4 ± 3.4 | 18.9 ± 2.2 | 66.5 ± 8.3 | 101.1 ± 5.6 | 183.6 ± 21.8 | 0.003 | 0.2023 | 0.512 | 0.049 |
| MGC81191/actn1 | 17.3 ± 9.2 | 68.1 ± 3.4 | 14.5 ± 3 | 62 ± 11.6 | 110.4 ± 5.5 | 140.4 ± 29.6 | 0.024 | 0.1444 | 0.136 | 0.455 |
| mecp2 | 17.2 ± 9 | 69.7 ± 3.4 | 12.9 ± 2.7 | 61.9 ± 10.7 | 112.9 ± 5.6 | 125.6 ± 26.6 | 0.017 | 0.1319 | 0.154 | 0.354 |
| mkrn2 | 20.5 ± 11.2 | 71.1 ± 4.3 | 8.4 ± 2.2 | 73.3 ± 14.1 | 115.1 ± 7.1 | 81.4 ± 21.5 | 0.397 | 0.1641 | 0.097 | 0.938 |
| mocs3 | 17.8 ± 11.8 | 73 ± 3.5 | 9.1 ± 1.4 | 63.8 ± 14.1 | 118.3 ± 5.7 | 88.2 ± 14.2 | 0.016 | 0.1346 | 0.114 | 0.877 |
| pura | 23.1 ± 8.7 | 68.1 ± 3.2 | 8.7 ± 2.2 | 82.7 ± 11 | 110.4 ± 5.2 | 84.3 ± 21.9 | 0.766 | 0.3536 | 0.19 | 0.907 |
| r3hdm2 | 27.3 ± 14.3 | 55.7 ± 4.3 | 16.8 ± 2 | 97.9 ± 18.1 | 90.2 ± 7 | 163.7 ± 19.7 | 0.067 | 0.9692 | 0.847 | 0.07 |
| rbfox2-b | 22.7 ± 10.4 | 65.9 ± 4.7 | 11.2 ± 3.1 | 81.3 ± 12.4 | 106.8 ± 7.7 | 109.4 ± 30.1 | 0.317 | 0.213 | 0.5 | 0.695 |
| slc12a2 | 22.8 ± 13.6 | 62.9 ± 5.1 | 14.2 ± 2.3 | 81.7 ± 16.3 | 101.9 ± 8.4 | 137.7 ± 23.2 | 0.118 | 0.4335 | 0.335 | 0.231 |
| tle1 | 20.5 ± 7.6 | 70.8 ± 2.9 | 8.6 ± 1.7 | 73.5 ± 9.6 | 114.6 ± 4.8 | 83.9 ± 17.4 | 0.073 | 0.2162 | 0.143 | 1 |
| vangl1 | 16.3 ± 8.3 | 73.9 ± 3.5 | 9.7 ± 1 | 58.3 ± 11.2 | 119.8 ± 5.8 | 94 ± 10.2 | 0.016 | 0.1624 | 0.099 | 0.582 |
| conMO Group 2 | 27.9 ± 16.1 | 61.7 ± 4.9 | 10.3 ± 2.7 | 100 ± 16.6 | 99.9 ± 8 | 100 ± 26.5 | - | - | - | - |
| cpeb1-a | 17.5 ± 13.9 | 70.7 ± 4.9 | 11.7 ± 2.1 | 80.9 ± 20.3 | 125.1 ± 8.7 | 53.7 ± 9.7 | 0.259 | 0.3787 | 0.149 | 0.17 |
| dio3 | 28.1 ± 22 | 51.8 ± 5.7 | 20 ± 3 | 130.2 ± 34 | 91.7 ± 10.1 | 91.4 ± 14 | 0.149 | 0.5433 | 0.703 | 0.909 |
| efna3 | 27 ± 9.5 | 57.3 ± 3.9 | 15.6 ± 1.6 | 125.1 ± 18 | 101.4 ± 7 | 71.3 ± 7.6 | 0.551 | 0.4211 | 0.92 | 0.651 |
| elk4 | 14.9 ± 6.6 | 60.7 ± 4.7 | 24.2 ± 3.7 | 69.1 ± 9.3 | 107.5 ± 8.4 | 110.9 ± 16.9 | 0.017 | 0.1075 | 0.511 | 0.108 |
| ephb1 | 26 ± 10.1 | 50.9 ± 5 | 23 ± 4 | 120.6 ± 21.1 | 90 ± 8.8 | 105.2 ± 18.2 | 0.044 | 0.5711 | 0.571 | 0.91 |
| epx | 18.5 ± 11 | 58 ± 3.5 | 23.3 ± 3.2 | 86 ± 13.6 | 102.7 ± 6.2 | 106.7 ± 14.9 | 0.083 | 0.4595 | 0.722 | 0.805 |
| fmr1-a | 20.3 ± 15.6 | 60.6 ± 6.9 | 19 ± 4.7 | 94.1 ± 25.6 | 107.2 ± 12.2 | 86.9 ± 21.4 | 0.582 | 0.8365 | 0.71 | 0.649 |
| fxr1-a | 21.4 ± 19.3 | 57.9 ± 6.4 | 20.5 ± 6.3 | 99.3 ± 28.3 | 102.6 ± 11.3 | 93.9 ± 29.1 | 0.041 | 0.7512 | 0.916 | 0.596 |
| gstp1 | 5.6 ± 4.9 | 85.4 ± 2.3 | 8.8 ± 1.5 | 26.2 ± 8.5 | 151.2 ± 4.1 | 40.3 ± 7.3 | < 0.0001 | 0.0057 | 0.001 | 0.024 |
| hdac6 | 8.6 ± 12 | 71.5 ± 9.1 | 19.7 ± 6.2 | 40.2 ± 22.6 | 126.5 ± 16.2 | 90.4 ± 28.7 | 0.001 | 0.0389 | 0.107 | 0.725 |
| hspa5 | 50.6 ± 17.6 | 36.9 ± 4.1 | 12.5 ± 4.5 | 234.3 ± 30.9 | 65.2 ± 7.2 | 57.1 ± 20.8 | < 0.0001 | 0.0021 | 0.018 | 0.204 |
| mmp9 | 10.6 ± 13.5 | 73.2 ± 8.2 | 16.1 ± 5.4 | 49.1 ± 25.5 | 129.5 ± 14.6 | 73.9 ± 25.1 | 0.023 | 0.0559 | 0.097 | 0.451 |
| prkaca | 11.2 ± 12.7 | 73.1 ± 7 | 15.6 ± 3.3 | 52.1 ± 24 | 129.3 ± 12.3 | 71.3 ± 15.3 | 0.062 | 0.1074 | 0.097 | 0.514 |
| wnt7b | 34.8 ± 22.4 | 53.6 ± 6 | 11.5 ± 5.3 | 161.3 ± 36.7 | 94.8 ± 10.7 | 52.6 ± 24.4 | 0.071 | 0.1731 | 0.836 | 0.083 |
| No morpholino | 14.1 ± 12.4 | 62.9 ± 3.8 | 22.8 ± 4.2 | 65.6 ± 15.9 | 111.3 ± 6.7 | 104.5 ± 19.4 | 0.359 | 0.1245 | 0.401 | 0.794 |
| conMO Combined | 21.5 ± 10.8 | 59.2 ± 3.7 | 15.9 ± 2.8 | 100 ± 15.1 | 100 ± 9.8 | 100 ± 20.3 | - | - | - | - |
Each control morpholino group (conMO group 1 and group 2) is compared to the experimental MO data acquired over the same period (experimental results grouped below each control group). All pSox2-bd::FP labeled cells in a tectal lobe were categorized as neurons, radial glial progenitors or unclassifiable/undifferentiated cells and the mean percent of each cell type and standard error of the mean (SEM) are given. The % of control values ± SEM is calculated by dividing the cell percentage of the experimental group by the control value. Pearson Chi-square values for differences in the distributions of cell populations between each control morpholino group (conMO groupl or conMO group 2) and experimental morpholino groups are given. Mann-Whitney Unpaired tests were conducted for differences in the percent of each cell type to the control values and the p-values are given. Significant differences from control values are in bold. The “conMO Combined” value averages all control MO values and is displayed in Figs. 7 and 8; it is not used for statistical comparisons.
Treatment with morpholinos against hspa5 also significantly limited proliferation in the tectum (Fig. 7C, Table 5), but our results suggest that hspa5 and gstp1 act through different mechanisms. MOs against hspa5 did not limit the numbers of progenitors or neurons on the first day of imaging (107.0±18.7% and 113.8±16.2% of control levels, respectively, p-values > 0.6; Fig. 7C and Table 2). By the 3rd day of the time lapse imaging experiment, 50.6±6.7% of the GFP+ cells in tecta electroporated with the hspa5 morpholino remained NPCs (arrow Fig. 7 C1–3), 234.3±30.9% of control values (p=0.002). The number of neurons (asterisk, Fig. 7C1–3) from the hspa5 MO treated animals was only 65.3±7.2% of control values, a significant reduction to just 36.9±4.1% of the total cell population (p=0.02). The pairwise comparisons failed to reveal the expected decrease in the NPC population and increase in differentiated neurons between day 1 and day 3 seen in controls (Wilcoxon Signed Rank test, p-values=0.16; Fig. 7C4 and C5 and Table 4). This resulted in an overall decrease in cell proliferation for the hspa5 morphants (Fig. 8A, and Tables 3 and 5). These data suggest that morpholinos against hspa5 prolonged the cell cycle and limited the differentiation of NPCs into neurons. In contrast, morpholinos against gstp1 also decreased neurogenesis but did so by promoting progenitors to exit the cell cycle, depleting the progenitor pool and increasing differentiation.
Fig. 8.
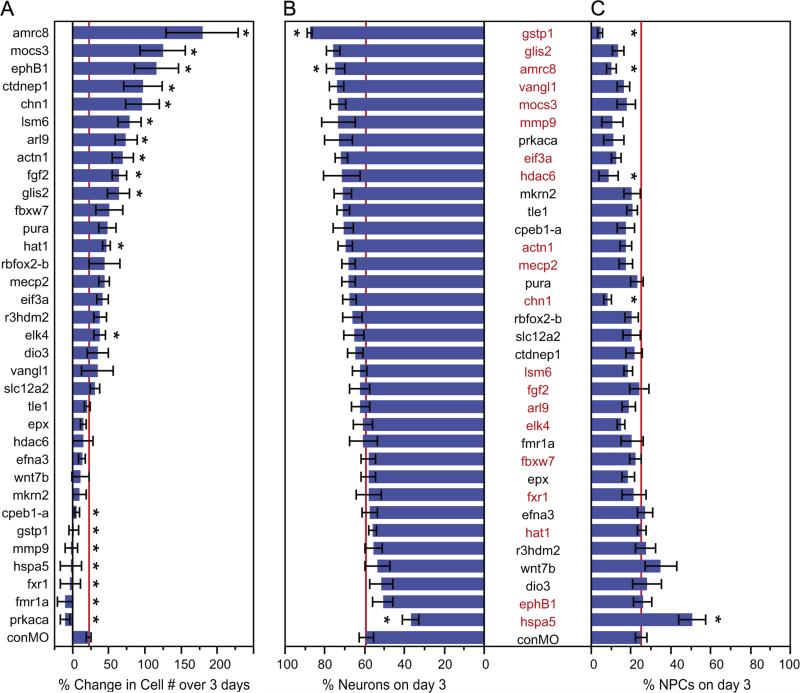
Morpholinos against candidate neurogenic genes generate a range of neurogenesis phenotypes. Summary of the in vivo imaging data showing the numbers of GFP-labeled cells generated over time (A), the proportion of the cells that were neurons (B) or NPCs (C) for each experimental condition. (A) The genes targeted with morpholinos are arranged by the magnitude of the change in cell number over 3 days. The asterisks indicate a significant decrease or increase compared to control morpholino conditions (red line). The mean values, SEMs and p-values are in Table 4. (B–C) The proportion of neurons (B) and NPCs (C) as a percentage of all cells counted on day 3. The genes targeted are listed along the y-axis and arranged by those that produced the greatest proportion of neurons. Asterisks indicate differences in the proportion of cell types between the experimental and matched control morpholino groups (Mann–Whitney U test, p <0.05). Gene symbols listed in red identify the morpholinos that resulted in a significant difference in the proportions of cell types compared to control (Pearson X-square test, p<0.05). The mean values, SEMs and p-values are in Table 2. Red lines on graphs indicate the mean control morpholino (conMO) values for the proportion of neurons or NPCs. The mean values, SEMs, and p-values are in Table 5.
A different proliferation phenotype was detected in animals electroporated with the morpholino directed against armc8. Like the hspa5 morphants, the initial proportions of NPCs (arrow, Fig. 7D1–3) and neurons (asterisk, Fig. 7D1–3) were not different on the first day of the experiment (99.1±11.9% and 82.8±25% of control values, respectively, p-values >0.2; Fig. 7D2 and Table 2). Comparing Fig. 7D1 and D3 reveals that, compared to control conditions, the armc8 morpholino increased cell proliferation by generating a higher proportion of differentiated neurons over the 48 h period. Pairwise comparisons of the armc8 morphants revealed significant increases in the proportions of neurons and decreases in NPC numbers between day 1 and day 3 (p-values <0.02; Table 4), similar to control MO results. The data from all armc8 morpholino treated animals (n=8) are summarized in Fig. 7D4 and D5, which show that the magnitude of the changes in cell proportions surpassed those found in the control animals. The animals treated with armc8 had significantly higher numbers of neurons on day 3 compared to control values (74.7±4.6% neurons on day 3; p=0.0037), which reduced the remaining proliferating cells to just 10.0±2.4% of the total cell population, significantly less than the control values (p <0.0001; Table 2 and Fig. 7D4 and D5). These data indicate that MOs against armc8 may act to shorten the cell cycle time and increase rates at which NPCs leave the progenitor pool and differentiate. Together these examples illustrate the value of our time-lapse imaging approach to test how genes affect cell proliferation and differentiation in the optic tectum based on direct observation of NPCs and their progeny. Similar graphs of the changing proportions cell types for all control animals and each set of the 34 experimental morpholino groups are provided in Supplementary Figs. 1–3.
Morpholinos generate a range of cell proliferation and neurogenesis phenotypes
The time lapse experiments and quantification shown in Fig. 7 give three examples that capture the range of cell proliferation and differentiation phenotypes that we measured from animals treated with the morpholinos designed against the 34 candidate genes. In Fig. 8A, the results of all experiments are summarized and the data are organized in the order of lowest proliferation (bottom of the graph) to highest levels of proliferation (top of the graph) with the control value as the bottom-most bar and red line in the graph (Fig. 8A). We found that 12 of the 34 morpholinos tested increased the proliferation rate, yielding higher numbers of GFP-positive cells on day 3 compared to control values. These are: actinin1 homolog (actn1 homolog, MGC81191); armc8; ADP-ribosylation factor-like 9 (arl9); chimerin 1 (chn1); CTD nuclear envelope phosphatase 1 (ctdnep1a/Dullard); ETS-domain protein (elk4); EPH receptor B1 (ephb1); fibroblast growth factor 2 (fgf2); GLIS family zinc finger 2 (glis2); histone deacetyltransferase 1 (hat1); LSM6 homolog - U6 small nuclear RNA associated (lsm6) and molybdenum cofactor synthesis 3 (mocs3). Treatment with these morpholinos produced a wide range of cell proliferation responses by the cells in the optic tectum. The smallest significant increase in cell number over 48 h was detected after electroporation of the elk4 MO (37.7±8.0% increase, p=0.05), or an increase to 181.7±38.7% of control values. The largest increase occurred with the armc8 MO, which resulted in a 179.1±49.8% increase in cell numbers (p=0.003), or 763.9±212.7% of control values (Figs. 7D, 8A and Table 5). Seven of the 34 morpholinos tested decreased the proliferation rate compared to controls: cytoplasmic polyadenylation element binding protein 1a (cpeb1-a); gstp1; fragile X mental retardation 1a (fmr1a); fragile X mental retardation auto-somal homolog 1 (fxr1); hspa5; matrix metallopeptidase 9 (mmp9) and cAMP-dependent protein kinase catalytic subunit alpha (prkaca). The prkaca MO inhibited cell proliferation most severely, decreasing the proliferation rate to −10.7±6.4% over 48 h, or 51.5±30.7% of control levels (p=0.003; Fig. 8A and Table 5). In contrast, the cpeb1 morpholino decreased the cell proliferation rate to 29.5±18.7% of the mean control levels, or an average increase in cell number of 6.2±3.8% over 48 h, which was the most modest decrease that was significantly different from control levels (p=0.03).
We also determined whether the morpholinos altered the fate of the labeled cells by comparing the proportion of NPCs and differentiated neurons that were generated by day 3 of the time lapse. Animals electroporated with pSox2-bd::FP alone or with control morpholinos had comparable proportions of neurons, NPCs and unclassified cells (Table 3). Figs. 8B and C summarize the proportion of neurons and NPCs seen 3 days after electroporating morpholinos into the tectum. Data are arranged such that morpholinos that produced in highest proportions of differentiated neurons are at the top and those that produced the lowest levels at the bottom of the graph. Control values are given in the bars at the bottom of the graph and with the red lines. 20 out of 34 morpholinos produced in a significant difference in the distributions of cell types compared to control animals (Pearson Chi-square value <0.05, names listed in red in Fig. 8B–C; data given in Table 3). We followed up these Pearson Chi-square analysis with Mann–Whitney U tests to determine which cell types differed in response to the morpholino treatments. Compared to the proportions of cell types found in control conditions, the proportions of NPCs were significantly different with the following 5 morpholino treatments: armc8, chn1, gstp1, hdac6 and hspa5 (marked with asterisks, Fig. 8C; Table 3). Morpholinos against armc8, gstp1, and hspa5 also altered the expected proportions of labeled neurons compared to controls. A summary of the differential expression analysis and in vivo imaging analysis is shown in Table 6.
Table 6.
Summary of screening results from microarray, NanoString and in vivo imaging studies for genes examined by morpholino treatments. 0 means no significant difference in the microarray, NanoString or in vivo analysis of neurogenesis phenotypes. 1 means a significant difference was detected in the assay(s) listed in the column. Concordance refers to the outcome detected in the microarray analysis comparing aNPC(vd) vs immature neurons and the NanoString analysis. If concordance = 1, the outcomes were the same. If concordance = 0, the outcomes were different. Nd = not determined.
| Gene | aNPCvd vs immature neuron | aNPC vs mature neuron | aNPC vs qNPC | Any array call | Nano-String call | Neuro-genesis pheno-type | Array call and neuro-genesis pheno-type | Nano String call and neuro-genesis phenotype | Differential expression by nano String or array | Differential expression and neuro-genesis phenotype | Concordance |
|---|---|---|---|---|---|---|---|---|---|---|---|
| actn1/MGC81191 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 |
| arl9 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 |
| armc8 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| chn1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| cpeb1-a | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| ctdnep1-a / Dullard | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| dio3 | 0 | 1 | 0 | 1 | nd | 1 | 1 | 0 | 1 | 1 | nd |
| efna3 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| eif3a | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 |
| elk4 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| ephb1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| epx | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 |
| fbxw7 | 1 | 1 | 0 | 1 | nd | 1 | 1 | 0 | 1 | 1 | nd |
| fgf2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 |
| fmr1-a | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 |
| fxrl-1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 |
| glis2 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 |
| gstp1 | 1 | 1 | 1 | 1 | nd | 1 | 1 | 0 | 1 | 1 | nd |
| hat1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| hdac6 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 |
| hspa5 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 |
| ism6 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| mecp2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 |
| mkrn2 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| mmp9 | 1 | 1 | 1 | 1 | nd | 1 | 1 | 0 | 1 | 1 | nd |
| mocs3 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| prkaca | nd | nd | nd | nd | 1 | 1 | nd | 1 | 1 | 1 | nd |
| pura | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| r3hdm2 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| rbfox2-b | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| slc12a2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| tle1/tle2 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 |
| vangl1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| wnt7b | 1 | 0 | 0 | 1 | nd | 0 | 0 | 0 | 1 | 0 | 0 |
| 9 | 11 | 10 | 18 | 15 | 26 | 13 | 10 | 26 | 19 | 13 |
Candidate gene analysis identifies subsets of mechanisms that regulate neurogenesis
The in vivo imaging data (Figs. 7 and 8, Supplementary Fig. 1–3, and Tables 2–6) indicate that the different morpholinos generate a range of phenotypes with respect to cell proliferation and differentiation. To determine whether the different neurogenesis phenotypes, for instance changes in the numbers of NPCs and neurons, show separate or nested realtionships, we generated a Venn diagram of the subsets of morpholino-induced phenotypes seen with the set of 34 genes (Fig. 9). Three genes targeted with the morpholino experiments altered numbers of neurons generated by day 3 (light blue, Fig. 9). All three of these genes were also among the set of 5 morpholinos where the proportions of NPCs were altered as well (purple set, Fig. 9). We found a reciprocal relationship between numbers of neurons and NPCs; when neuron numbers were significantly decreased (i.e., hspa5; Figs. 7 and 8), NPCs were significantly decreased; when neuronal numbers were increased (i.e., gstp1 and armc8; Figs. 7 and 8), NPCs were significantly decreased (Table 4 and Fig. 8) compared to control levels.
Fig. 9.

Candidate gene sets defined by neurogenesis phenotypes. Categories of neurogenesis phenotypes from morpholinos are shown as colored ellipses where the area of each ellipse is proportional to the number of genes in that category. Of the 34 candidate genes (red) tested with morpholino treatment, 24 significantly altered the proportions of cell types (blue circle, Pearson's Chi-square) and 19 significantly altered the proliferation rate (yellow, Mann–Whitney U test). The morpholinos against 15 candidate genes altered both the cell types generated and the proliferation rate. The overlap between these two categories is shown in green. Among these 15 genes, 5 generated significant differences in the proportions of NPCs (purple, Mann–Whitney U test) and of those, 3 also had significant differences in the number of neurons that were generated (light blue, Mann–Whitney U test). One of the 5 (purple) changed both proliferation rate and NPC number (peach).
When morpholinos alter proliferation rates, they were also likely to change the ratio of cell types imaged on day 3. Twenty genes targeted with morpholinos produced a significant change in the proportions of cell types generated compared to controls (dark blue set in Fig. 9, the genes indicated with red font in Fig. 8, and Table 5). All but two of the morpholinos that resulted in significant changes to the proportions of cell types on day 3 also altered the cell proliferation rates (yellow set). Of the 19 genes that altered proliferation rates (yellow set, Fig. 9), 15 were among those that showed significant changes to the proportions of cell types generated compared to controls (the overlap indicated with green, Fig. 9). We found that only morpholinos against hdac6 (peach color, Fig. 9) altered the proportion of a cell type (a significant decrease in the number of NPCs) without altering the level of cell proliferation in the tectum. This analysis shows the nested relationship of the neurogenesis phenotypes and suggests mechanistic pathways governing neurogenesis.
Discussion
Summary of major observations
Many studies designed to identify genes regulating neurogenesis are conducted under conditions in which neural stem or progenitor cells are removed from their native environment in the intact animal. Such an experimental design precludes the opportunity to detect the participation of genes that might be uniquely regulated by events in the intact system. Our previous studies in the tadpole visual system demonstrated that visual deprivation increases cell proliferation within the optic tectum, while stimulation of the developing visual circuits in the optic tectum drives newly generated cells to differentiate into neurons (Bestman et al., 2012; Sharma and Cline, 2010). Here we used in vivo manipulations of neural circuit activity to bias populations of Sox2-expressing GFP-labeled NPCs toward different fates (neurons or NPCs), followed by and differential expression analysis of transcripts from the enriched cell populations in an effort to identify a broad range of neurogenic transcripts and signaling pathways. Bioinformatic analysis of the microarray data indicates that experimental conditions designed to enrich for NPC or neuron populations consistently identified signaling pathways related to cell cycle regulation and cell differentiation. In vivo time-lapse imaging of neurogenesis in the optic tectum demonstrated that morpholinos against 24 of 34 candidates regulated cell proliferation, cell differentiation or both in the intact animal (Fig. 10). Our high success rate at identifying candidates that when knocked down altered neurogenesis, validates the logic of the screen. The candidates that we tested by morpholino treatment appear to fall within diverse signaling pathways, suggesting that additional genes within these signaling pathways would be worth testing for in vivo neurogenic effects.
Fig. 10.
Summary of neurogenic outcomes under control conditions and with candidate gene morpholinos. (A) Diagram of NPC fates summarized from in vivo imaging experiments. NPCs can either differentiate into neurons or divide and generate NPCs or neurons. (B) Cartoons of the proportions of NPCs and neurons observed on day 1 and day 3 for control animals and animals electroporated with morpholinos against several candidate genes, which represent a range of neurogenic outcomes found in the present study. In control animals, the proportion of NPCs decreases and the proportion of neurons increases over the observation period. Morpholinos against armc8 result in an exaggerated increase in neurons, gstp1 morpholinos result in a rapid shift in the proportion of NPCs to neurons, whereas hspa5 morpholinos result in an increase in the proportion of NPCs and a decrease in the proportion of neurons.
Logic of the microarray screen and validation of the logic
We were interested in using gene expression profiling to identify candidate transcripts that might regulate cell proliferation and neurogenesis in vivo. To do this we took advantage of the X. laevis retinotectal system, where we transfected cells in vivo and harvested tectal cell populations enriched for NPCs or differentiated neurons. Optic tectal cells were collected from tadpoles at different stages of development and with different visual experience, and RNA was isolated only from cells expressing our pSox2-bd::FP cell-specific reporter, which is expressed in Sox2-expressing NPCs and their lineages. By comparing the cell groups in the manner described at the bottom of Fig. 1, we made 3 independent comparisons, each designed to reveal transcripts involved in cell proliferation and neurogenesis pathways. NanoString analysis of tectal cell populations enriched for aNPCs and Immature Neurons demonstrated approximately 50% concordance between the Nano-String and microarray data.
A critical aspect to our experimental strategy was the NPC-specific reporter we used to isolate cells for differential expression analysis and to image NPCs in the optic tectum. It is widely recognized that neural stem cells have heterogeneous responses to extrinsic signals which affects their proliferative capacity, cell differentiation and lineages (Alvarez-Buylla et al., 2008; Bonaguidi et al., 2011, 2012; Carney et al., 2012; Encinas et al., 2006; Giachino and Taylor, 2009; Lugert et al., 2010; Maisel et al., 2007; Park et al., 2012; Song et al., 2012; Vergano-Vera et al., 2009). By harvesting and imaging only cells labeled by pSox2bd::GFP expression, we limited the heterogeneity of the cell populations that we analyzed. A second critical aspect of our strategy was that intact animals were reared under conditions known to affect NPC proliferation and fate to enrich for specific cell populations. A third element of our experimental design was by conducting multiple microarray comparisons in parallel, in addition to performing the NanoString analysis, identified transcripts that are differentially expressed in actively dividing NPCs compared to neurons.
One gene profiling comparison between aNPCvd and Immature Neurons was based on our observation that visual deprivation increased NPC proliferation and expanded the progenitor pool, whereas visual experience increased neuronal differentiation (Bestman et al., 2012; Sharma and Cline, 2010). Studies in other systems have also suggested that early brain activity regulates stem cell fate and neuronal differentiation (LoTurco et al., 1995; Nacher and McEwen, 2006; Reynolds et al., 2008). We anticipated that transcripts that are differentially expressed under these conditions would play a regulatory role in either maintaining NPCs in a proliferative state or changing their fate to generate neurons. The second microarray comparison was based on our in vivo studies demonstrating that optic tectal neurons differentiate and integrate into the functional visual circuit within 5 days (Bestman et al., 2012; Chiu et al., 2008; Sharma and Cline, 2010). We therefore compared transcripts from cells collected 1 or 5 days after labeling to identify differentially expressed transcripts between NPCs and differentiated neurons, similar to previous reports (Bhattacharya et al., 2009; Carney et al., 2012; Falk et al., 2007; Geschwind and Rakic, 2013; Karsten et al., 2003; Marei et al., 2011; Parker et al., 2005). We then focused our attention on transcripts that were differentially expressed in both sets of comparisons, based on the idea that both comparisons would distinguish transcripts expressed in NPCs from those expressed in neurons. The third microarray comparison was based on observations that rates of proliferation of Sox2-expressing NPCs decrease over development (Sharma and Cline, 2010). We compared relatively active and quiescent NPCs isolated from stage 47 and 49 tadpole optic tecta, anticipating that the differential expression analysis would identify transcripts that play a role in maintaining NPCs in a proliferative or quiescent state. We selected genes for follow up analysis by in vivo imaging based on their fold-change values and p values in the microarray and NanoString analyses. Some transcripts were chosen based on deduced functions from the literature. Of the 27 differentially expressed transcripts, morpholinos against 18 (66%) altered neurogenesis. Nano-String identified 16 differentially-expressed transcripts, of which morpholinos against 11 (69%) generated a neurogenesis phenotype. Similarly, 65% (13/20) of differentially expressed transcripts identified by microarrays had a neurogenic phenotype (Table 6).
Several bioinformatic analyses that we performed validated our strategy to identify differentially expressed genes in NPCs and neurons. All three microarray comparisons and the NanoString analysis identified transcripts involved in processes governing cell proliferation and differentiation. Of the 759 genes that were differentially expressed in multiple microarray comparisons, 87% (659) were enriched for either cell proliferation or cell differentiation. These relationships are illustrated by the network generated by Gephi (Fig. 3A), which tightly clustered the aNPC transcript groups together. In each of the microarray comparisons (Fig. 2B), and in the subset of transcripts that were upregulated across multiple aNPC groups (Fig. 3B), the DAVID bioinformatic analysis identified an abundance of transcripts associated with the nucleosome and chromatin assembly. RNA binding proteins were also significantly higher in aNPCs. In addition, the analysis identified gene clusters that are associated with cellular metabolism and oxygen transport, which are gene families that have known functions in cell cycle control and differentiation (Mochizuki et al., 2009). Analysis with MetaCore software showed that cell cycle and cell fate pathways are up-regulated in aNPCs, consistent with the DAVID analysis. A range of cell cycle and cell fate pathways are enriched in aNPCs, including transcripts involved in developmental processes (e.g. notch and wnt signaling), the cell cycle (e.g. cyclins and histones), and those that are involved in both the immune response and cell proliferation (e.g. complement signaling proteins).
Together, our analyses indicate that the strategy to enrich populations of NPCs and differentiated neurons from intact brain was successful. This approach identified known regulators of NPCs and differentiating neurons (e.g., vimentin, Musashi1, Emx2, GLAST, sox and pou transcription factors, neuroD, and HuC). Notably, the bioinformatic analysis of the microarray data indicates that NPCs in Xenopus optic tecta utilize a broad range of regulatory pathways in neurogenesis. Furthermore, our in vivo analysis of neurogenesis in animals treated with morpholinos indicates that a strategy of rapid selection of candidate genes by differential gene expression combined with sensitive in vivo morphogenetic assays effectively identifies candidate neurogenic genes for further study.
Activity-dependent regulation of neurogenesis
Growing evidence indicates that neural activity regulates NPC proliferation and differentiation (LoTurco et al., 1995; Nacher and McEwen, 2006; Reynolds et al., 2008), although the mechanisms by which changes in neuronal circuit activity affect NPC proliferation and differentiation remain unclear. Neuronal activity is well known to induce hundreds of genes in neurons, many of which play a role in synaptic plasticity, and some may indirectly affect NPC functions. This study was designed to take advantage of in vivo cellular interactions within a functional circuit that occur with visual deprivation and developmental time to maximize differences in NPC and neuronal populations in the harvested cell groups. However, the differentially-expressed transcripts identified in NPCs are not necessarily directly regulated by activity in NPCs. We note that aNPCs from the different microarray comparisons share hundreds of upregulated transcripts, but aNPCs and neurons share few differentially-expressed transcripts. Further studies are required to determine how changes in sensory input affect changes in gene expression in NPCs, and to determine how candidate genes affect neurogenesis.
Differential contributions of cell proliferation and differentiation to neurogenesis
Neurogenesis includes multiple distinct cellular events, each regulated by distinct processes: cell proliferation, cell survival, and differentiation. Our in vivo time-lapse imaging studies have shown that optic tectal NPCs fall into several categories. A minority of NPCs are either quiescent or symmetrically dividing; a majority of NPCs divide asymmetrically and generate neurons (Bestman et al., 2012). Many NPCs exhibit a protracted delay after cell division before differentiating (Bestman et al., 2012), suggesting that NPCs may respond to cues directing them to either exit the cell cycle and differentiate or retain a progenitor fate. We anticipate that distinct neurogenic regulatory genes affect these different cellular events.
Our analysis demonstrated that 24 of the 34 genes targeted with morpholinos altered cell proliferation, and the majority of these also changed the proportions of GFP+ neurons and NPCs. Among these 24 candidate neurogenic regulatory genes, we found a range of outcomes with respect to cell fate and proliferative capacity (Fig. 10). For example, morpholinos against gstp1 significantly increased the number of differentiated neurons and reduced the number of NPCs compared to controls. This decreased total cell proliferation over the imaging period, because the neural progenitor pool was depleted. In contrast, morpholinos against hspa5 also limited total cell number, but the proportion of NPCs increased and the proportion of neurons decreased, suggesting that hspa5 regulates neurogenesis by limiting cell proliferation and decreasing differentiation. These examples reflect three general mechanisms that control neurogenesis: the regulation of progenitor proliferation, the regulation of cell cycle exit, and the initiation of differentiation. It is also possible that individual candidate genes affect several different cellular processes contributing to neurogenesis. For instance, we have recently demonstrated that fxr1a, which encodes the Xenopus homolog of FMRP, regulates neurogenesis by controlling both NPC survival and neuronal differentiation (Faulkner et al., 2015). Finally, overlapping cellular mechanisms may be affected by several of the genes targeted in our study, as suggested below.
Candidates identify categories of cellular responses affecting neurogenesis
Several candidate genes that were first flagged in the microarray comparisons, and then shown to have a neurogenesis phenotype in the in vivo imaging assays, play a role in regulating cytoskeletal dynamics. For instance, histone deacetylase 6 (HDAC6), a cytoskeletal scaffold protein expressed in both neural stem cells and neurons (Valenzuela-Fernandez et al., 2008), is one of the candidates identified and validated by our two-tiered screen. Following morpholino treatment, hdac6 morphant animals had significantly fewer GFP-labeled NPCs at all timepoints compared to control animals (Tables 1 and 2), resulting in an overall decrease in cell proliferation (Fig. 8A and Table 4). Unlike control animals, hdac6 morphants had stable numbers of neurons and NPCs over the three days of in vivo imaging (Table 3). HDAC6 may affect cell proliferation through either its deacetylase activity or its ability to regulate protein ubiquitination and turnover. HDAC6 deacetylates several cytoskeletal proteins, including tubulin, a major component of microtubules, and cortactin, which regulates actin polymerization, suggesting that HDAC6 may affect cell proliferation by regulating the microtubule-based and actin-based cytoskeleton (Gao et al., 2007; Valenzuela-Fernandez et al., 2008; Zhang et al., 2007). In neurons, HDAC6 regulates axon and dendrite outgrowth (Ageta-Ishihara et al., 2013; Kim et al., 2009), neurodegeneration, and response to injury (Rivieccio et al., 2009; Simoes-Pires et al., 2013), likely via its capacity to regulate cytoskeletal dynamics and protein homeostasis. Because electroporation distributes morpholinos throughout the optic tectum, it is possible that morpholinos against hdac6 decrease proliferation indirectly by modifying neuron or circuit function.
A second feature common to several of the candidates that we tested is their potential role in protein homeostasis. As mentioned above, HDAC6 contains an ubiquitin binding domain and may stabilize polyubiquinated proteins (Valenzuela-Fernandez et al., 2008). Similarly, ARMC8 is part of a highly conserved complex that regulates protein degradation through both proteasomal/polyubiquitin-dependent degradation and endocytosis of targets followed by lysosome-mediated degradation (Tewari et al., 2010). Although relatively little is known about ARMC8 function in NPCs and neurons, evidence from other systems suggests it may bind micro-tubules and thereby regulate microtubule dynamics and cell division (Kobayashi et al., 2007; Tewari et al., 2010). HSPA5, also known as BiP and GRP78, is an endoplasmic reticulum chaperone protein that associates with newly synthesized proteins and facilitates protein folding (Dudek et al., 2009; Zoghbi, 2005). HSPA5 is involved in the unfolded protein response (UPR), an indicator of cell stress in which aberrantly folded proteins are degraded. Recently, HSPA5 has been implicated in Marinesco–Sjogren syndrome, a rare inherited syndrome characterized by cerebellar ataxia and developmental delay (Anttonen et al., 2005; Inaguma et al., 2014; Senderek et al., 2005; Zoghbi, 2005) characterized by defective cortical neuron migration and axon growth. HSPA5 may also affect stem cell proliferation and neurodegeneration (Fan, 2012; Paschen, 2003; Prinsloo et al., 2009). A fourth candidate that plays a role in protein homeostasis is the detoxification protein, GSTpi, encoded by gstp1. GSTpi is the most highly expressed GST in the CNS, where it is concentrated in neurogenic regions. Several studies suggest that GSTpi plays a role in neurogenesis, through mechanisms independent of its detoxification functions (De Luca et al., 2003; Le Belle et al., 2011; Walton et al., 2012). Neural stem cells have high endogenous levels of reactive oxygen (ROS) and markers for oxidative stress (Le Belle et al., 2011; Walton et al., 2012) and modest increases in ROS may increase proliferation, while low ROS levels correlate with NPC quiescence (Le Belle et al., 2011). The data suggest that the energy-intensive process of cell division produces ROS and induces expression of proteins that are classically considered pathological stress proteins (Walton et al., 2012). We find that the GSTpi morpholino increase neuron numbers and deplete the progenitor pool (Fig. 10). This is consistent with the idea that GSTpi and ROS are dynamically regulated in the developing brain to control specific cellular signaling pathways and functional outcomes (Townsend et al., 2009; Walton et al., 2012).
Several candidates may affect neurogenesis through transcriptional effects. For instance, ARMC8 regulates beta-catenin transactivation and nuclear signaling indirectly by enhancing degradation of its inactivating binding partner, alpha-catenin (Suzuki et al., 2008). ARMC8 knockdown is therefore predicted to increase alpha-catenin and increase proliferation (Stocker and Chenn, 2009). Consistent with this, we observed an increase in neurogenesis with the armc8 morpholino. ELK4, or ETS-like transcription factor, (a.k.a. serum response factor accessory protein 1, SAP-1), belongs to a family of transcription factors that is targeted by extracellular signaling pathways and known as key regulators of neural developmental events in a variety of species, including Xenopus (Janesick et al., 2013; Remy and Baltzinger, 2000; Willardsen et al., 2014). ELK4 may act as a transcriptional repressor or activator, depending on post-translational modification (Kaikkonen et al., 2010). In addition, ELK4 has been shown to enhance expression of anti-apoptotic proteins in glioma, thereby increasing proliferation (Day et al., 2011). We find that morpholinos against elk4 increased cell proliferation by 182% of the control proliferation rate (Table 6). A third candidate that may control neurogenesis through regulating transcription is Glis2 (also known as NKL) (Hosking et al., 2007; Lamar et al., 2001; Lichti-Kaiser et al., 2012). Glis2 can act as a transcriptional activator or repressor, depending on its binding partners (Hosking et al., 2007; Vasanth et al., 2011). Glis2 (in Xenopus, mouse, and chick) is expressed in committed neural progenitors and neurons (Lamar et al., 2001). Expressing Glis2 in Xenopus and chick CNS promotes neuronal differentiation, likely by activating differentiation genes downstream of proneural genes (Lamar et al., 2001). Morpholinos against Glis2 increased the proliferation rate by 272% of the control rate (Fig. 7A and Table 6), consistent with a role in directing NPCs to exit the cell cycle.
The final category of candidates includes extracellular pro-teases, like matrix metalloprotease 9 (MMP9), a secreted Zinc-dependent extracellular endopeptidase (Wlodarczyk et al., 2011). MMP9 regulates the pericellular environment by local proteolysis of specific substrates, including extracellular matrix (ECM) components, beta-dystroglycan, beta 1 integrin, and neuroligins (Lee et al., 2014; Michaluk et al., 2007; Peixoto et al., 2012; Wang et al., 2008; Wlodarczyk et al., 2011). The ECM is thought to limit structural dynamics of cells as well as extracellular signaling capacity. We found that morpholinos against mmp9 stopped cell proliferation, yielding no net increase in GFP-labeled cells over the time lapse period. MMP9 plays a role in cell proliferation, neuronal differentiation and plasticity, and neurodegeneration (Barkho et al., 2008; Dziembowska et al., 2012; Kaplan et al., 2014; Marei et al., 2011; Meighan et al., 2007; Nagy et al., 2006; Okulski et al., 2007; Peixoto et al., 2012; Szklarczyk et al., 2002). It is interesting to point out that MMP9 synthesis is increased in neurons downstream of NMDA receptor activity (Dziembowska et al., 2012), and might affect NPCs in a non-cell autonomous manner.
In conclusion, we have carried out a 2-tiered screen to identify and validate candidate neurogenic genes based on a combinatorial analysis of differential expression followed by a high content time-lapse in vivo imaging protocol. That several candidates have known roles in neurogenesis and brain development is evidence that our strategy was capable of identifying neurogenic genes. While here we used our gene profiling results to guide the selection of candidate genes, the in vivo imaging protocol could also be used to understand the contributions of novel genes that are identified through clinical genomics. We found that the genes we identified participate in diverse cellular events and signaling pathways, suggesting that our strategy was not biased toward particular classes of genes or types of cellular signaling events. The diversity of gene families and pathways identified in this study also suggests that further analysis will identify more candidates within the different pathways. Our data identified potentially novel neurogenic regulators, demonstrating the utility of using Xenopus as a vertebrate experimental system for gene discovery. Finally, several candidate genes have been implicated in human disease, suggesting that Xenopus will be a valuable experimental system to probe mechanisms related to human brain health and disease. Further studies are required to elucidate the mechanisms by which specific candidates affect neurogenesis, for instance the series of experiments described in our recent study that examined the role of FMRP in neurogenesis (Faulkner et al., 2015).
Materials and methods
Animal rearing
All animal protocols were approved by the Institutional Animal Care and Use Committees of Cold Spring Harbor Laboratory (approval # 05-02-04), The Marine Biological Laboratory at Woods Hole, MA (approval # 08-07C), or the Scripps Research Institute at La Jolla, CA, (approval # 08-0083). Fertilized eggs were acquired from hormone-induced matings of albino X. laevis frogs in our colony or purchased from commercial sources (Nasco, Fort Atkinson, WI), Xenopus Express (Brooksville, FL), or Xenopus One (Dexter, MI). Tadpoles were housed at 23 °C with a 12 h light: 12 h dark diurnal cycle until used for experiments. Tadpoles were anesthetized for in vivo experiments by placing them in 0.01% MS-222 (Sigma; all chemicals were purchased from Sigma unless noted), and terminally anesthetized in 0.2% MS-222 before brains were dissected to harvest labeled cells (see below).
Animals used for microarray experiments were divided into 5 treatment groups. All animals were raised in 12 h Light: 12 h Dark (L:D) conditions. At stage 46 (5 days post fertilization, dpf) or stage 48 (10 dpf) (Nieuwkoop and Faber, 1956), tadpole optic tecta were electroporated with pSox2-bd::tGFP. The pSox2-bd::FP plasmid is described in detail in Bestman et al. (2012). Briefly, the Sox2/Oct3-4 transcription factor binding domain and FGF4 minimal promoter were cloned upstream of gal4/UAS and a fluorescent protein (FP), in this case tGFP (Bestman et al., 2012). Tadpoles were reared in one of three visual experience conditions: normal 12 h light:12 h dark conditions; visual deprivation, in which animals were placed in a dark chamber; or enhanced visual experience, in which animals were placed in a chamber in which they received a simulated motion stimulus from an array of LEDs (Sin et al., 2002). Depending upon the experimental group, after either 1 or 5 days under these conditions, the animals were anesthetized and tGFP-positive cells were harvested manually, as described below. These rearing conditions produced the 5 following cell groups: active NPCs (aNPCs) isolated from the stage 46 animals one day after electroporation; Mature Neurons isolated from the stage 46 tad-poles 5 days after electroporation; Immature Neurons isolated from stage 46 animals that were exposed to an enhanced visual environment for the previous 24 h; aNPCvd isolated from st46 tadpoles that had been visually deprived for previous 24 h; and qNPCs isolated from the stage 49 tadpoles 1 day after electro-poration at stage 48.
Plasmid constructs and in vivo tectal cell transfection
DNA plasmids and morpholinos were electroporated into tectal cells using bulk electroporation, a process by which macromolecules are injected into the tectal ventricle and driven into tectal cells using voltage pulses across the midbrain (Sin et al., 2002). For microarray studies, stage 46 or stage 48 tadpoles (Nieuwkoop and Faber, 1956) were anesthetized and electroporated with 2 μg/μl pSox2-bd::tGFP. For the in vivo imaging experiments, stage 46 tadpoles were anesthetized and co-electroporated with pSox2-bd:: FP plasmid and antisense morpholino oligonucleotides (described below). The fluorophores used in the pSox2-bd:FP plasmid for the in vivo imaging experiments were turboGFP (Evrogen), a fast maturing green fluorescent protein, or Kaede (MBL International), a fluorophore whose emission can be irreversibly converted from green to red fluorescence after exposure to a UV light source. In some experiments, we also co-electroporated the pSox2-bd:turboGFP with pSox2-bd:turboRFPnls. TurboRFPnls is a fast maturing red fluorescent protein (Evrogen) tagged with a nuclear localization sequence (nls) to label cell nuclei.
Microarray analysis
Harvesting GFP-expressing cells for RNA isolation/purification for microarray analysis
pSox2-bd::eGFP-expressing cells were harvested from 5 different treatment groups of tadpoles, which differed according to their developmental stage and visual experience rearing conditions, as described above and in Fig. 1. Tadpoles were terminally anesthetized in 0.2% MS-222 and their midbrains were dissected out and placed in 1.5 ml microcentrifuge tubes containing 270 μl of amphibian phosphate buffered saline (aPBS; 113 mM sodium chloride, 8 mM sodium phosphate dibasic, 1.5 mM potassium phosphate monobasic, pH 7.7) with 0.1% EDTA (Sigma). Once 8 midbrains were dissected, 30 μl of 2.5% Trypsin-EDTA (Invitrogen) was added to yield a final working concentration of 0.25% Trypsin-EDTA. After 15 min incubation at room temperature, the reaction was stopped with 300 μl of Defined Trypsin Inhibitor (Invitrogen). The tissue was gently triturated with a large bore fire-polished glass Pasteur pipette and transferred into a 15 ml Falcon tube containing 4 parts L15-Leibovitz medium (Invitrogen), 5 parts APBS, and 1 part 10% Bovine Calf Serum (Invitrogen) in a total volume of 10 ml. After 15 min on ice, the solution was triturated again using a smaller bore fire-polished glass Pasteur pipette. The dissociated cell mixture was divided into 3 wells of a 4-well rectangular cell culture plate (Nunc; well capacity 22 ml). The fourth well of the plate contained media and served as the receptacle for the harvested cells. The plate was placed on the stage of a Zeiss Axiovert 200 microscope equipped with fluorescence and an oil-filled microinjector to hold a cell-picking micropipette mounted on a 3-axis hydraulic micromanipulator (both, Narishige). Micro-pipettes (1 mm outer diameter, 0.5 mm inner diameter glass, WPI) were pulled using a Sutter P-97 puller, and scored with a light swipe from a second micropipette so that they could be broken to produce a clean, 50 μm diameter tip (Sutter Pipette Cookbook, (Osterele, 2012). After the cells settled for 15 min in the culture plate, eGFP-expressing cells were aspirated into the micropipette and ejected into the 4th well of the plate, containing media. All cells were collected within 90 min after the tissue was first dissociated with trypsin. The sorted cells were then pulled into a new micropipette, ejected into 1.5 mL microcentrifuge tube, and centrifuged gently to pellet the cells. Excess media was removed and the samples were frozen at −80 °C. Typically, 8 tadpoles yielded 100–200 FP-positive cells. For each experimental condition, we repeated the cell dissociation/harvesting procedure 12 times to yield at least 1200 cells per condition.
RNA extraction, amplification, preparation and hybridization for microarray analysis
Harvested cells from each tadpole rearing condition were pooled and then divided into 5 replicates that were prepared simultaneously as follows. RNA was extracted using the PicoPure RNA Isolation Kit (Applied Biosystems) and immediately amplified using the WT-Ovation Pico Kit (NuGEN) according to manufacturer's instructions, except that we used 100% rather than 80% ethanol during the RNA cleaning steps. The cDNA was fragmented and labeled using the Encore Biotin Module (NuGEN). RNA quality was analyzed before and after the amplification with a Bioanalyzer (Agilent). Labeled cDNA products were hybridized to the X. laevis GeneChip 2.0 (Affymetrix) according to the NuGEN FL-Ovation manual. Microarrays were rinsed and read using the methods recommended by Affymetrix.
Microarray analysis
A GeneChip Scanner 3000 (Affymetrix) and GeneChip Command Console and Expression Console Software (Affymetrix) were used to read the microarray chips. Data were exported as CEL files by Affymetrix Expression Console Software. Hybridization quality across the chips was evaluated, and regions with edge effects or bubbles were excluded from the analysis. Raw intensity data were evaluated and processed by software developed by Tim Tully and Philip Cheung (Dart Neuroscience LLC) as follows: (1) no background subtraction; (2) normalization using the geometric mean across the whole gene chip to reduce the effect of uneven hybridization across individual arrays; (3) data were subjected to a Box– Cox transformation to normalize residuals (error variance) across the replicates; (4) filter to exclude the individual probes affected by the hybridization artifacts and outlier data points for single probes; (5) data were subjected to a Box-Cox transformation after removing filtered probes to generate a more accurate transformation; (6) the 5 microarray replicates for each of the experimental conditions were analyzed to derive gene expression values. Calls are made based on either the p-value and/or powerT, which incorporates the pooled variance of all 5 samples and reduced the false positive hit (type I error).
We generated an updated version of the .CDF file (annotation file matching the probes on the GeneChip to GeneBank mRNAs) to incorporate the recent updates of the X. laevis genome data based on work performed by the BrainArray group at the University of Michigan (http://brainarray.mbni.med.umich.edu/Brainarray/Data base/CustomCDF/genomic_curated_CDF.asp) (Dai et al., 2005). We aligned the nucleotide sequence of each probe against the X. laevis genome. Probes were removed from the analysis if they did not match 100% to the X. laevis genome or if they matched more than one transcript. In addition, in step 4 stated above, transcripts were removed from the list of potential gene calls if the number of probes identifying a transcript did not pass threshold. The updated GeneChip annotation enabled us to screen for 7127 transcripts in the X. laevis genome based on 105,838 probes. The numbers of genes and probes we include in our analysis are different from the Affymetrix-claimed 32,400 probe sets for 29,900 transcripts on the GeneChip, because of updates to the X. laevis genome data. For the arrays analyzed in the study, 7103–7115 transcripts per array passed all the criteria for inclusion in the analysis. Transcripts that exhibited differential expression between 2 rearing conditions, were identified based on p-value <0.05 and Power T. We selected 34 candidate genes for further validation using antisense morpholino oligonucleotides (GeneTools, LLC) to inhibit their translation. Morpholinos were designed by GeneTools, and the sequences for the experimental and control morpholinos used in the study are given in Table 1
NanoString
Late stage 46 animals were electroporated with pSox2-bd:: turboGFP and reared in the dark (visual deprivation) or with enhanced visual experience for 24 h to generate samples enriched for aNPCvd and Immature Neurons, respectively. 100 midbrains were collected from animals reared in each condition and dissociated with aPBS. Approximately 40,000 Sox2+ve-turboGFP cells were collected using Fluorescence Activated Cell Sorting (FACSAria II, BD Biosciences, USA). Total RNA was extracted following the mirVana miRNA kit protocol (Life Technologies, USA), followed by DNase treatment to remove genomic DNA and cleanup using RNeasy mini kit (Qiagen, USA). Total RNA had RIN >8, measured with a Bioanalyser. 2ng of total RNA was amplified to 2–3 μg of double-stranded cDNA, using the Ovation RNA-Seq System V2 (NuGEN, USA). The amplified cDNA was purified, using Agencourt AMPure XP beads, quantified by NanoDrop, and its quality was confirmed with the Bioanalyser. 200 ng of cDNA was used as input for the NanoString nCounter Gene Expression system according to the manufacturer's protocol. 4 biological replicates were used for each condition.
The digital output of the number of transcripts was analyzed, using nSolver Analysis Software 2.0. Background and hybridization were normalized with spike-in negative and positive controls, provided by the manufacturer; reference genes, GapdH (NM_001087098.1), actb (NM_001088953.1), and rps13 (NM_001086882.1) were selected for normalization between different biological replicates.
Welch's t-test was used to determine the significance of differential expression of each of the 95 transcripts between the 2 conditions in each replicate. A p-value <0.05 was required for the transcripts to be called as significantly different between the 2 conditions.
Concordance analysis was performed among 4 replicates to determine whether the transcript expression levels were statistically different between the aNPCvd and Immature Neuron samples based on the following criteria: if 4 out of 4 replicates have p-value <0.05, and 3–4 replicates show differential expression in the same direction, or if 2–3 replicates out of 4 have p-value <0.05, and all 4 show differential expression in the same direction. Otherwise, the expression level of transcripts was considered nonsignificant.
Bioinformatic analysis
The gene networks were plotted using the Gephi program (version 0.8.2; Bastian et al., 2009) using the Force Atlas layout with the following parameters to generate a stable gene network: 0.1 attraction strength, 0.2 auto stabilize function, 10000 autostab strength, 30 gravity, 10000 maximum displacement, 10,0000 repulsion strength, adjusted by sizes and attraction distribution. Pathway analyses were performed with DAVID, the Database for Annotation, Visualization and Integrated Discovery (http://david.abcc.ncifcrf.gov; (Huang et al., 2009) and MetaCore (Thomson Reuters, NY) to investigate the biological and functional implications of the differentially-regulated gene expression seen in the different rearing conditions. DAVID was used to compare the frequency of gene ontology terms in a transcript list to the expected frequency of the background. The transcripts from the Affymetrix X. laevis 2.0 chips were used as the background. We set DAVID EASE Scores (a modified Fisher exact p-value) to p <0.05, and used the Functional Annotation Clustering algorithm to categorize genes with Benjamini false discovery rates of p <0.05.
MetaCore (Thomson Reuters Ltd.; http://www.genego.com/metacore.php) mines a privately curated integrated knowledge database and is designed for pathway analysis of human gene lists. Human homologs of the X. laevis transcripts identified by searching oligonucleotides on the Affimetrix 2.0 Xenopus microarray were used as input for MetaCore. MetaCore maps the input genes in a list onto genes in the built-in functional ontologies, such as pathway maps and map folders, and networks, and ranks the map folders or pathway maps based on their p-values of hypergeo-metric distribution. The p-value is calculated as the probability of a particular mapping of a list to an ontology compared to chance, considering the size of the ontology and the gene list from the comparison. p-value <0.05 was used as the cut-off to ensure the non-randomness of the selected entities in any given ontology. In addition to p-value, a False Discovery Rate (FDR) <0.05 was also applied to minimize the occurrence of type I errors in the multiple comparisons. Two major ontologies were used to describe the differentially expressed genes from three comparisons, map folders and canonical pathway maps. Map folders are a collection of pathway maps, grouped hierarchically into folders based on the main biological processes. Canonical pathway maps have multiple sequential steps of interactions, defining a signaling mechanism. Map folders provide an overview on the major biological processes of the genes of interest from each comparison list. Canonical pathway maps provide detailed information on the cascades in the signaling mechanism. In addition, individual canonical pathway maps can be ranked independently of the hierarchical ranking of the map folders, in which they are categorized. These two different ontologies provide insights into the biological processes of the genes enriched in either cell proliferation or differentiation.
In vivo time-lapse imaging and data analysis
We selected 34 candidate genes for analysis of their roles in cell proliferation and neuronal differentiation (Table 1). Of these 27/34 showed differential expression by microarray or NanoString analysis (Table 6). The remaining morpholinos were included in the candidate list based on predicted effects on neurogenesis from the literature or based on initial microarray comparisons, which subsequent analysis using an updated CDF file and more stringent inclusion criteria suggested were not differentially expressed. Morpholinos initially designed against elk4 or elk4-b were subsequently predicted to interact with both transcripts.
To prepare animals for in vivo time-lapse imaging, stage 46 tadpoles were bulk electroporated with 0.1 mM antisense morpholino oligonucleotide (GeneTools, Philomath, OR) and 2 μg/μl pSox2-bd::FP (where FP was Kaede or turboGFP) alone or combined with pSox2-bd::turboRFPnls. Morpholinos were resuspended in water to make 1 mM stock solutions that were diluted to 0.1 mM working concentrations. Animals were returned to normal rearing conditions after electroporation and time-lapse imaging protocols began 24–36 h later when the tadpoles were stage 47. For imaging, tadpoles were anesthetized in 0.01% MS-222 and placed in a custom-built chamber with a coverslip placed in contact with the skin directly over the brain. We monitored blood flow in the CNS to ensure the animals were healthy. Tadpoles with approximately 20 sparsely labeled cells per optic tectal lobe were selected for time-lapse imaging. Complete 3D confocal stacks (180 μm depth at 1 μm intervals) of all labeled cells in the tectum were acquired once a day over 3 days, with an Ultraview VoX, Yokogawa spinning disk confocal system (Perkin-Elmer), as described (Bestman et al., 2012). After each imaging session, tad-poles were placed in Steinberg's solution in a 6-well tissue culture dish and returned to normal 12 h light: 12 h dark conditions.
We used the automated functions of Volocity 5 software (Improvision/Perkin Elmer) to identify and count cells in the 3D confocal stacks, and verified results of automated analysis by visual inspection. Analysis was also conducted using the FIJI imaging processing package of Image J (Schindelin et al., 2012; Schneider et al., 2012), in which FP-labeled cells were identified and counted. The numbers of FP-expressing cells at day 1 and day 3 were counted and the proliferation rates were calculated as the change in FP-expressing cell numbers between day 3 and day 1, normalized to the number of labeled cells on day 1. In addition, each cell was assigned an identity based on its morphological features described previously (Bestman et al., 2012; Wu and Cline, 2003; Wu et al., 1999). To summarize, we designated the cell as a neural progenitor cell when it had a radial process that ended on the pial surface with endfoot enlargement. Neural progenitors lack axons or dendritic arbors characteristic of neurons. Young neurons with few branches are distinguished from neural progenitors by the outgrowth of axons along the surface of the tectum. These data were used to determine the proportion of neurons and neural progenitors present in the final image for each experimental condition.
The time-lapse imaging experiments were conducted in two blocks at the Marine Biological Laboratory and The Scripps Research Institute. Imaging data using the control morpholinos were comparable between the 2 blocks of experiments, with respect to the proliferation rate over 48 h (23.5%±5.5 and 20.8±4.1%; p=0.12, Mann–Whitney U test) and the proportions of neurons, radial glia and unidentifiable cell types (Pearson's Chi-square test, p=0.06). Nevertheless, the experimental datasets were compared to the corresponding control data collected during the same period. The experimental data and the corresponding control data for each block of experiments are provided in Tables 3–5. The control data shown in Figs. 7A4–5 and 8 are combined from both sets.
Morpholinos injected into early stage Zebrafish embryos can result in off target effects and apoptosis by activating p53, although the mechanisms underlying p53 activation remain unclear (Robu et al., 2007). Our in vivo imaging in this and previous studies indicate that targeted electroporation of control or experimental morpholinos into the optic tectum produce a variety of phenotypes with respect to neurogenesis and synaptic plasticity (Bestman and Cline, 2008; Chiu et al., 2008; Ewald et al., 2008; Sharma and Cline, 2010; Shen et al., 2009; Shen et al., 2014) and do not appear to damage cells. This could be due to more targeted delivery of morpholinos, differences in morpholino concentrations in cells or incomplete knockdown in our studies, or greater sensitivity of early developmental events to off target p53 activation in Zebrafish. In addition, we have previously demonstrated that MO treatments can be replicated by dominant negative constructs (Chiu et al., 2008) and that titration of morpholino-insensitive expression constructs can rescue phenotypes seen with morpholino treatment (Faulkner et al., 2015). A recent report suggests that Zebrafish morphant embryos and genetic knockdown by site-specific nucleases do not produce comparable outcomes, based on assays of vascular development (Kok et al., 2015; Stainier et al., 2015). The phenotypic differences between site-specific nucleases and morpholino-mediated knockdown could be due to a variety of as yet unconfirmed but biologically interesting effects, such as hypomorphic effects due to exon skipping, compensation by family members, or different degrees of knockdown in the morphant and mutant fish (Stainier et al., 2015). Nevertheless, our study was designed as a screen for candidate neurogenic regulatory genes and candidates must be further evaluated with a battery of mechanistic studies. The studies mentioned above in Zebrafish emphasize the importance of follow up analysis, including a full range of gene and protein expression manipulations, as we describe in our analysis of fmr1a in neurogenesis (Faulkner et al., 2015).
Statistical tests of time-lapse data
Pearson's Chi-square test was used to determine whether there were significant differences in the proportions of cell types generated between control MO conditions and experimental MO conditions. When significant differences were detected, we used a Mann–Whitney Unpaired test to determine which cell types (neurons, neural progenitors or unclassified cells) were different significantly between the groups. Mann–Whitney Unpaired tests were also used to identify significant differences in the proliferation rates between experimental groups and the control Morpholino conditions. p≤0.05 were considered significant. Wilcoxon Signed Rank tests were used to detect pairwise changes in the cells types identified at day one and day 3. Graphs show means ± standard error of the mean. All values for data summarized in the figures are presented in Tables 3–6.
Supplementary Material
Acknowledgments
We thank members of the Cline Lab for helpful discussions and Melissa Lau for assistance editing the manuscript. We also thank Dr. Monica Boyle at Dart NeuroScience, LLC for help with the NanoString experiments. This work was supported by grants from the US National Institutes of Health (EY011261 and NS076006), Dart NeuroScience, LLC, and the US Department of Defense (W81XWH-12-1-0207) to HTC, an endowment from the Hahn Family Foundation to HTC, a fellowship from the California Institute for Regenerative Medicine (CIRM) Interdisciplinary Stem Cell Training Program at TSRI to L-CH and a fellowship from FRAXA Research Foundation to JEB.
Footnotes
Appendix A. Supplementary Information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2015.03.010.
References
- Ageta-Ishihara N, Miyata T, Ohshima C, Watanabe M, Sato Y, Hamamura Y, Higashiyama T, Mazitschek R, Bito H, Kinoshita M. Septins promote dendrite and axon development by negatively regulating microtubule stability via HDAC6-mediated deacetylation. Nat. Commun. 2013;4:2532. doi: 10.1038/ncomms3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Buylla A, Kohwi M, Nguyen TM, Merkle FT. The heterogeneity of adult neural stem cells and the emerging complexity of their niche. Cold Spring Harb. Symp. Quant. Biol. 2008;73:357–365. doi: 10.1101/sqb.2008.73.019. [DOI] [PubMed] [Google Scholar]
- Anttonen AK, Mahjneh I, Hamalainen RH, Lagier-Tourenne C, Kopra O, Waris L, Anttonen M, Joensuu T, Kalimo H, Paetau A, et al. The gene disrupted in Marinesco-Sjogren syndrome encodes SIL1, an HSPA5 cochaperone. Nat. Genet. 2005;37:1309–1311. doi: 10.1038/ng1677. [DOI] [PubMed] [Google Scholar]
- Barkho BZ, Munoz AE, Li X, Li L, Cunningham LA, Zhao X. Endogenous matrix metalloproteinase (MMP)-3 and MMP-9 promote the differentiation and migration of adult neural progenitor cells in response to chemokines. Stem Cells. 2008;26:3139–3149. doi: 10.1634/stemcells.2008-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks.. Proceedings of the International AAAI Conference on Weblogs and Social Media..2009. [Google Scholar]
- Bestman JE, Cline HT. The RNA binding protein CPEB regulates dendrite morphogenesis and neuronal circuit assembly in vivo. Proc. Natl. Acad. Sci. USA. 2008;105:20494–20499. doi: 10.1073/pnas.0806296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestman JE, Cline HT. Morpholino studies in Xenopus brain development. Methods Mol. Biol. 2014;1082:155–171. doi: 10.1007/978-1-62703-655-9_11. [DOI] [PubMed] [Google Scholar]
- Bestman JE, Lee-Osbourne J, Cline HT. In vivo time-lapse imaging of cell proliferation and differentiation in the optic tectum of Xenopus laevis tadpoles. J. Comp. Neurol. 2012;520:401–433. doi: 10.1002/cne.22795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya B, Puri S, Puri RK. A review of gene expression profiling of human embryonic stem cell lines and their differentiated progeny. Curr. Stem Cell Res. Ther. 2009;4:98–106. doi: 10.2174/157488809788167409. [DOI] [PubMed] [Google Scholar]
- Bonaguidi MA, Song J, Ming GL, Song H. A unifying hypothesis on mammalian neural stem cell properties in the adult hippocampus. Curr. Opin. Neurobiol. 2012;22(5):754–761. doi: 10.1016/j.conb.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaguidi MA, Wheeler MA, Shapiro JS, Stadel RP, Sun GJ, Ming GL, Song H. In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell. 2011;145:1142–1155. doi: 10.1016/j.cell.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackley KI, Grantham J. Activities of the chaperonin containing TCP-1 (CCT): implications for cell cycle progression and cytoskeletal organisation. Cell Stress Chaperones. 2009;14:23–31. doi: 10.1007/s12192-008-0057-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney TD, Miller MR, Robinson KJ, Bayraktar OA, Osterhout JA, Doe CQ. Functional genomics identifies neural stem cell sub-type expression profiles and genes regulating neuroblast homeostasis. Dev. Biol. 2012;361:137–146. doi: 10.1016/j.ydbio.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapouton P, Jagasia R, Bally-Cuif L. Adult neurogenesis in non-mammalian vertebrates. BioEssays. 2007;29:745–757. doi: 10.1002/bies.20615. [DOI] [PubMed] [Google Scholar]
- Charvet CJ, Striedter GF. Causes and consequences of expanded subventricular zones. Eur. J. Neurosci. 2011;34:988–993. doi: 10.1111/j.1460-9568.2011.07818.x. [DOI] [PubMed] [Google Scholar]
- Cheung AFP, Pollen AA, Tavare A, DeProto J, Molnár Z. Comparative aspects of cortical neurogenesis in vertebrates. J. Anat. 2007;211:164–176. doi: 10.1111/j.1469-7580.2007.00769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu SL, Chen CM, Cline HT. Insulin receptor signaling regulates synapse number, dendritic plasticity, and circuit function in vivo. Neuron. 2008;58:708–719. doi: 10.1016/j.neuron.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover JC, Notti RQ. The neural stem cell niche. Cell Tissue Res. 2008;331:211–224. doi: 10.1007/s00441-007-0503-6. [DOI] [PubMed] [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daveau M, Benard M, Scotte M, Schouft MT, Hiron M, Francois A, Salier JP, Fontaine M. Expression of a functional C5a receptor in regenerating hepatocytes and its involvement in a proliferative signaling pathway in rat. J. Immunol. 2004;173:3418–3424. doi: 10.4049/jimmunol.173.5.3418. [DOI] [PubMed] [Google Scholar]
- Day BW, Stringer BW, Spanevello MD, Charmsaz S, Jamieson PR, Ensbey KS, Carter JC, Cox JM, Ellis VJ, Brown CL, et al. ELK4 neutralization sensitizes glioblastoma to apoptosis through downregulation of the anti-apoptotic protein Mcl-1. Neuro-oncology. 2011;13:1202–1212. doi: 10.1093/neuonc/nor119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca A, Favaloro B, Carletti E, Sacchetta P, Di Ilio C. A novel amphibian Pi-class glutathione transferase isoenzyme from Xenopus laevis: importance of phenylalanine 111 in the H-site. Biochem. J. 2003;373:539–545. doi: 10.1042/BJ20030261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek J, Benedix J, Cappel S, Greiner M, Jalal C, Muller L, Zimmermann R. Functions and pathologies of BiP and its interaction partners. Cell. Mol. Life Sci. 2009;66:1556–1569. doi: 10.1007/s00018-009-8745-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowska M, Milek J, Janusz A, Rejmak E, Romanowska E, Gorkiewicz T, Tiron A, Bramham CR, Kaczmarek L. Activity-dependent local translation of matrix metalloproteinase-9. J. Neurosci. 2012;32:14538–14547. doi: 10.1523/JNEUROSCI.6028-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JS, Smith JC. Controlling morpholino experiments: don't stop making antisense. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- Encinas JM, Vaahtokari A, Enikolopov G. Fluoxetine targets early progenitor cells in the adult brain. Proc. Natl. Acad. Sci. USA. 2006;103:8233–8238. doi: 10.1073/pnas.0601992103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald RC, Van Keuren-Jensen KR, Aizenman CD, Cline HT. Roles of NR2A and NR2B in the development of dendritic arbor morphology in vivo. J. Neurosci. 2008;28:850–861. doi: 10.1523/JNEUROSCI.5078-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone C, Filippis C, Granzotto M, Mallamaci A. Emx2 expression levels in NSCs modulate astrogenesis rates by regulating EgfR and Fgf9. Glia. 2015;63:412–422. doi: 10.1002/glia.22761. [DOI] [PubMed] [Google Scholar]
- Falk A, Karlsson TE, Kurdija S, Frisen J, Zupicich J. High-throughput identification of genes promoting neuron formation and lineage choice in mouse embryonic stem cells. Stem Cells. 2007;25:1539–1545. doi: 10.1634/stemcells.2006-0485. [DOI] [PubMed] [Google Scholar]
- Fan GC. Role of heat shock proteins in stem cell behavior. Prog. Mol. Biol. Transl. Sci. 2012;111:305–322. doi: 10.1016/B978-0-12-398459-3.00014-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner RL, Wishard TJ, Thompson CK, Liu H-H, Cline HT. FMRP regulates neurogenesis in vivo in Xenopus laevis tadpoles. eNeuro. 2015;1:51–60. doi: 10.1523/ENEURO.0055-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BL, Hersman MN, Darlington RB. Patterns of vertebrate neurogenesis and the paths of vertebrate evolution. Brain Behav. Evol. 1998;52:232–242. doi: 10.1159/000006566. [DOI] [PubMed] [Google Scholar]
- Gaiano N, Nye JS, Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26:395–404. doi: 10.1016/s0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- Gao YS, Hubbert CC, Lu J, Lee YS, Lee JY, Yao TP. Histone deacetylase 6 regulates growth factor-induced actin remodeling and endocytosis. Mol. Cell. Biol. 2007;27:8637–8647. doi: 10.1128/MCB.00393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Rakic P. Cortical evolution: judge the brain by its cover. Neuron. 2013;80:633–647. doi: 10.1016/j.neuron.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giachino C, Taylor V. Lineage analysis of quiescent regenerative stem cells in the adult brain by genetic labelling reveals spatially restricted neurogenic niches in the olfactory bulb. Eur. J. Neurosci. 2009;30:9–24. doi: 10.1111/j.1460-9568.2009.06798.x. [DOI] [PubMed] [Google Scholar]
- Götz M, Huttner WB. The cell biology of neurogenesis. Nat. Revi. Mol. Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Gu Y, Filippi MD, Cancelas JA, Siefring JE, Williams EP, Jasti AC, Harris CE, Lee AW, Prabhakar R, Atkinson SJ, et al. Hematopoietic cell regulation by Rac1 and Rac2 guanosine triphosphatases. Science. 2003;302:445–449. doi: 10.1126/science.1088485. [DOI] [PubMed] [Google Scholar]
- Hardwick LJ, Philpott A. Nervous decision-making: to divide or differentiate. Trends Genet. 2014;30(6):254–261. doi: 10.1016/j.tig.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes GL. The long-term effects of neonatal seizures. Clin. Perinat. 2009;36:901–914. vii–viii. doi: 10.1016/j.clp.2009.07.012. [DOI] [PubMed] [Google Scholar]
- Hosking CR, Ulloa F, Hogan C, Ferber EC, Figueroa A, Gevaert K, Birchmeier W, Briscoe J, Fujita Y. The transcriptional repressor Glis2 is a novel binding partner for p120 catenin. Mol. Biol. Cell. 2007;18:1918–1927. doi: 10.1091/mbc.E06-10-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R, Lempicki RA. Extracting biological meaning from large gene lists with DAVID. Curr. Protoc. Bioinform. 2009;11 doi: 10.1002/0471250953.bi1311s27. Chapter 13:Unit 13.11. [DOI] [PubMed] [Google Scholar]
- Inaguma Y, Hamada N, Tabata H, Iwamoto I, Mizuno M, Nishimura YV, Ito H, Morishita R, Suzuki M, Ohno K, et al. SIL1, a causative cochaperone gene of Marinesco-Sojgren syndrome, plays an essential role in establishing the architecture of the developing cerebral cortex. EMBO Mol. Med. 2014;6:414–429. doi: 10.1002/emmm.201303069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janesick A, Abbey R, Chung C, Liu S, Taketani M, Blumberg B. ERF and ETV3L are retinoic acid-inducible repressors required for primary neurogenesis. Development. 2013;140:3095–3106. doi: 10.1242/dev.093716. [DOI] [PubMed] [Google Scholar]
- Kaikkonen S, Makkonen H, Rytinki M, Palvimo JJ. SUMOylation can regulate the activity of ETS-like transcription factor 4. Biochim. Biophys. Acta. 2010;1799:555–560. doi: 10.1016/j.bbagrm.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Kaplan A, Spiller KJ, Towne C, Kanning KC, Choe GT, Geber A, Akay T, Aebischer P, Henderson CE. Neuronal matrix metalloproteinase-9 is a determinant of selective neurodegeneration. Neuron. 2014;81:333–348. doi: 10.1016/j.neuron.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsten SL, Kudo LC, Jackson R, Sabatti C, Kornblum HI, Geschwind DH. Global analysis of gene expression in neural progenitors reveals specific cell-cycle, signaling, and metabolic networks. Dev. Biol. 2003;261:165–182. doi: 10.1016/s0012-1606(03)00274-4. [DOI] [PubMed] [Google Scholar]
- Kim AH, Puram SV, Bilimoria PM, Ikeuchi Y, Keough S, Wong M, Rowitch D, Bonni A. A centrosomal Cdc20-APC pathway controls dendrite morphogenesis in postmitotic neurons. Cell. 2009;136:322–336. doi: 10.1016/j.cell.2008.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N, Yang J, Ueda A, Suzuki T, Tomaru K, Takeno M, Okuda K, Ishigatsubo Y. RanBPM, Muskelin, p48EMLP, p44CTLH, and the armadillo-repeat proteins ARMC8alpha and ARMC8beta are components of the CTLH complex. Gene. 2007;396:236–247. doi: 10.1016/j.gene.2007.02.032. [DOI] [PubMed] [Google Scholar]
- Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, van Impel A, Kirchmaier BC, Peterson-Maduro J, Kourkoulis G, Male I, et al. Reverse genetic screening reveals poor correlation between Morpholino-induced and mutant phenotypes in Zebrafish. Dev. Cell. 2015;32:97–108. doi: 10.1016/j.devcel.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Noctor S, Martinez-Cerdeno V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat. Rev. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- Lamar E, Kintner C, Goulding M. Identification of NKL, a novel Gli-Kruppel zinc-finger protein that promotes neuronal differentiation. Development. 2001;128:1335–1346. doi: 10.1242/dev.128.8.1335. [DOI] [PubMed] [Google Scholar]
- Le Belle JE, Orozco NM, Paucar AA, Saxe JP, Mottahedeh J, Pyle AD, Wu H, Kornblum HI. Proliferative neural stem cells have high endogenous ROS levels that regulate self-renewal and neurogenesis in a PI3K/Akt-dependant manner. Cell Stem Cell. 2011;8:59–71. doi: 10.1016/j.stem.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Lee EJ, Song YS, Kim E. Long-term depression-inducing stimuli promote cleavage of the synaptic adhesion molecule NGL-3 through NMDA receptors, matrix metalloproteinases and presenilin/gamma-secretase. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2014;369:20130158. doi: 10.1098/rstb.2013.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichti-Kaiser K, ZeRuth G, Kang HS, Vasanth S, Jetten AM. Gli-similar proteins: their mechanisms of action, physiological functions, and roles in disease. Vitam. Horm. 2012;88:141–171. doi: 10.1016/B978-0-12-394622-5.00007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilja T, Heldring N, Hermanson O. Like a rolling histone: epigenetic regulation of neural stem cells and brain development by factors controlling histone acetylation and methylation. Biochim. Biophys. Acta. 2013;1830:2354–2360. doi: 10.1016/j.bbagen.2012.08.011. [DOI] [PubMed] [Google Scholar]
- LoTurco JJ, Owens DF, Heath MJ, Davis MB, Kriegstein AR. GABA and glutamate depolarize cortical progenitor cells and inhibit DNA synthesis. Neuron. 1995;15:1287–1298. doi: 10.1016/0896-6273(95)90008-x. [DOI] [PubMed] [Google Scholar]
- Lugert S, Basak O, Knuckles P, Haussler U, Fabel K, Gotz M, Haas CA, Kempermann G, Taylor V, Giachino C. Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell. 2010;6:445–456. doi: 10.1016/j.stem.2010.03.017. [DOI] [PubMed] [Google Scholar]
- Maisel M, Herr A, Milosevic J, Hermann A, Habisch HJ, Schwarz S, Kirsch M, Antoniadis G, Brenner R, Hallmeyer-Elgner S, et al. Transcription profiling of adult and fetal human neuroprogenitors identifies divergent paths to maintain the neuroprogenitor cell state. Stem Cells. 2007;25:1231–1240. doi: 10.1634/stemcells.2006-0617. [DOI] [PubMed] [Google Scholar]
- Marei HE, Althani A, Afifi N, Michetti F, Pescatori M, Pallini R, Casalbore P, Cenciarelli C, Schwartz P, Ahmed AE. Gene expression profiling of embryonic human neural stem cells and dopaminergic neurons from adult human substantia nigra. PloS One. 2011;6:e28420. doi: 10.1371/journal.pone.0028420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meighan PC, Meighan SE, Davis CJ, Wright JW, Harding JW. Effects of matrix metalloproteinase inhibition on short- and long-term plasticity of schaffer collateral/CA1 synapses. J. Neurochem. 2007;102:2085–2096. doi: 10.1111/j.1471-4159.2007.04682.x. [DOI] [PubMed] [Google Scholar]
- Michaluk P, Kolodziej L, Mioduszewska B, Wilczynski GM, Dzwonek J, Jaworski J, Gorecki DC, Ottersen OP, Kaczmarek L. Beta-dystroglycan as a target for MMP-9, in response to enhanced neuronal activity. J. Biol. Chem. 2007;282:16036–16041. doi: 10.1074/jbc.M700641200. [DOI] [PubMed] [Google Scholar]
- Mochizuki M, Kwon YW, Yodoi J, Masutani H. Thioredoxin regulates cell cycle via the ERK1/2-cyclin D1 pathway. Antioxid. Redox Signal. 2009;11:2957–2971. doi: 10.1089/ars.2009.2623. [DOI] [PubMed] [Google Scholar]
- Molnar Z. Evolution of cerebral cortical development. Brain Behav. Evol. 2011;78:94–107. doi: 10.1159/000327325. [DOI] [PubMed] [Google Scholar]
- Morest DK, Silver J. Precursors of neurons, neuroglia, and ependymal cells in the CNS: What are they? Where are they from? How do they get where they are going?. Glia. 2003;43:6–18. doi: 10.1002/glia.10238. [DOI] [PubMed] [Google Scholar]
- Nacher J, McEwen BS. The role of N-methyl-d-asparate receptors in neurogenesis. Hippocampus. 2006;16:267–270. doi: 10.1002/hipo.20160. [DOI] [PubMed] [Google Scholar]
- Nagy V, Bozdagi O, Matynia A, Balcerzyk M, Okulski P, Dzwonek J, Costa RM, Silva AJ, Kaczmarek L, Huntley GW. Matrix metalloproteinase-9 is required for hippocampal late-phase long-term potentiation and memory. J. Neurosci. 2006;26:1923–1934. doi: 10.1523/JNEUROSCI.4359-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis (Daudin); A Systemical and Chronological Survey of the Development from Fertilized Egg Till the End of Metamorphosis. North-Holland Publishing Co.; Amsterdam: 1956. [Google Scholar]
- Okano H, Kawahara H, Toriya M, Nakao K, Shibata S, Imai T. Function of RNA-binding protein Musashi-1 in stem cells. Exp. Cell Res. 2005;306:349–356. doi: 10.1016/j.yexcr.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Okulski P, Jay TM, Jaworski J, Duniec K, Dzwonek J, Konopacki FA, Wilczynski GM, Sanchez-Capelo A, Mallet J, Kaczmarek L. TIMP-1 abolishes MMP-9-dependent long-lasting long-term potentiation in the prefrontal cortex. Biol. Psychiatry. 2007;62:359–362. doi: 10.1016/j.biopsych.2006.09.012. [DOI] [PubMed] [Google Scholar]
- Osterele A. P-1000 & P-97 Pipette Cookbook. Sutter Instruments; 2012. 2011. [Google Scholar]
- Park JH, Choi MR, Park KS, Kim SH, Jung KH, Chai YG. The characterization of gene expression during mouse neural stem cell differentiation in vitro. Neurosci. Lett. 2012;506:50–54. doi: 10.1016/j.neulet.2011.10.046. [DOI] [PubMed] [Google Scholar]
- Parker MA, Anderson JK, Corliss DA, Abraria VE, Sidman RL, Park KI, Teng YD, Cotanche DA, Snyder EY. Expression profile of an operationally-defined neural stem cell clone. Exp. Neurol. 2005;194:320–332. doi: 10.1016/j.expneurol.2005.04.018. [DOI] [PubMed] [Google Scholar]
- Paschen W. Endoplasmic reticulum: a primary target in various acute disorders and degenerative diseases of the brain. Cell Calcium. 2003;34:365–383. doi: 10.1016/s0143-4160(03)00139-8. [DOI] [PubMed] [Google Scholar]
- Peixoto RT, Kunz PA, Kwon H, Mabb AM, Sabatini BL, Philpot BD, Ehlers MD. Transsynaptic signaling by activity-dependent cleavage of neuroligin-1. Neuron. 2012;76:396–409. doi: 10.1016/j.neuron.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny LH, Nicolis SK. Sox2 roles in neural stem cells. Int. J. Biochem. Cell Biol. 2010;42:421–424. doi: 10.1016/j.biocel.2009.08.018. [DOI] [PubMed] [Google Scholar]
- Pierfelice T, Alberi L, Gaiano N. Notch in the vertebrate nervous system: an old dog with new tricks. Neuron. 2011;69:840–855. doi: 10.1016/j.neuron.2011.02.031. [DOI] [PubMed] [Google Scholar]
- Prinsloo E, Setati MM, Longshaw VM, Blatch GL. Chaperoning stem cells: a role for heat shock proteins in the modulation of stem cell self-renewal and differentiation? BioEssays. 2009;31:370–377. doi: 10.1002/bies.200800158. [DOI] [PubMed] [Google Scholar]
- Remy P, Baltzinger M. The Ets-transcription factor family in embryonic development: lessons from the amphibian and bird. Oncogene. 2000;19:6417–6431. doi: 10.1038/sj.onc.1204044. [DOI] [PubMed] [Google Scholar]
- Reynolds A, Brustein E, Liao M, Mercado A, Babilonia E, Mount DB, Drapeau P. Neurogenic role of the depolarizing chloride gradient revealed by global overexpression of KCC2 from the onset of development. J. Neurosci. 2008;28:1588–1597. doi: 10.1523/JNEUROSCI.3791-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivieccio MA, Brochier C, Willis DE, Walker BA, D'Annibale MA, McLaughlin K, Siddiq A, Kozikowski AP, Jaffrey SR, Twiss JL, et al. HDAC6 is a target for protection and regeneration following injury in the nervous system. Proc. Natl. Acad. Sci. USA. 2009;106:19599–19604. doi: 10.1073/pnas.0907935106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelshorn DW, Schneider A, Kuschinsky W, Weber D, Kruger C, Dittgen T, Burgers HF, Sabouri F, Gassler N, Bach A, et al. Expression of hemoglobin in rodent neurons. J. Cereb. Blood Flow Metab.: Off. J. Int. Soc. Cereb. Blood Flow Metab. 2009;29:585–595. doi: 10.1038/jcbfm.2008.152. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to Image J: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonthaler HB, Guinea-Viniegra J, Wagner EF. Targeting inflammation by modulating the Jun/AP-1 pathway. Ann. Rheum. Dis. 2011;70(S1):i109–112. doi: 10.1136/ard.2010.140533. [DOI] [PubMed] [Google Scholar]
- Senderek J, Krieger M, Stendel C, Bergmann C, Moser M, Breitbach-Faller N, Rudnik-Schoneborn S, Blaschek A, Wolf NI, Harting I, et al. Mutations in SIL1 cause Marinesco-Sjogren syndrome, a cerebellar ataxia with cataract and myopathy. Nat. Genet. 2005;37:1312–1314. doi: 10.1038/ng1678. [DOI] [PubMed] [Google Scholar]
- Sharma P, Cline HT. Visual activity regulates neural progenitor cells in developing Xenopus CNS through musashi1. Neuron. 2010;68:442–455. doi: 10.1016/j.neuron.2010.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- Shen W, Da Silva JS, He H, Cline HT. Type A GABA-receptor-dependent synaptic transmission sculpts dendritic arbor structure in Xenopus tadpoles in vivo. J. Neurosci. 2009;29:5032–5043. doi: 10.1523/JNEUROSCI.5331-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Liu HH, Schiapparelli L, McClatchy D, He HY, Yates JR, 3rd, Cline HT. Acute synthesis of CPEB is required for plasticity of visual avoidance behavior in Xenopus. Cell Rep. 2014;6:737–747. doi: 10.1016/j.celrep.2014.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Pires C, Zwick V, Nurisso A, Schenker E, Carrupt PA, Cuendet M. HDAC6 as a target for neurodegenerative diseases: what makes it different from the other HDACs? Mol. Neurodegener. 2013;8:7. doi: 10.1186/1750-1326-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin WC, Haas K, Ruthazer ES, Cline HT. Dendrite growth increased by visual activity requires NMDA receptor and Rho GTPases. Nature. 2002;419:475–480. doi: 10.1038/nature00987. [DOI] [PubMed] [Google Scholar]
- Song J, Zhong C, Bonaguidi MA, Sun GJ, Hsu D, Gu Y, Meletis K, Huang ZJ, Ge S, Enikolopov G, et al. Neuronal circuitry mechanism regulating adult quiescent neural stem-cell fate decision. Nature. 2012;489:150–154. doi: 10.1038/nature11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainier DY, Kontarakis Z, Rossi A. Making sense of anti-sense data. Dev. Cell. 2015;32:7–8. doi: 10.1016/j.devcel.2014.12.012. [DOI] [PubMed] [Google Scholar]
- Stocker AM, Chenn A. Focal reduction of alphaE-catenin causes premature differentiation and reduction of beta-catenin signaling during cortical development. Dev. Biol. 2009;328:66–77. doi: 10.1016/j.ydbio.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Ueda A, Kobayashi N, Yang J, Tomaru K, Yamamoto M, Takeno M, Ishigatsubo Y. Proteasome-dependent degradation of alpha-catenin is regulated by interaction with ARMc8alpha. Biochem. J. 2008;411:581–591. doi: 10.1042/BJ20071312. [DOI] [PubMed] [Google Scholar]
- Szklarczyk A, Lapinska J, Rylski M, McKay RD, Kaczmarek L. Matrix metalloproteinase-9 undergoes expression and activation during dendritic remodeling in adult hippocampus. J. Neurosci. 2002;22:920–930. doi: 10.1523/JNEUROSCI.22-03-00920.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantin D. Oct transcription factors in development and stem cells: insights and mechanisms. Development. 2013;140:2857–2866. doi: 10.1242/dev.095927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari R, Bailes E, Bunting KA, Coates JC. Armadillo-repeat protein functions: questions for little creatures. Trends Cell Biol. 2010;20:470–481. doi: 10.1016/j.tcb.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Tremblay M, Fugere V, Tsui J, Schohl A, Tavakoli A, Travencolo BAN, Costa L. d.F., Ruthazer ES. Regulation of Radial Glial Motility by Visual Experience. J. Neurosci. 2009;29:14066–14076. doi: 10.1523/JNEUROSCI.3542-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend DM, Manevich Y, He L, Hutchens S, Pazoles CJ, Tew KD. Novel role for glutathione S-transferase pi. Regulator of protein S-Glutathionylation following oxidative and nitrosative stress. J. Biol. Chem. 2009;284:436–445. doi: 10.1074/jbc.M805586200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uy BR, Simoes-Costa M, Koo DE, Sauka-Spengler T, Bronner ME. Evolutionarily conserved role for SoxC genes in neural crest specification and neuronal differentiation. Dev. Biol. 2015;397:282–292. doi: 10.1016/j.ydbio.2014.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela-Fernandez A, Cabrero JR, Serrador JM, Sanchez-Madrid F. HDAC6: a key regulator of cytoskeleton, cell migration and cell-cell interactions. Trends Cell Biol. 2008;18:291–297. doi: 10.1016/j.tcb.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Vasanth S, ZeRuth G, Kang HS, Jetten AM. Identification of nuclear localization, DNA binding, and transactivating mechanisms of Kruppel-like zinc finger protein Gli-similar 2 (Glis2). J. Biol. Chem. 2011;286:4749–4759. doi: 10.1074/jbc.M110.165951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergano-Vera E, Mendez-Gomez HR, Hurtado-Chong A, Cigudosa JC, Vicario-Abejon C. Fibroblast growth factor-2 increases the expression of neurogenic genes and promotes the migration and differentiation of neurons derived from transplanted neural stem/progenitor cells. Neuroscience. 2009;162:39–54. doi: 10.1016/j.neuroscience.2009.03.033. [DOI] [PubMed] [Google Scholar]
- Walton NM, Shin R, Tajinda K, Heusner CL, Kogan JH, Miyake S, Chen Q, Tamura K, Matsumoto M. Adult neurogenesis transiently generates oxidative stress. PloS One. 2012;7:e35264. doi: 10.1371/journal.pone.0035264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanet A, Arnould T, Renard P. Mitochondrial involvement in stemness and stem cell differentiation. In: Lou PH, Peterson N, editors. Cellular Bioenergetics in Health and Disease: New Perspective in Mitochondrial Biology. Research Signpost; Kerala, India: 2012. pp. 195–215. [Google Scholar]
- Wang XB, Bozdagi O, Nikitczuk JS, Zhai ZW, Zhou Q, Huntley GW. Extracellular proteolysis by matrix metalloproteinase-9 drives dendritic spine enlargement and long-term potentiation coordinately. Proc. Natl. Acad. Sci. USA. 2008;105:19520–19525. doi: 10.1073/pnas.0807248105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willardsen M, Hutcheson DA, Moore KB, Vetter ML. The ETS transcription factor Etv1 mediates FGF signaling to initiate proneural gene expression during Xenopus laevis retinal development. Mech. Dev. 2014;131:57–67. doi: 10.1016/j.mod.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarczyk J, Mukhina I, Kaczmarek L, Dityatev A. Extracellular matrix molecules, their receptors, and secreted proteases in synaptic plasticity. Dev. Neurobiol. 2011;71:1040–1053. doi: 10.1002/dneu.20958. [DOI] [PubMed] [Google Scholar]
- Wu GY, Cline HT. Time-lapse in vivo imaging of the morphological development of Xenopus optic tectal interneurons. J. Comp. Neurol. 2003;459:392–406. doi: 10.1002/cne.10618. [DOI] [PubMed] [Google Scholar]
- Wu GY, Zou DJ, Rajan I, Cline H. Dendritic dynamics in vivo change during neuronal maturation. J. Neurosci. 1999;19:4472–4483. doi: 10.1523/JNEUROSCI.19-11-04472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yuan Z, Zhang Y, Yong S, Salas-Burgos A, Koomen J, Olashaw N, Parsons JT, Yang XJ, Dent SR, et al. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol. Cell. 2007;27:197–213. doi: 10.1016/j.molcel.2007.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi HY. SILencing misbehaving proteins. Nat. Genet. 2005;37:1302–1303. doi: 10.1038/ng1205-1302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.