Abstract
Background:
Viral load measurements are commonly used to monitor HCV infection in patients with chronic diseases or determining the number of HCV-genomes in serum samples of patients after sustained virological response. However, in some patients, HCV viral load in serum samples is too low to be detected by PCR, especially after treatment.
Objectives:
The aim of this study was to develop a highly specific, sensitive, and reproducible in-house quantitative PCR using specific primers and probe cited in highly conservative region of HCV genome that allows simultaneous detection of HCV genotypes 1 - 4.
Materials and Methods:
In this study, three sets of primer pairs and a TaqMan probe for amplification and detection of selected region within 5’-non-coding (5’NCR) of four HCV genotypes were used. Using plasmid containing 5’NCR region of HCV, standard curve, threshold, and threshold cycle (CT) values were determined. Real-time and nested PCR were performed on HCV genotypes 1 - 4 extracted from plasma and peripheral blood mononuclear cells (PBMCs) samples collected from patients with chronic HCV infection.
Results:
The lower limit detection of this in-house HCV real-time RT-PCR was determined as 100 RNA copies/mL. Inter- and intra-assay coefficient of variation (CV) of this in-house HCV real-time RT-PCR ranged from 0.9% to 1.8% and 1.76% to 3.94%, respectively. The viral load of the genotyped samples ranged from 2.0 × 106 ± 0.31 to 2.7 × 105 ± 0.46 copies/mL in serum samples and 5 × 102 ± 0.36 to 4.0 × 103 ± 0.51 copies/106 cells/mL of PBMCs.
Conclusions:
The quite sensitive in-house TaqMan real time RT-PCR assay was able to detect and quantify all four main HCV genotypes prevailing around all geographical regions of Iran.
Keywords: Hepatitis C Virus, Mononuclear Leukocytes, Real-Time Polymerase Chain Reaction
1. Background
Hepatitis C virus (HCV), a positive strand RNA virus, is still a major health problem worldwide (1). It frequently leads to chronic infection contributing to liver cirrhosis and hepatocellular carcinoma. Based on HCV genotype, antiviral therapy is successful in approximately 50 to 70% of infected patients (2, 3). However, sustained virological response (SVR) contributes to negative findings for HCV RNA in serum for six months after termination of treatment (2). Recurrence of infection is defined as reappearance of HCV RNA in serum (4).
On the other hand, hepatitis C virus is not a strictly hepatotropic virus and there is evidence showing its ability to replicate in peripheral blood mononuclear cells (PBMCs). Several reports have been published regarding detection of HCV RNA in PBMCs. The infected cells were reported to contain HCV RNA-negative strand, which is a viral replicative intermediate and viral genomic sequences were often found to be distinct from those found in serum and liver (1, 5, 6).
Presence of HCV RNA years after apparent successful treatment has been reported by Pham et al. (7) who were able to amplify viral sequence from follow-up sera or PBMC in 11 of 11 SVR patients for up to five years after therapy. It has been shown that in patients with SVR after IFN or IFN/ribavirin therapy, small quantities of HCV RNA may persist in liver or PBMCs for up to nine years. This continuous presence of HCV RNA could present a potential risk for transmission or infection reactivation (8, 9).
Viral load measurements are commonly used to monitor HCV infection in patients with chronic diseases or determining the number of HCV-genomes in serum samples of patients after SVR. Recently, assays based on real-time reverse transcriptase polymerase chain reaction (RT-PCR) have been introduced in routine diagnosis and rapidly replaced the assays based on standard RT-PCR and signal amplification (7, 10).
In patients with undetectable level, still test might be positive for viral hepatitis. This is because viral load tests have a limit to how few viruses they can detect. Tests vary in terms of sensitivity. For instance, while some viral load tests can detect as little as 50 IU (equal to 100 vg copies/mL), others can only go as low as 615 IU/mL. After HCV infection treatment and SVR, HCV RNA may not be detectable in serum samples of patients (11, 12). During the remission period it is possible that HCV replicates in PBMCs and hepatitis recurs in patients. During the remission period, HCV viral load in serum samples is too low to be detected by PCR, therefore PBMCs would be a valuable source for follow-up of patients after SVR (8, 13).
2. Objectives
In the present study, a highly specific, sensitive, and reproducible in-house quantitative PCR was described using specific primers and probe cited in highly conservative 5’-non-coding region (5’NCR) of HCV genome that allows simultaneous detection of four main HCV genotypes in the region. Moreover, the sensitivity of assay was investigated for detection of HCV RNA in peripheral blood mononuclear cell and plasma samples of patients with current chronic hepatitis who had not received any treatment.
3. Materials and Methods
3.1. Design of Primers and TaqMan Probe
PCR primers that target highly conserved sites of the 5’NCR region were selected for their robustness. In this study, three sets of primer pairs and a TaqMan probe were used for amplification and detection of HCV genotypes in clinical samples. Two sets of published primer pairs (14), an in-house designed primer pair, BK1 and BK2, and a TaqMan probe for amplification and detection of selected region within 5’NCR of HCV genome were also used (Table 1). The primers and TaqMan probes used in this study were designed using the Primer3 and Generunner software and synthesized by Bioneer Corporation (South Korea). The TaqMan probe sequence was selected within primer pair BK1 and BK2 and cited at positions 166 - 187, which was designed to be perfectly complementary to the target sequence. The TaqMan probe was labeled with 6-carboxyfluorescein (6-FAM) and the quencher Carboxytetramethylrhodamine (TAMRA) at the 5’ and 3’ ends, respectively.
Table 1. Primer Sequences Used in the Study.
| Primer Name | Primer Sequence and the Probe Sequences | Primers and the Probe/Sequence Position on the Genome |
|---|---|---|
| BKP7 | 5’-CACTCCCCTGTGAGGAACTACTGTCT-3’ | Outer forward primer/38 - 63 |
| BKP8 | 5’-TGGTGCACGGTCTACGAGACCTCCC -3’ | Outer reverse primer/319 - 343 |
| S130 | 5’-CGGGAGAGCCATAGTGGTCTGCG-3’ | Inner forward primer/130 - 152 |
| AS311 | 5’-CTCGCAAGCACCCTATCAGGCAGTA -3’ | Inner reverse primer/287 - 311 |
| Bk1 | 5’-CGG GAG AGC CAT AGT GGT -3’ | Forward primer for real time PCR/130 - 147 |
| Bk2 | 5’-CAA GCA CCC TAT CAG GCA -3’ | Reverse primer for real time PCR/290 - 307 |
| Probe | 5’-CAA GGC CTT TCG CGA CCC AA -3’ | TaqMan probe/264 - 283 |
3.2. Clinical Specimens
To validate practical application of developed in-house HCV real-time RT-PCR, the presence of HCV RNA was determined in both sero-positive and -negative plasma specimens collected from 38 patients with definitive diagnosis of HCV infection and 14 normal healthy donors, respectively. Patients were already known to have positive results for HCV antibody and RNA. They all were new cases and had received no treatment on the time of sample collection. To assess whether the developed real time RT-PCR would be able to detect the HCV genotypes 1 to 4 which are dominant in the region, HCV genotypes were determined for all the plasma samples using a commercially available kit (Sacace Biotechnologies Srl, Caserta, Italy). There were 17 genotype 1, 2 genotype 2, 17 genotype 3 and 2 genotype 4. All the blood samples were identified as HIV-1 antibody negative by anonymous testing with ELISA method (anti-HIV Tetra ELISA, Biotest Co. Germany). Blood samples were centrifuged at 805 × g for 10 minutes and the plasma was recovered. PBMCs were isolated from the remaining volume by 3:1 dilution with phosphate buffer saline (PBS) and Ficoll-PaqueTM PLUS (1.077 g/mL, Amersham Biosciences, Freiburg, Germany) density gradient centrifugation (1578 × g, 30 minutes, 25°C). Harvested cells (1 × 106 cells/mL) from the interface and plasma samples were stored at -80°C until RNA extraction.
3.3. RNA Extraction
Total RNA from plasma and PBMC extracted with Viral RNA PrepMate™ kit (Bioneer. South Korea). In brief, the total RNA was extracted by mixing 100 μL plasma or PBMC (105 cells/100 μL) with lysis buffer and incubated at 4°C. Chloroform was then added and after centrifugation at 13000 × g for 15 minutes, RNA-containing aqueous phase was precipitated in isopropanol. RNA precipitates were then washed once in 75% ethanol and re-suspended in 20 μL of RNase-free water.
3.4. Reverse Transcription
One tenth of total RNA (2 µL) was subjected to reverse transcription with specific primer (anti-sense) AS343 for one hour at 42°C, followed by 5 minutes inactivation of enzyme at 99°C. The resulting cDNA was used as a template for PCR amplification.
3.5. Preparation of In-House HCV RNA Standard Control
To prepare HCV RNA control, cDNA was amplified by PCR with the primer pair of S38/AS343. The amplicons were then separated on a 3% agarose gel and purified with silica bead dna gel extraction kit (Fermentas, Lithuania). The purified PCR product was then cloned into pJET1.2 vector using the CloneJET™ PCR Cloning Kit (Fermentas, Lithuania). The recombinant plasmid covering HCV 5’NCR cDNA was transformed in to E. coli DH5α; afterwards purified with GeneJET™ Plasmid Miniprep Kit (Fermentas, Lithuania). The restriction digestion was performed using SmlI enzyme to achieve the 910 base pair fragment containing T7 transcription and HCV 5’-NCR and purified by phenol chloroform extraction method and alcohol precipitation. T7 transcription kit (Fermentas, Lithuania) was used to transcribe HCV 5’NCR cDNA into specific RNA and then purified with phenol-chloroform extraction and alcohol precipitation.
3.6. Real Time PCR Assay
To prepare a standard curve, a set of six dilutions of HCV 5’NCR RNA control (102 - 107 copies) of HCV RNA were transcribed into cDNA. These samples have been serially diluted in an HIV, HCV, and HBV negative serum to generate a set of six standards, which were then amplified by real-time PCR. The Bio Rad IQ5DH5α Detection System was used for real-time analysis. Thermal cycling program was designed as follows; initial denaturation at 95°C for 3 minutes followed by 45 cycles of 95°C for 15 seconds and 59°C for 60 seconds. Fluorescent measurements were recorded during each extension step. Data was automatically analyzed by the system and amplification plots were generated at the end of each PCR run. For each PCR reaction, 2 µL of cDNA template was added to 48 μL of PCR master mixture (200 μM dNTP, 500 nM of each primer, 2.5 U of Maxima® Hot Start Taq DNA Polymerase, 400 nM of TaqMan probe, 1.5 mM MgCl2 and 1x PCR buffer) respectively for each PCR reaction.
All amplification reactions were performed in triplicate. A standard curve was generated with 10-fold dilutions of HCV 5’NCR RNA control (102 - 107 copies/mL) that had been pre-quantitated by a Nano drop spectrophotometer.
3.7. Nested RT-PCR
The S38/AS343 outer and AS130/AS311 inner primers were used for qualitative analysis of HCV RNA. The first PCR reaction conditions were as follows: denaturation at 94°C for 5 minutes, followed by 35 cycles at 94°C for 30 seconds, 58°C for 40 seconds and 72°C for 1 minute. The second PCR was performed with 1 μL of the first PCR product in the same conditions. PCR products were then visualized on 1% agarose gel and stained with fluorescent dye ethidium bromide.
3.8. Quantification of HCV RNA in Plasma and PBMCs
Real-time and nested PCR were performed on RNA extracted from plasma and PBMCs samples collected from patients with chronic HCV infection. An outcome of interest in this part of study was to determine if the primers sets were able to detect various HCV genotypes in the samples both quantitatively and qualitatively.
4. Results
4.1. Sensitivity and Reproducibility of Real-Time RT-PCR
The analytical sensitivity of real-time RT-PCR was determined using dilution series of HCV 5’NCR RNA transcripts containing 0, 102, 103, 104, 105, 106, and 107 molecules and tested five times, each in triplicate. The amplification plots obtained using the in-house standards and the standard curve are shown in Figure 1. Sensitivity was determined as 100 RNA copies per reaction mixture. To determine the reproducibility of the assay, RNA transcripts with different dilutions were tested with quadruplicates of each dilution in each run ((intra-assay) in different days (inter-assay)). Tables 2 and 3 indicated the mean coefficient of variation (CV) of Ct values and input copy number of HCV RNA within a run and in different days (inter-assay). The intra-assay coefficient of variation (CV) of this in-house HCV real-time RT-PCR ranged from 0.9 to 1.8% (Table 3). The inter-assay CV of this in-house HCV real-time RT-PCR ranged from 1.76% to 3.94% (Table 2). No significant differences observed between the results on each day and between the results on the respective days. Four sero-positive specimens were run by real-time RT-PCR four times (Table 4). To analyze the variance of repeated runs, a selection of isolates from each of the four genotypes tested on separate days.
Figure 1. A, Amplification Plot of 10-Fold Serial Dilutions (102 - 107 Copies/mL) of the HCV 5’NCR RNA Control; B, The Standard Curve of 10-Fold Serial Dilutions of HCV 5’NCR RNA Control With a Correlation Coefficient (R2) of 0.995.
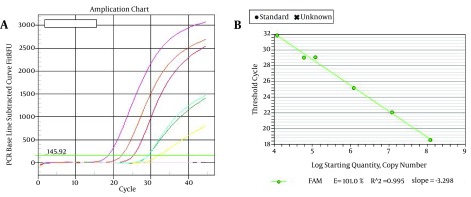
Table 2. Inter-Assay Reproducibility of HCV Real-Time RT-PCR.
| Day | Input Copy Numbers of HCV 5’NCR RNA Control | ||||
|---|---|---|---|---|---|
| Threshold Cycle (Ct) | |||||
| 103 | 104 | 105 | 106 | 107 | |
| 1 | 29.21 | 26.47 | 22.06 | 19.88 | 15.28 |
| 2 | 29.50 | 26.61 | 23.30 | 19.23 | 15.91 |
| 3 | 30.10 | 26.03 | 21.54 | 19.06 | 15.12 |
| 4 | 29.43 | 25.40 | 22.21 | 19.41 | 14.79 |
| 5 | 28.65 | 27.23 | 20.87 | 20.37 | 16.32 |
| Values a | 29.39 ± 0.52 | 26.34 ± 0.68 | 21.99 ± 0.89 | 19.61 ± 0.50 | 15.48 ± 0.61 |
| CV b | 1.76 | 2.58 | 4.04 | 2.54 | 3.94 |
a Data are presented as mean ± SD.
b Abbreviation: coefficient of variation.
Table 3. Intra-Assay Accuracy of HCV Real-Time RT-PCR.
| Tube | Input Copy Numbers of HCV 5’NCR RNA Control | ||||
|---|---|---|---|---|---|
| Threshold Cycle (Ct) | |||||
| 103 | 104 | 105 | 106 | 107 | |
| 1 | 28.70 | 26.15 | 21.22 | 19.76 | 15.30 |
| 2 | 29.12 | 26.28 | 21.05 | 19.98 | 15.06 |
| 3 | 28.54 | 25.83 | 21.46 | 19.49 | 15.53 |
| 4 | 28.61 | 26.38 | 21.90 | 19.51 | 15.74 |
| Values a | 28.74 ± 0.26 | 26.40 ± 0.30 | 21.40 ± 0.36 | 19.68 ± 0.23 | 15.40 ± 0.29 |
| CV b | 0.90 | 1.13 | 1.68 | 1.16 | 1.88 |
a Data are presented as mean ± SD.
b Abbreviation: coefficient of variation.
Table 4. Reproducibility of HCV Real-Time RT-PCR Assay Runs on Four Plasma Samples.
| Mean of 5 Runs | Mean of Threshold Cycles (Ct) | |||
|---|---|---|---|---|
| Sample 1 | Sample 2 | Sample 3 | Sample 4 | |
| Values a | 30.45 ± 0.17 | 28.91 ± 0.32 | 23.54 ± 0.20 | 18.36 ± 0.15 |
| CV b | 0.55 | 1.10 | 0.84 | 0.81 |
a Data are presented as mean ± SD.
b Abbreviation: coefficient of variation.
4.2. Application to Clinical Samples
Different HCV genotypes tested in this study were genotype 1 (n = 17), genotype 2 (n = 2), genotype 3 (n = 17), and genotype 4 (n = 2). Real-time and nested RT-PCR were performed to determine whether the major HCV genotypes in the region could be identified by the designed primers and probe. HCV positive samples of genotypes 1 - 4 were quantified using in-house TaqMan assay. Genotyping was performed as described before. RNA extracted from plasma and PBMCs of patients infected with different HCV genotypes were introduced into real time PCR assay and all four genotypes were identified by the same primer sets and probe. The viral load of genotyped samples ranged from 2.0 × 106 ± 0.31 to 2.7 × 105 ± 0.46 copies/mL in serum samples and 5 × 102 ± 0.36 to 4.0 × 103 ± 0.51 copies/106 cells/mL of PBMCs. Nested PCR was able to detect all four HCV genotypes in the samples as well. Overall, the results indicated that HCV RNA load in the sera was higher than PBMC. Moreover, none of the control group had positive result by either real time or nested PCR assay.
5. Discussion
Currently, TaqMan technology is widely used for detection and quantification of viral genomes. In this study, an in-house real-time PCR assay was developed using TaqMan technology with good accuracy for quantification of HCV RNA in plasma and PBMCs samples from patients with chronic hepatitis C virus infection. The sensitivity of the assay was outstanding with a good dynamic range for detection of HCV RNA. There are varieties of commercial assays available for quantitation of HCV RNA routinely used in diagnostic laboratories. However, most of them have a limited ratio between the largest and smallest values of HCV RNA quantity. Therefore, to obtain the accurate results, clinical samples may need to be diluted and retested. Although for the calibration of the assay, WHO standard of HCV RNA was not used, instead an in-house standard with transcript of recombinant plasmid covering HCV 5’NCR cDNA was used. Overall, the sensitivity of the assay was about 100 copies/tube of HCV RNA, which is equal to 50 IU/tube. Furthermore, all four HCV genotypes could be detected within a single tube using a single set of primers and a target-specific probe. Yang et al. (14) used a molecular beacon probe with four nucleotide mismatches, which make it difficult to determine all the HCV genotypes properly by real time PCR. To overcome the problem, following alignments to reference sequences and BLAST analysis, a new set of primer and a TagMan probe were designed. Using TagMan probe instead of molecular beacon for setting up a real time PCR for detection of all genotypes is feasible and straightforward in the assay.
TaqMan quantitative HCV assay showed over a range of 5 × 102 to 2 × 106 copies of HCV RNA in a clinical sample, which was comparable to other published HCV real-time PCR quantitative assays (15-17). TaqMan HCV quantitative test exhibited good reproducibility for both triplicate assays and independent tests. As expected, the intra-assay CV was lower than the inter-assay CV. The probe sequence was chosen so that it would hybridize to a sequence within its target amplicon conserved among most clinical types. The primers and probe designed for this assay are specific for HCV and do not contain any perfect match sequences for any other virus, bacteria or genomic DNA. It has been reported that HCV RNA could be detected in peripheral blood mononuclear cells of patients with HCV infections (18, 19).
In some patients, HCV RNA may persist in PBMCs even after sustained virologic response (SVR) to antiviral therapy (20). Thus, it is possible that infectious viral HCV RNA could not be cleared from PBMCs, rather than serum. Therefore, it may be a better marker of assessing HCV treatment failure or relapse. The developed TaqMan real time PCR was evaluated to determine the sensitivity of the assay for detection and quantification of HCV RNA in PBMCs of the same patients whose sera were tested for HCV-RNA. Simultaneously, HCV RNA was detected in freshly isolated peripheral blood mononuclear cells of patients as well as serum samples. However, copy number of HCV-RNA in PBMCs was lower than sera, which is still important when assessing virological response to treatment.
In conclusion, developed in-house TaqMan real time RT-PCR assay was able to detect and quantify HCV genotypes 1 - 4 prevailing in all geographical regions of Iran. The assay was quite sensitive for detection of low copies of HCV RNA in peripheral blood mononuclear cell and plasma samples, which is important when assessing virological response to treatment.
Footnotes
Authors’ Contributions:Study concept and design: Abbas Behzad-Behbahani, Seiied Alireza Taghavi and Ali Farhadi. Sampling and executive procedure: Bahman Khalvati Fahlyani, Mohammad Ali Okhovat, Reza Ranjbaran, Gholam Reza Rafiei Dehbidi, Saeede Salehi and Setareh Adibzadeh. Administrative, technical and material support: Farzaneh Aboualizadeh and Parniyan Alavi. Drafting of the manuscript: Abbas Behzad-Behbahani and Negin Nikouyan. Arash Shakibzadeh: technical and material support.
Funding/Support:This study was supported by a grant 4344 from Shiraz university of medical sciences, Shiraz,IR Iran.
References
- 1.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5(6):453–63. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 2.Lindsay KL. Introduction to therapy of hepatitis C. Hepatology. 2002;36(5 Suppl 1):S114–20. doi: 10.1053/jhep.2002.36226. [DOI] [PubMed] [Google Scholar]
- 3.Lau DT, Kleiner DE, Ghany MG, Park Y, Schmid P, Hoofnagle JH. 10-Year follow-up after interferon-alpha therapy for chronic hepatitis C. Hepatology. 1998;28(4):1121–7. doi: 10.1002/hep.510280430. [DOI] [PubMed] [Google Scholar]
- 4.Pearlman BL, Traub N. Sustained virologic response to antiviral therapy for chronic hepatitis C virus infection: a cure and so much more. Clin Infect Dis. 2011;52(7):889–900. doi: 10.1093/cid/cir076. [DOI] [PubMed] [Google Scholar]
- 5.Laskus T, Radkowski M, Wang LF, Jang SJ, Vargas H, Rakela J. Hepatitis C virus quasispecies in patients infected with HIV-1: correlation with extrahepatic viral replication. Virology. 1998;248(1):164–71. doi: 10.1006/viro.1998.9269. [DOI] [PubMed] [Google Scholar]
- 6.Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5(9):558–67. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 7.Pham TN, MacParland SA, Mulrooney PM, Cooksley H, Naoumov NV, Michalak TI. Hepatitis C virus persistence after spontaneous or treatment-induced resolution of hepatitis C. J Virol. 2004;78(11):5867–74. doi: 10.1128/JVI.78.11.5867-5874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radkowski M, Gallegos-Orozco JF, Jablonska J, Colby TV, Walewska-Zielecka B, Kubicka J, et al. Persistence of hepatitis C virus in patients successfully treated for chronic hepatitis C. Hepatology. 2005;41(1):106–14. doi: 10.1002/hep.20518. [DOI] [PubMed] [Google Scholar]
- 9.Radkowski M, Horban A, Gallegos-Orozco JF, Pawelczyk A, Jablonska J, Wilkinson J, et al. Evidence for viral persistence in patients who test positive for anti-hepatitis C virus antibodies and have normal alanine aminotransferase levels. J Infect Dis. 2005;191(10):1730–3. doi: 10.1086/429834. [DOI] [PubMed] [Google Scholar]
- 10.Hadinedoushan H, Salmanroghani H, Amirbaigy MK, Akhondi-Meybodi M. Hepatitis C virus genotypes and association with viral load in yazd, central province of iran. Hepat Mon. 2014;14(3):e28895. doi: 10.5812/hepatmon.11705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mackay IM, Arden KE, Nitsche A. Real-time PCR in virology. Nucleic Acids Res. 2002;30(6):1292–305. doi: 10.1093/nar/30.6.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott JD, Gretch DR. Molecular diagnostics of hepatitis C virus infection: a systematic review. JAMA. 2007;297(7):724–32. doi: 10.1001/jama.297.7.724. [DOI] [PubMed] [Google Scholar]
- 13.Mehta SH, Cox A, Hoover DR, Wang XH, Mao Q, Ray S, et al. Protection against persistence of hepatitis C. Lancet. 2002;359(9316):1478–83. doi: 10.1016/S0140-6736(02)08435-0. [DOI] [PubMed] [Google Scholar]
- 14.Yang JH, Lai JP, Douglas SD, Metzger D, Zhu XH, Ho WZ. Real-time RT-PCR for quantitation of hepatitis C virus RNA. J Virol Methods. 2002;102(1-2):119–28. doi: 10.1016/s0166-0934(02)00007-1. [DOI] [PubMed] [Google Scholar]
- 15.Castelain S, Descamps V, Thibault V, Francois C, Bonte D, Morel V, et al. TaqMan amplification system with an internal positive control for HCV RNA quantitation. J Clin Virol. 2004;31(3):227–34. doi: 10.1016/j.jcv.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Morandi L, Ferrari D, Lombardo C, Pession A, Tallini G. Monitoring HCV RNA viral load by locked nucleic acid molecular beacons real time PCR. J Virol Methods. 2007;140(1-2):148–54. doi: 10.1016/j.jviromet.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 17.Daniel HD, David J, Grant PR, Garson JA, Chandy GM, Abraham P. Whole blood as an alternative to plasma for detection of hepatitis C virus RNA. J Clin Microbiol. 2008;46(11):3791–4. doi: 10.1128/JCM.00045-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blackard JT, Kemmer N, Sherman KE. Extrahepatic replication of HCV: insights into clinical manifestations and biological consequences. Hepatology. 2006;44(1):15–22. doi: 10.1002/hep.21283. [DOI] [PubMed] [Google Scholar]
- 19.Boisvert J, He XS, Cheung R, Keeffe EB, Wright T, Greenberg HB. Quantitative analysis of hepatitis C virus in peripheral blood and liver: replication detected only in liver. J Infect Dis. 2001;184(7):827–35. doi: 10.1086/323391. [DOI] [PubMed] [Google Scholar]
- 20.MacParland SA, Pham TN, Guy CS, Michalak TI. Hepatitis C virus persisting after clinically apparent sustained virological response to antiviral therapy retains infectivity in vitro. Hepatology. 2009;49(5):1431–41. doi: 10.1002/hep.22802. [DOI] [PubMed] [Google Scholar]


