Abstract
Long noncoding RNAs (lncRNAs) are emerging as an integral part of the regulatory information encoded in the genome. LncRNAs possess the unique capability to interact with nucleic acids and proteins and exert discrete effects on numerous biological processes. Recent studies have delineated multiple lncRNA pathways that control metabolic tissue development and function. The expansion of the regulatory code that links nutrient and hormonal signals to tissue metabolism gives new insights into the genetic and pathogenic mechanisms underlying metabolic disease. This review discusses lncRNA biology with a focus on its role in the development, signaling, and function of key metabolic tissues.
Keywords: lncRNA, energy metabolism, signaling, metabolic disease, transcription, brown fat
The homeostatic control of nutrient and energy metabolism in mammals is governed by reciprocal signaling between the tissues that primarily serve regulatory functions, such as the pancreatic islets and the central nervous system, and major metabolic tissues including adipose tissues, skeletal muscle, and the liver. These tissues acquire their highly specialized regulatory functions and metabolic activities during development and exhibit an amazing degree of plasticity in adulthood. For example, the different types of skeletal muscle fibers are characterized by varying oxidative capacity and contractile function, whereas adipocytes from white and brown fat have nearly opposite roles in fuel storage and oxidation. Disruption of energy homeostasis underlies the pathogenesis of major metabolic disorders, including obesity, type 2 diabetes, dyslipidemia, and non-alcoholic fatty liver disease. The protein factors that control metabolic tissue development, signaling, and function have been extensively investigated. Recent work on long noncoding RNAs (lncRNAs) has added a new dimension to the regulatory networks that impinge on metabolic homeostasis and disease [1, 2].
LncRNAs: emerging regulators of diverse biological processes
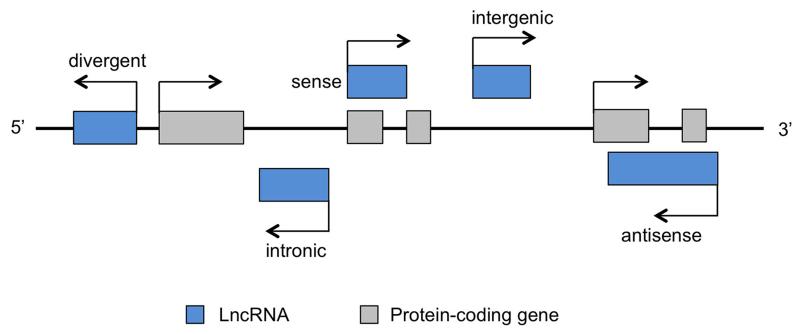
Long noncoding RNAs refer to a class of RNA transcripts that lack identifiable open reading frames and thus protein-coding potential [3, 4]. LncRNAs are commonly defined as transcripts longer than 200 nucleotides, as compared to other shorter noncoding RNA species, such as microRNA (miRNA), small interfering RNA (siRNA), and piwi-interacting RNA (piRNA). Perhaps the best-known lncRNAs are ribosomal RNAs and X inactive specific transcript (Xist), which play critical roles in protein translation and X chromosome inactivation, respectively. Large-scale discovery of lncRNAs became feasible initially with tiling microarray [5, 6] and full-length cDNA sequencing [7]. More recently, epigenome analysis [8] and whole transcriptome RNA-sequencing (RNA-Seq) have identified even greater numbers of lncRNAs [9, 10]. Despite this, significant challenges remain to accurately annotate lncRNA genes, as illustrated in a recent study showing that a transcript originally classified as a muscle lncRNA indeed encodes a small protein in the cell [11]. The coding potential of RNA transcripts can be predicted using bioinformatic tools such as PhyloCSF, which is based on comparative genomic analysis of the coding probability of nucleotides across multiple species [12]. Other approaches, such as Coding Potential Calculator (CPC), PORTRIAT and Coding Potential Assessment Tool (CPAT), are also capable of assessing coding potential when lineage specificity limits cross-species alignments [13-15]. Recently, ribosomal profiling analysis of ribosome occupancy on RNA transcripts has provided experimental evidence that lncRNAs lack the capacity to encode proteins [16]. Based on their location relative to nearby protein-coding genes, lncRNAs can be categorized into sense, antisense, intronic, divergent, and intergenic groups (Figure 1). It was recently estimated that the human genome produces thousands of lncRNAs as a result of pervasive transcription from intergenic regions [17]. The widespread transcriptional activity beyond the 2% of the genome encoding proteins was also supported by global analysis of chromatin marks, which revealed that many of the intergenic lncRNAs were marked by characteristic histone signature that marks transcriptionally active chromatin domains [8].
Figure 1. Different classes of lncRNAs.
LncRNA genes can be classified into divergent, intronic, sense, antisense, and intergenic groups according to their location relative to the nearby protein-coding genes.
The expression of lncRNAs exhibits remarkable tissue specificity and is highly regulated during development and in response to physiological signals [18-21]. Like mRNA transcripts, most lncRNAs are transcribed by RNA polymerase II and undergo further steps of processing, including splicing and polyadenylation. LncRNAs have been observed in the nucleus, the cytosol, or both [22], consistent with their roles in regulating diverse biological processes including transcription [23-25], cell differentiation [26, 27], tissue development [28, 29], and tumorigenesis/metastasis (Box 1) [18, 30, 31]. In the nucleus, lncRNAs may function as transcriptional coactivators through direct interaction with transcription factors [32, 33]. However, several lncRNAs have also been found to impair the assembly of transcriptional complexes, leading to inhibition of gene expression [34, 35]. The gene silencing activity of LncRNAs can be attributed to their recruitment of repressive chromatin-remodeling complexes [23, 36], such as the Polycomb Repressive Complex 2 (PRC2) and the SWItch/Sucrose Non-Fermentable (SWI/SNF) complexes. Recent work demonstrated that lncRNAs facilitate the recruitment of PRC2 to chromatin, likely through association with multiple components of the PRC2 complex, including the Suppressor of zeste 12 homolog (SUZ12), Enhancer of zeste homolog 2 (EZH2), and Jumonji, AT rich interactive domain 2 (JARID2) [37, 38]. Some plant lncRNAs engage the SWI/SNF chromatin-remodeling complexes to induce transcriptional silencing via a separate lncRNA-binding protein [39]. Interestingly, the lncRNAs H19 and lincRNA-RoR exert their post-transcriptional effects on gene expression by serving as molecular sponges for microRNAs [40, 41].
Box 1. Molecular and genetic tools for probing lncRNA biology.
The expanding role of lncRNAs in biological regulation has spurred the development of experimental tools to dissect the molecular and functional aspects of lncRNA biology. To globally analyze chromatin occupancy by lncRNAs, Chromatin Isolation by RNA Purification (ChIRP) was developed to enrich endogenous lncRNA and its associated genomic targets [105]. Using chemical cross-linking and tiling biotinylated antisense DNA oligonucleotides for hybridization, this method is capable of generating high-resolution maps of lncRNA binding sites on native chromatin. Using a similar approach, the ChIRP-mass spectrometry (ChIRP-MS) method was developed for identifying endogenous protein factors that associate with specific lncRNA by affinity purification followed by liquid chromatography (LC)-MS/MS [106]. RNA antisense purification (RAP) and capture hybridization analysis of RNA targets (CHART) are alternative methods for selective purification RNA complexes in order to map chromatin binding sites and interacting proteins [107, 108].
LncRNAs form extensive secondary structure through intramolecular base pairing. The presence of distinct structural motifs is critical for lncRNAs to assume their biological functions. Selective 2′-hydroxyl acylation and primer extension (SHAPE) provides a method for selective acylation of 2′-hydroxyls at single-stranded RNA regions but not at double-stranded regions [109]. This approach enables the interrogation of RNA structure at single-nucleotide resolution. To globally study RNA structure, parallel analysis of RNA structure (PARS) was developed using V1 and S1 RNA nucleases, which are specific for double- and single-strand RNA domains, respectively. Selective digestion of distinct RNA domains followed by deep sequencing provides a powerful tool for genome-wide interrogation of RNA structure [110]. More recently, domain-specific ChIRP (dChIRP) was successfully used for identifying the functional domain architecture of lncRNAs [111].
For functional analyses of lncRNAs, both gain- and loss-of-function expression constructs have been routinely used in mammalian cells and in mice [112-114]. Traditional RNAi and antisense oligonucleotides (ASO) can be used to efficiently suppress lncRNA expression [115]. With the development of CRISPR-Cas9 gene editing tools, the activation of endogenous lncRNA genes becomes feasible using a specific guide RNA and a Cas9 fused to a transcriptional activation domain [116]. More recently, a method combining CRISPR-Cas9 directed RNA targeting, called CRISPR-display, was developed to direct RNA domains, including natural lncRNAs, to specific genomic loci [117]. This method allows multiplexed targeting of various RNA modules to different locations in the genome.
Regulation of brown and beige adipocyte differentiation by lncRNAs
Adipose tissues play multifaceted roles in energy storage and expenditure, endocrine signaling, and immune-metabolic crosstalk. Compared to white adipocytes, brown adipocytes contain high mitochondrial content and express uncoupling protein 1 (UCP1), which dissipates chemical energy through heat production. Recent studies demonstrated that metabolically active brown adipose tissue (BAT) is present in adult humans [42-45], raising the prospect that augmenting brown fat abundance and/or function may provide an effective treatment of obesity and its associated metabolic disorders [46, 47]. While sharing key molecular and metabolic characteristics with the classical rodent BAT, brown fat in adult human appears to contain both classical and brown-like adipocytes [48-51], the latter has been termed beige or brite adipocytes [52-54]. In rodents, brown/beige fats appear to have distinct developmental origins [54-57]. The determination, differentiation, and metabolic functions of thermogenic adipocytes are under the control of a growing list of extracellular signaling cues, transcription factors and cofactors, and microRNAs [58-61].
The extent to which, lncRNAs are involved in the regulation of brown/beige fat development and function remained unexplored until recently. Using whole transcriptome RNA-Seq, a cluster of lncRNAs that exhibited differential expression during adipogenesis was identified, several of which appeared to be required for adipocyte differentiation [20]. Among these, linc-RAP-1 (Firre) physically interacts with heterogeneous nuclear ribonucleoprotein (hnRNP) U [62]. Using a microarray platform containing probesets that interrogate both protein-coding and lncRNA transcripts, lncRNA expression in brown and white adipose tissues upon inguinal white fat browning and during brown adipocyte differentiation was analyzed [33]. A cluster of 21 lncRNAs was identified as enriched in brown fat, highly induced during brown adipogenesis, and inducible during browning of white fat in response to the adrenergic agonist CL-316,243. Among this cluster of differentially regulated lncRNAs, brown fat lncRNA1 (Blnc1) was identified as a novel lncRNA that promotes brown and beige adipocyte differentiation and function. The Blnc1 RNA transcript is primarily localized in the nuclear compartment, suggesting that it may play a role in transcriptional regulation.
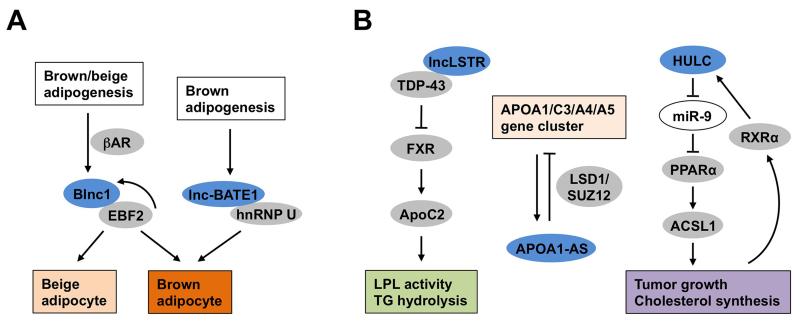
Gain- and loss-of-function studies established Blnc1 as a potent activator of thermogenic adipocyte differentiation. Notably, transplantation of preadipocytes transduced with a recombinant Blnc1 retrovirus in nude mice resulted in the formation of fat pads reminiscent of brown fat, with Ucp1 expression reaching approximately one third of the levels observed in the endogenous brown fat. Whether Blnc1 is absolutely required for the development of brown adipose tissue and browning of white fat remains to be investigated using mice lacking Blnc1. Despite its stimulatory effects on brown and beige preadipocyte differentiation, Blnc1 failed to promote differentiation of 3T3-L1 and C3H10T1/2 progenitor cells into Ucp1-positive adipocytes, suggesting that Blnc1 may act during early adipogenesis and in mature adipocytes to exert its stimulatory effects on the thermogenic gene program. Mechanistically, Blnc1 physically interacts with early B-cell factor 2 (EBF2), a transcription factor recently found to regulate adipocyte differentiation and brown fat development [63, 64], and forms a ribonucleoprotein transcriptional complex to stimulate the expression of genes involved in fuel oxidation and uncoupled respiration. EBF2 also regulates the expression of Blnc1, forming a feedforward regulatory loop that likely serves as a potent switch for thermogenic adipocyte differentiation (Figure 2A).
Figure 2. Regulation of thermogenic adipocyte differentiation and hepatic metabolism by lncRNAs.
(A) Regulation of brown and beige adipogenesis by lncRNAs Blnc1 and lnc-BATE1. Blnc1 is highly induced during brown and beige adipocyte differentiation and physically interacts with EBF2 to promote thermogenic adipocyte differentiation through a feedforward regulatory loop. Lnc-BATE1 is enriched in brown fat, associates with hnRNP U, and is required for brown adipogenesis. (B) Regulation of hepatic metabolism by lncRNAs. LncLSTR regulates hydrolysis of plasma triglycerides by LPL through modulating ApoC2 expression in the liver. APOA1-AS is an antisense transcript that originates from and regulates the expression of genes within the APOA1/C3/A4/A5 cluster. HULC modulates lipid synthesis and tumor growth through its regulation of PPARα-ACSL1 expression. LncRNAs were shown in blue, microRNAs were shown in white, proteins were shown in grey. The abbreviations are as the following: Brown fat lncRNA 1 (Blnc1), Early B-cell factor 2 (EBF2), β-adrenergic receptor (βAR), Heterogeneous nuclear ribonucleoprotein (hnRNP) U, Liver-specific triglyceride regulator (lncLSTR), TAR DNA-binding protein 43 (TDP-43), Farnesoid X receptor (FXR), Lipoprotein lipase (LPL), Antisense transcript of ApoA1 (ApoA1-AS), Lysine-specific demethylase 1 (LSD1), Suppressor of zeste 12 homolog (SUZ12), Highly Up-regulated in Liver Cancer (HULC), Peroxisome proliferator-activated receptor α (PPARα), Acyl-CoA Synthetase Long-Chain Family Member 1 (ACSL1) and Retinoid X Receptor α (RXRα).
More recently, RNA-Seq analysis of transcriptomes in different adipose tissues identified a cluster of brown fat-enriched lncRNAs [65]. Among these, Lnc-BATE1 was found to be induced during brown adipocyte differentiation and expressed at higher levels in BAT than WAT. Knockdown of Lnc-BATE1 by siRNA impaired differentiation of brown adipocytes, as revealed by decreased expression of brown fat markers and mitochondrial genes. Overexpression of Lnc-BATE1, however, failed to augment brown adipogenesis, suggesting that the levels of this lncRNA may not be limiting in thermogenic gene expression. Although lnc-BATE1 appeared to be equally distributed between the cytosol and nucleus, it physically interacts with the nuclear matrix factor hnRNP U, a factor required for brown adipocyte differentiation (Figure 2A). Interestingly, hnRNP U also interacts with Firre, another lncRNA involves in adipogenesis. Several profiling studies have been described to explore how lncRNA expression is regulated during thermogenic adipocyte development [66-68]. However, the significance of candidate lncRNAs in adipocyte biology remains unknown. It is likely that Blnc1 and Lnc-BATE1 are only a tip of iceberg that illustrates the important role of lncRNA regulators in brown and beige adipocyte development. In addition to lncRNA discovery, future work is needed to address the role of these lncRNAs in adipocyte metabolism and metabolic physiology.
LncRNAs and hepatic metabolism
Liver plays a central role in coordinating diverse metabolic processes, including glucose and lipid metabolism, bile acid synthesis, detoxification of xenobiotic compounds, and the secretion of numerous plasma proteins. Hepatic metabolism is highly regulated by nutritional, hormonal, and circadian signals to maintain whole body nutrient and energy homeostasis. Not surprisingly, dysregulation of hepatic metabolism has been implicated in the pathogenesis of several metabolic disorders in metabolic syndrome, such as hyperglycemia, dyslipidemia, and non-alcoholic fatty liver disease. Analysis of the transcriptome architecture of the liver by RNA-Seq revealed a large set of transcripts exhibiting circadian regulation, many of which were mapped to genomic loci corresponding to lncRNAs [21, 69]. The diurnal regulation of genome-wide transcriptional activities was further supported by rhythmic changes in chromatin marks and RNA polymerase II enrichment. Interestingly, the Period circadian clock 2 (Per2) locus was found to produce an antisense transcript (asPer2) that reached its peak levels in opposite phase to the Per2 transcript peaks [21, 69]. Per2 encodes one of the core circadian clock proteins, and while the functional significance of asPer2 in circadian biology remains to be clarified, it is reminiscent of another antisense transcript of the Neurospora clock component antisense frequency (Frq) [70].
Recent studies have demonstrated that lncRNAs are important regulators of hepatic metabolism and plasma lipid homeostasis (Figure 2B). Analysis of microarray data covering a panel of mouse tissues led to the identification of liver-specific triglyceride regulator (lncLSTR), a liver-enriched lncRNA [71]. In vivo knockdown of lncLSTR lowered plasma triglyceride levels through induction of Apolipoprotein C2 (ApoC2), which promotes lipoprotein lipase-mediated catabolism of triglyceride-rich lipoproteins. Further mechanistic studies indicated that lncLSTR physically interacts with TAR DNA-binding protein 43 (TDP-43), a transcriptional repressor, to attenuate the expression of ApoC2 through a bile acid-mediated transcriptional regulatory pathway. In a separate study, an antisense transcript of ApoA1 (ApoA1-AS) was identified as an lncRNA that negatively regulates ApoA1 expression [72]. Knockdown of ApoA1-AS increased ApoA1 gene expression, likely because of the recruitment of histone-modifying enzymes Lysine-specific demethylase 1 (LSD1) and SUZ12 to the ApoA1 promoter. Oligonucleotides targeting ApoA1-AS significantly increased ApoA1 mRNA expression in hepatic cell lines and African green monkeys, illustrating the possibility that lncRNAs may serve as targets for RNA-based therapeutic intervention. Dysregulation of lncRNA expression has also been observed in hepatocellular carcinoma (HCC). For example, the expression of the lncRNA Highly Up-regulated in Liver Cancer (HULC) was elevated in HCC [73]. HULC appeared to facilitate tumor cell growth in part through its induction of Acyl-CoA Synthetase Long-Chain Family Member 1 (ACSL1) and disruption of circadian clock function.
Despite the emerging role of lncRNAs in the regulation of liver clock and metabolism, their significance in liver biology awaits further studies using gain- and loss-of-function mouse models. In addition, how these lncRNAs interface with hormonal and nutritional signaling pathways in the liver remains an important unanswered question. It is possible that some lncRNAs may play a dominant role in nutrient signaling and that their expression levels dictate the downstream metabolic response. Alternatively, other lncRNAs may serve a more permissive function to facilitate metabolic adaptation in the liver.
Regulation of skeletal and cardiac muscle development and function by lncRNAs
Skeletal muscle is an important metabolic tissue, as it plays a major role in postprandial glucose disposal by increasing glucose uptake in response to circulating insulin. Skeletal myofibers generate ATP through a combination of glycolysis and mitochondrial oxidative phosphorylation to support the energetic demand of muscle contraction. Not surprisingly, muscle energy metabolism has significant implications in whole body energy homeostasis. Impaired muscle insulin action is an early hallmark of the metabolic derangements in metabolic syndrome. As such, skeletal muscle development and metabolism have been a major focus of numerous studies.
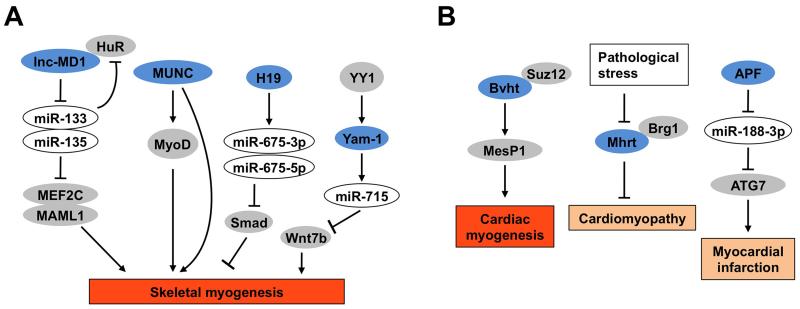
Several lncRNAs have been shown to regulate skeletal muscle development (Figure 3A). Linc-MD1 is a muscle-specific lncRNA that promotes skeletal myocyte differentiation by serving as a sponge for microRNAs, including miR-133 and miR-135 [27]. The inactivation of these microRNAs relieved their inhibitory effects on Mastermind-like protein 1 (MAML1) and Myocyte-specific enhancer factor 2C (MEF2C), two pro-myogenic transcriptional regulators, leading to increased myogenesis. A subsequent study demonstrated that linc-MD1 interacts with the RNA-binding protein HuR, which regulates the accumulation of linc-MD1 during myocyte differentiation [74]. Similarly, MyoD upstream noncoding (MUNC) and H19 are two lncRNAs that also promote muscle differentiation. MUNC induces MyoD expression and myogenic genes expression through MyoD-dependent and independent mechanisms [75], whereas H19 induces skeletal myocyte differentiation through its induction of miR-675-3p and miR-675-5p, two microRNAs generated within the H19 locus [76]. For both MUNC and H19, in vivo knockdown studies showed that they play an important role in progenitor cell differentiation and muscle regeneration. In another study, a group of lncRNAs regulated by Yin Yang 1 (YY1), a transcription factor that represses multiple muscle genes were analyzed. Among these lncRNAs, YY-1 associated muscle lincRNA (Yam-1) expression was downregulated during differentiation and acted as an inhibitor of myogenesis through its cis regulation of miR-715/Wnt7b signaling [77].
Figure 3. Regulation of skeletal and cardiac muscle development and function by lncRNAs.
(A) Regulation of skeletal myogenesis by lncRNAs. Lnc-MD1 serves as a sponge for microRNAs that target myogenic regulators MEF2C and MAML1. MUNC regulates myogenesis through MyoD-dependent and independent mechanisms. The H19 lncRNA transcript induces two microRNAs that antagonize the inhibitory effects of Smad on myocyte differentiation. Yam-1 is a target gene of the transcription factor YY1 that negatively regulates skeletal myogenesis. (B) Regulation of cardiac myocyte development and function by lncRNAs. Bvht promotes cardiovascular lineage commitment and is required for activation of a core cardiovascular gene network through the regulation of MesP1. Mhrt is cardiac-specific lncRNA transcripts that responds to pathologic stress in the heart and plays a protective role in cardiomyopathy. APF promotes autophagic cell death and myocardial infarction through its regulation of ATG7. LncRNAs were shown in blue, microRNAs were shown in white, proteins were shown in grey. The abbreviations are as the following: Myocyte-specific enhancer factor 2C (MEF2C), Mastermind-like protein 1 (MAML1), MyoD upstream noncoding (MUNC), Yin Yang 1 (YY1), Brave heart (Bvht), Suppressor of zeste 12 homolog (SUZ12), Mesoderm posterior 1 (MesP1), Myosin heavy-chain-associated RNA transcript (Mhrt), Brahma-related gene 1 (Brg1), Autophagy-promoting factor (APF) and Autophagy Related 7 (ATG7).
Cardiac muscle is extremely metabolically active and undergoes significant changes in its energy metabolism in disease states. Similar to skeletal myogenesis, lncRNAs also play an important regulatory role in cardiomyocyte differentiation and function (Figure 3B). Braveheart (Bvht) was identified as a heart-associated lncRNA that is essential for the progression from nascent mesoderm toward mature cardiomyocytes. Through interaction with PRC2 component SUZ12, Bvht epigenetically regulates mesoderm posterior 1 (MesP1), a master regulator of multipotent cardiovascular progenitors, and activates a global cardiovascular gene network during cardiomyocyte differentiation [78]. LncRNAs also regulate the function and homeostasis of mature cardiomyocytes. Pathological stresses resulted in the inhibition of the lncRNA myosin heavy-chain-associated RNA transcript (Mhrt) expression in mice [79]. Moreover, transgenic expression of Mhrt protects the heart from hypertrophic response to stress stimuli, showing a functional role for Mhrt in cardiomyocyte biology. Mhrt was also downregulated in various types of myopathic hearts in humans, suggesting a potentially conserved role of this lncRNA in protection against cardiomyopathy. Another study demonstrated that the lncRNA autophagy-promoting factor (APF) regulates autophagy and myocardial cell death by targeting miR-188-3p and autophagy related 7 (ATG7) [80]. Knockdown of APF significantly reduced myocardial infarction sizes following ischemia/reperfusion-induced injury. Together, these studies underscore an important role of lncRNAs in skeletal and cardiac muscle development and function (Figure 3).
LncRNAs and islet function
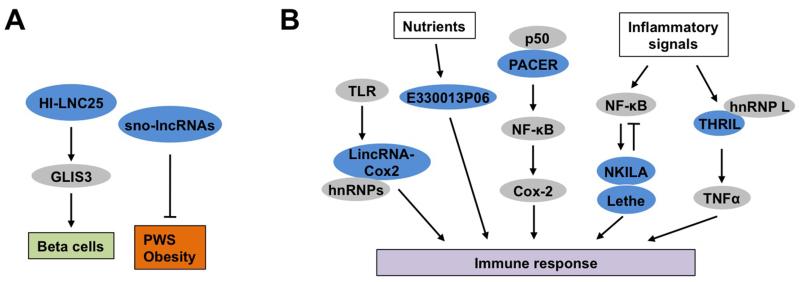
Pancreatic islets serve a critical role in metabolic homeostasis through the secretion of key endocrine hormones such as insulin and glucagon. Recent RNA-Seq studies revealed an extensive collection of intergenic and antisense lncRNAs in mouse and human islets [81, 82]. Many of them exhibited highly tissue-specific and regulated expression patterns during β cell differentiation and maturation. Interestingly, islet lncRNAs frequently map near chromatin domains containing islet-specific coding genes, suggesting that protein-coding and lncRNA genes may share common regulatory elements to direct their expression in pancreatic islets. While lncRNA expression profiles in islets exhibit notable species specificity [83], HI-LNC25 was identified as a β cell-specific lncRNA conserved between mouse and human. HI-LNC25 regulates the expression of GLI-similar 3 (GLIS3) (Figure 4A), a susceptibility gene for type 1 and type 2 diabetes, suggesting that HI-LNC25 may potentially play a role in the development of diabetes [82]. The expression of a subset of islet lncRNAs was altered in type 2 diabetes, whereas some lncRNAs were mapped to genetic loci that influence the susceptibility to diabetes. Given that many disease-associated genetic variations are mapped to intergenic regions, it is possible that the metabolic consequences of some of the variants may be due to their influences on lncRNA expression and/or function.
Figure 4. Regulation of other metabolically relevant cell types by lncRNAs.
(A) HI-LNC25 is a β cell-specific lncRNA that is required for maintaining the expression of GLIS3. The sno-lncRNAs originated from the PWS locus modulate energy balance through their actions in the central nervous system. (B) Regulation of immune response by lncRNAs. LincRNA-Cox2 and PACER are two lncRNAs originated from the Cox2 gene locus that regulate cytokine signaling. E330013P06 is regulated by nutrients in macrophages and promotes inflammatory signaling. NKILA and Lethe are both cytokine-inducible lncRNAs that serve as negative feedback regulator of NF-κB. THRIL mediates the induction of TNFα gene expression in macrophages in response to proinflammatory stimuli. LncRNAs and protein were shown in blue and grey, respectively. The abbreviations are as the following: GLI-similar 3 (GLIS3), Prader-Willi syndrome (PWS), Toll-like receptor (TLR), Heterogeneous nuclear ribonucleoprotein (hnRNP), p50-associated COX-2 extragenic RNA (PACER), Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), NF-κB interacting lncRNA (NKILA), LncRNA pseudogene (Lethe), Tumor necrosis factor α (TNFα) and TNFα and hnRNP L related immunoregulatory LincRNA (THRIL).
Regulation of immune response by lncRNAs
Chronic low-grade inflammation has emerged as an important pathogenic link between obesity and metabolic disease [84-88]. Obesity-associated adipose tissue inflammation is characterized by a robust shift of adipose tissue macrophages from alternatively activated to classically activated subtypes [89, 90]. This shift from anti-inflammatory to proinflammatory macrophage polarization is likely an early event during the development of insulin resistance. Accumulating evidences suggest that lncRNAs play an important role in modulating multiple aspects of immune responses [91] (Figure 4B). Using RNA-Seq, the expression profiles of bone marrow-derived macrophages isolated from control and leptin receptor-deficient (db/db) obese mice were analyzed [92]. The lncRNA transcript E330013P06 was found to be significantly elevated in macrophages from db/db and diet-induced insulin resistant mouse models. Overexpression of E330013P06 in macrophage cells augmented cytokine signaling and proinflammatory gene expression, whereas RNAi knockdown elicited opposite effects. Whether E330013P06 plays a role in obesity-associated adipose tissue inflammation and metabolic dysregulation remains currently unknown. Nevertheless, these findings illustrate the dysregulation of the non-coding genome in obesity and its potential contribution to the pathogenesis of metabolic disorders.
LincRNA-Cox2 was discovered as a member of a cluster of lncRNAs that was stimulated in response to inflammatory stimuli through the activation of Toll-like receptors in bone marrow derived macrophages (BMDM) [93]. Interestingly, lincRNA-Cox2 exerts both the activation and repression on different target genes, in part through its interaction with hnRNPs. Another lncRNA, p50-associated COX-2 extragenic RNA (PACER) originated from the upstream promoter region of the COX-2 gene locus was found to interact with p50, a repressive subunit of nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), upon induction of COX-2 expression [94]. This PACER/p50 association leads to the assembly of active NF-κB transcriptional complexes that stimulate Cox-2 gene expression and the inflammatory response. In two separate studies, cytokine-inducible NF-κB interacting lncRNA (NKILA) and a pseudogene lncRNA (Lethe) were identified to play a negative feedback regulatory role in proinflammatory cytokine signaling [95, 96]. NKILA inhibits IκB phosphorylation, leading to suppression of NF-κB activation, whereas Lethe attenuates DNA binding and transcriptional function of the NF-κB subunit RelA. Further, a panel of lncRNAs were identified as differentially regulated in response to innate activation in the macrophage cell line THP-1 [97]. Among these, Tumor necrosis factor α (TNFα) and hnRNP L related immunoregulatory LincRNA (THRIL) interacts with hnRNP L and regulates the expression of TNFα and other genes involved in immune response. Together, these studies demonstrate that lncRNAs target multiple cytokine signaling and inflammatory response pathways. It would be of great interest to explore the potential pathogenic involvements of these lncRNAs in the pathogenesis of metabolic disease.
Potential role of LncRNAs in metabolic disease
Dysregulation of lncRNA gene expression has been implicated in several human diseases, such as Facioscapulohumeral muscular dystrophy [98], the HELLP Syndrome (Hemolysis, elevated liver enzymes, low platelets) [99], and the Angelman Syndrome [100]. The latter is a single-gene neurological disorder caused by maternal deficiency of the imprinted gene ubiquitin protein ligase E3A (UBE3A) in part through lncRNA-mediated gene silencing. Remarkably, antisense oligonucleotides targeting the UBE3A antisense transcript lowered its expression level, leading to reactivation of Ube3a gene expression and amelioration of cognitive deficits associated with the disease [100]. A compelling example of the involvements of lncRNAs in metabolic disease is Prader-Willi syndrome (PWS). PWS is a genetic disorder that causes childhood obesity and various neurological symptoms [101]. PWS results from loss of expression of paternally expressed genes located on the PWS region on chromosome 15 (15q11-q13), which contains multiple paternally expressed noncoding RNAs [102]. Interestingly, these lncRNAs are processed into a unique class of lncRNAs that are flanked by C/D box containing small nucleolar RNAs (snoRNAs) at the 5’ and 3’ ends (sno-lncRNAs) [103]. These PWS sno-lncRNAs appeared to accumulate near their sites of synthesis in the nucleus. Deletion of the host gene for these sno-lncRNAs increased energy expenditure in mice, likely due to altered expression of diurnally regulated genes in the brain, including the core clock and metabolic genes [104] (Figure 4A). The exact mechanisms through which the PWS lncRNAs regulate energy balance remain unknown at present.
Concluding remarks
LncRNAs are emerging as an important class of regulatory factors that control the development and function of metabolic tissues. Acting in concert with protein factors and other non-protein regulators, such as microRNAs, lncRNAs provide an unorthodox link through which genetic information is transmitted to influence biological processes in the cell. The discovery and functional study of individual lncRNAs are set to expand our horizon on the genetic mechanisms of metabolic homeostasis and disease. Several challenges and opportunities arise from the study of lncRNA biology in metabolic control (Outstanding Questions). First, the annotation of lncRNAs remains challenging and incomplete at present. Unlike protein-coding genes, lncRNAs exhibit a relatively low degree of nucleotide sequence conservation across species. In addition, lncRNAs may encode micropeptides that have important biological functions. As such, our ability to predict the structure of lncRNA molecules and their biological function remains limited. Second, lncRNAs likely exert their biological effects through diverse mechanisms, many of which remain to be discovered. A critical question in the context of metabolic regulation is how lncRNAs interface with the classical nutrient and hormonal signaling pathways to control the development and function of various metabolic cell types. Given the highly regulated nature of lncRNA gene expression, it is likely that certain lncRNAs may emerge as “master” regulators of tissue development and energy metabolism. Finally, significant efforts are needed to deconvolute the role of lncRNA pathways in metabolic physiology and disease. Investigating highly conserved lncRNAs will arguably provide biological insights into the basic principles of lncRNA biology. In addition, the knowledge on conserved lncRNA pathways may prove relevant for human disease conditions.
OUTSTANDING QUESTIONS BOX.
How can we comprehensively annotate lncRNA genes and accurately predict their molecular functions? To what extent are lncRNA sequence and function conserved among different species?
How are lncRNAs integrated with hormonal and nutrient signals to control metabolic tissue development, plasticity, and function? How do lncRNAs and protein factors work in concert to exert effects on epigenetic modification and metabolic gene expression?
What are the physiological roles of lncRNAs in the regulation of glucose and lipid metabolism and whole body energy balance? What is the extent to which altered lncRNA expression contributes to metabolic disorders in animal models and humans? A major challenge is to develop tools to target lncRNAs to alter the course of metabolic disease progression.
TRENDS BOX.
LncRNAs exhibit tissue-specific and highly regulated expression patterns and are frequently dysregulated in disease states;
LncRNAs regulate diverse biological processes through the formation of lncRNA-protein and lncRNA-miRNA complexes to control gene expression and function;
LncRNAs regulate metabolic tissue development and function, including adipogenesis, hepatic lipid metabolism, islet function, and energy balance;
LncRNAs are important regulators of skeletal and cardiac muscle development and immune response;
Acknowledgments
We would like to thank other members of the lab for discussion. This work was supported by the National Institutes of Health (DK077086 and DK095151) and American Heart Association. We apologize to colleagues whose relevant work was not cited here due to space limitation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Knoll M, et al. Long non-coding RNAs as regulators of the endocrine system. Nature reviews. Endocrinology. 2015;11:151–160. doi: 10.1038/nrendo.2014.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kornfeld JW, et al. Regulation of metabolism by long, non-coding RNAs. Front Genet. 2014;5:57. doi: 10.3389/fgene.2014.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rinn JL, et al. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cech TR, et al. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Kapranov P, et al. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- 6.Rinn JL, et al. The transcriptional activity of human Chromosome 22. Genes & development. 2003;17:529–540. doi: 10.1101/gad.1055203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maeda N, et al. Transcript annotation in FANTOM3: mouse gene catalog based on physical cDNAs. PLoS genetics. 2006;2:e62. doi: 10.1371/journal.pgen.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & development. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson DM, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin MF, et al. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic acids research. 2007;35:W345–349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arrial RT, et al. Screening non-coding RNAs in transcriptomes from neglected species using PORTRAIT: case study of the pathogenic fungus Paracoccidioides brasiliensis. BMC bioinformatics. 2009;10:239. doi: 10.1186/1471-2105-10-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, et al. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic acids research. 2013;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guttman M, et al. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hangauer MJ, et al. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS genetics. 2013;9:e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun L, et al. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci U S A. 2013;110:3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vollmers C, et al. Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metab. 2012;16:833–845. doi: 10.1016/j.cmet.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 24.Mercer TR, et al. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 25.Wang KC, et al. Molecular mechanisms of long noncoding RNAs. Molecular cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guttman M, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cesana M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Batista PJ, et al. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu W, et al. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012;13:971–983. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panzitt K, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 31.Ji P, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 32.Feng J, et al. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes & development. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao XY, et al. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol Cell. 2014;55:372–382. doi: 10.1016/j.molcel.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martianov I, et al. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaneko S, et al. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell. 2014;53:290–300. doi: 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cifuentes-Rojas C, et al. Regulatory interactions between RNA and polycomb repressive complex 2. Mol Cell. 2014;55:171–185. doi: 10.1016/j.molcel.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Y, et al. A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Mol Cell. 2013;49:298–309. doi: 10.1016/j.molcel.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kallen AN, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 42.Nedergaard J, et al. Unexpected evidence for active brown adipose tissue in adult humans. Am J Physiol Endocrinol Metab. 2007;293:E444–452. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- 43.Cypess AM, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Marken Lichtenbelt WD, et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 45.Virtanen KA, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 46.Enerback S. Human brown adipose tissue. Cell metabolism. 2010;11:248–252. doi: 10.1016/j.cmet.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 47.Nedergaard J, et al. The changed metabolic world with human brown adipose tissue: therapeutic visions. Cell metabolism. 2010;11:268–272. doi: 10.1016/j.cmet.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 48.Lidell ME, et al. Evidence for two types of brown adipose tissue in humans. Nat Med. 2013;19:631–634. doi: 10.1038/nm.3017. [DOI] [PubMed] [Google Scholar]
- 49.Cypess AM, et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat Med. 2013;19:635–639. doi: 10.1038/nm.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jespersen NZ, et al. A classical brown adipose tissue mRNA signature partly overlaps with brite in the supraclavicular region of adult humans. Cell metabolism. 2013;17:798–805. doi: 10.1016/j.cmet.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 51.Sharp LZ, et al. Human BAT possesses molecular signatures that resemble beige/brite cells. PLoS One. 2012;7:e49452. doi: 10.1371/journal.pone.0049452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petrovic N, et al. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J Biol Chem. 2010;285:7153–7164. doi: 10.1074/jbc.M109.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schulz TJ, et al. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:143–148. doi: 10.1073/pnas.1010929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu J, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berry R, et al. Characterization of the adipocyte cellular lineage in vivo. Nat Cell Biol. 2013;15:302–308. doi: 10.1038/ncb2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seale P, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Timmons JA, et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4401–4406. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harms M, et al. Brown and beige fat: development, function and therapeutic potential. Nat Med. 2013;19:1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- 59.Peirce V, et al. The different shades of fat. Nature. 2014;510:76–83. doi: 10.1038/nature13477. [DOI] [PubMed] [Google Scholar]
- 60.Trajkovski M, et al. MicroRNA networks regulate development of brown adipocytes. Trends Endocrinol Metab. 2013;24:442–450. doi: 10.1016/j.tem.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu J, et al. Adaptive thermogenesis in adipocytes: is beige the new brown? Genes & development. 2013;27:234–250. doi: 10.1101/gad.211649.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hacisuleyman E, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jimenez MA, et al. Critical role for Ebf1 and Ebf2 in the adipogenic transcriptional cascade. Mol Cell Biol. 2007;27:743–757. doi: 10.1128/MCB.01557-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rajakumari S, et al. EBF2 determines and maintains brown adipocyte identity. Cell Metab. 2013;17:562–574. doi: 10.1016/j.cmet.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alvarez-Dominguez JR, et al. De Novo Reconstruction of Adipose Tissue Transcriptomes Reveals Long Non-coding RNA Regulators of Brown Adipocyte Development. Cell Metab. 2015 doi: 10.1016/j.cmet.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen J, et al. Differential lncRNA expression profiles in brown and white adipose tissues. Mol Genet Genomics. 2015;290:699–707. doi: 10.1007/s00438-014-0954-x. [DOI] [PubMed] [Google Scholar]
- 67.You LH, et al. Transcriptome analysis reveals the potential contribution of long noncoding RNAs to brown adipocyte differentiation. Mol Genet Genomics. 2015 doi: 10.1007/s00438-015-1026-6. [DOI] [PubMed] [Google Scholar]
- 68.Zhang J, et al. Distinct expression profiles of LncRNAs between brown adipose tissue and skeletal muscle. Biochem Biophys Res Commun. 2014;443:1028–1034. doi: 10.1016/j.bbrc.2013.12.092. [DOI] [PubMed] [Google Scholar]
- 69.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kramer C, et al. Role for antisense RNA in regulating circadian clock function in Neurospora crassa. Nature. 2003;421:948–952. doi: 10.1038/nature01427. [DOI] [PubMed] [Google Scholar]
- 71.Li P, et al. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab. 2015;21:455–467. doi: 10.1016/j.cmet.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Halley P, et al. Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Rep. 2014;6:222–230. doi: 10.1016/j.celrep.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cui M, et al. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer research. 2015;75:846–857. doi: 10.1158/0008-5472.CAN-14-1192. [DOI] [PubMed] [Google Scholar]
- 74.Legnini I, et al. A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol Cell. 2014;53:506–514. doi: 10.1016/j.molcel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mueller AC, et al. MUNC, a long noncoding RNA that facilitates the function of MyoD in skeletal myogenesis. Mol Cell Biol. 2015;35:498–513. doi: 10.1128/MCB.01079-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dey BK, et al. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes & development. 2014;28:491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu L, et al. Genome-wide survey by ChIP-seq reveals YY1 regulation of lincRNAs in skeletal myogenesis. The EMBO journal. 2013;32:2575–2588. doi: 10.1038/emboj.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Klattenhoff CA, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Han P, et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514:102–106. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang K, et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nature communications. 2015;6:6779. doi: 10.1038/ncomms7779. [DOI] [PubMed] [Google Scholar]
- 81.Ku GM, et al. Research resource: RNA-Seq reveals unique features of the pancreatic beta-cell transcriptome. Mol Endocrinol. 2012;26:1783, 1792. doi: 10.1210/me.2012-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moran I, et al. Human beta cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012;16:435, 448. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Benner C, et al. The transcriptional landscape of mouse beta cells compared to human beta cells reveals notable species differences in long non-coding RNA and protein-coding gene expression. BMC Genomics. 2014;15:620. doi: 10.1186/1471-2164-15-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gregor MF, et al. Inflammatory mechanisms in obesity. Annu Rev Immunol. 2011;29:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 85.Lumeng CN, et al. Inflammatory links between obesity and metabolic disease. The Journal of clinical investigation. 2011;121:2111–2117. doi: 10.1172/JCI57132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Odegaard JI, et al. Pleiotropic actions of insulin resistance and inflammation in metabolic homeostasis. Science. 2013;339:172–177. doi: 10.1126/science.1230721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Osborn O, et al. The cellular and signaling networks linking the immune system and metabolism in disease. Nat Med. 2012;18:363–374. doi: 10.1038/nm.2627. [DOI] [PubMed] [Google Scholar]
- 88.Sun S, et al. Mechanisms of inflammatory responses in obese adipose tissue. Annu Rev Nutr. 2012;32:261, 286. doi: 10.1146/annurev-nutr-071811-150623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lumeng CN, et al. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 2007;117:175–184. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lumeng CN, et al. Increased inflammatory properties of adipose tissue macrophages recruited during diet-induced obesity. Diabetes. 2007;56:16–23. doi: 10.2337/db06-1076. [DOI] [PubMed] [Google Scholar]
- 91.Heward JA, et al. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014;35:408–419. doi: 10.1016/j.it.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reddy MA, et al. Regulation of inflammatory phenotype in macrophages by a diabetes-induced long noncoding RNA. Diabetes. 2014;63:4249–4261. doi: 10.2337/db14-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Carpenter S, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789, 792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Krawczyk M, et al. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-kappaB complexes. Elife. 2014;3:e01776. doi: 10.7554/eLife.01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu B, et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 96.Rapicavoli NA, et al. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife. 2013;2:e00762. doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li Z, et al. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A. 2014;111:1002–1007. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cabianca DS, et al. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149:819–831. doi: 10.1016/j.cell.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.van Dijk M, et al. HELLP babies link a novel lincRNA to the trophoblast cell cycle. J Clin Invest. 2012;122:4003–4011. doi: 10.1172/JCI65171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Meng L, et al. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015;518:409–412. doi: 10.1038/nature13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Farooqi IS, et al. Monogenic obesity in humans. Annu Rev Med. 2005;56:443–458. doi: 10.1146/annurev.med.56.062904.144924. [DOI] [PubMed] [Google Scholar]
- 102.Sahoo T, et al. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat Genet. 2008;40:719–721. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yin QF, et al. Long noncoding RNAs with snoRNA ends. Mol Cell. 2012;48:219–230. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 104.Powell WT, et al. A Prader-Willi locus lncRNA cloud modulates diurnal genes and energy expenditure. Hum Mol Genet. 2013;22:4318–4328. doi: 10.1093/hmg/ddt281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chu C, et al. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chu C, et al. Systematic discovery of xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Engreitz J, et al. RNA antisense purification (RAP) for mapping RNA interactions with chromatin. Methods in molecular biology. 2015;1262:183–197. doi: 10.1007/978-1-4939-2253-6_11. [DOI] [PubMed] [Google Scholar]
- 108.Simon MD, et al. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci U S A. 2011;108:20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wilkinson KA, et al. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nature protocols. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 110.Wan Y, et al. Genome-wide mapping of RNA structure using nuclease digestion and high-throughput sequencing. Nature protocols. 2013;8:849–869. doi: 10.1038/nprot.2013.045. [DOI] [PubMed] [Google Scholar]
- 111.Quinn JJ, et al. Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat Biotechnol. 2014;32:933–940. doi: 10.1038/nbt.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yildirim E, et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152:727–742. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tian D, et al. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sleutels F, et al. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 115.Gutschner T, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer research. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Maeder ML, et al. CRISPR RNA-guided activation of endogenous human genes. Nature methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shechner DM, et al. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nature methods. 2015 doi: 10.1038/nmeth.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]