Abstract
A key aspect of the control of gene expression is the differential rates of mRNA translation and degradation, including alterations due to extracellular inputs. Surprisingly, multiple examples now argue that Hsp70 protein chaperones and their associated Hsp40 partners modulate both mRNA degradation and translation. Hsp70 proteins affect mRNA metabolism by a variety of mechanisms including regulating nascent polypeptide chain folding, activating signal transduction pathways, promoting clearance of stress granules, and controlling mRNA degradation in an mRNA-specific manner. Taken together, these observations highlight the general principle that mRNA metabolism is coupled to the proteostatic state of the cell, often as assessed by the presence of unfolded or misfolded proteins.
Keywords: Hsp70, translation, mRNA decay, stress granules
The growing appreciation of Hsp70 protein function in post-transcriptional mRNA regulation
Control of the mRNA transcriptome is crucial for proper gene regulation. Within the nucleus, the mRNA transcriptome is controlled through modulation of transcription, splicing, and nuclear export. In the cytosol, mRNA function can be controlled through subcellular localization, as well as through changes in translation and decay. Certain stimuli and cellular states strongly regulate mRNA function through some or all of these mechanisms. In particular, stress responses dramatically alter the translation, degradation, and localization of mRNA [1}[2,3]). Finally, during stress the localization of mRNAs is controlled through accumulation of untranslated mRNAs either in stress granules, which are messenger ribonucleoprotein particle (mRNP) assemblies that are thought to contain mRNAs stalled in early phases of translation initiation [4], or alternatively in P-bodies, which are assemblies of untranslating mRNAs, translation repressors and components of the mRNA degradation machinery [5]. Since P-bodies and stress granules assemble in part through interactions between the prion domains or intrinsically disordered domains of RNA-binding proteins [6–9], proteins that modulate protein-protein interactions could be important in controlling mRNP granule assembly and/or disassembly.
A key set of proteins involved in protein folding and rearrangements of either aggregates or protein complexes are the Hsp70 proteins. Initially discovered as a class of 70KDa proteins induced during heat stress to deal with the increased load of misfolded proteins, it is now known that there are also constitutively expressed Hsp70 proteins [10,11]. Herein, we refer to both of these classes as Hsp70 proteins. Hsp70 proteins interact with numerous protein folding states (unfolded, misfolded, terminally aggregated) to promote protein refolding and/or to target misfolded proteins for degradation [12], and play roles in rearranging cellular protein assemblies. For example, Hsp70 proteins are known to play a role in the disassembly of clathrin coats [13] and in the disassembly/clearance of stress granules [14].
Hsp70 proteins typically function with Hsp40 proteins. Hsp40 proteins greatly increase the ATPase activity of Hsp70 proteins and also direct client protein specificity [12,15]. The ATPase cycle of Hsp70 proteins oscillates between the ATP-bound state (low substrate affinity) and ADP-bound state (high substrate affinity), which is promoted by Hsp40 proteins stimulating ATP hydrolysis by Hsp70. Importantly, different Hsp70:Hsp40 pairings elicit distinct outcomes for their client proteins [12], and therefore different Hsp40 proteins can provide specificity to the more generic Hsp70 proteins. Hsp70 function is also controlled by nucleotide exchange factors that promote the exchange of ADP for ATP, and thereby promote substrate release.
In the past 15 years, it has become increasingly clear that Hsp70 proteins play important roles in mRNA metabolism. In this review, we describe the diversity of mechanisms by which Hsp70 proteins impact mRNA metabolism. An important general principle is that by coupling mRNA translation or degradation to Hsp70 activity, it allows the cell to coordinate aspects of mRNA metabolism with the proteostatic state of the cell.
Effects of Hsp70/Hsp40 complexes on translation
Early evidence that Hsp70/Hsp40 proteins could affect translation came from three main types of observations. First, it was observed in yeast that conditional inactivation of Hsp70 or the Sis1 Hsp40 family member led to a decrease in translation as assessed by 35S amino acid incorporation or polysome profiling [16,17]. Similarly, inhibition of Hsp70 function with chemical inhibitors or dominant negative alleles in mammalian cells also generally inhibited translation [18,19]. Second, Hsp70 and Hsp40 proteins are observed to associate with polysomes [17,19,20]. In yeast, there is even a dedicated Hsp70 protein, encoded by the Ssb1 and Ssb2 genes, that specifically interacts with ribosomes [20]. Finally, in yeast a conditional loss of function allele of Sis1 (an Hsp40 family member) is rescued by the same suppressor mutations that also rescue deletion of Pab1, a known promoter of translation initiation [17]. From these results, it was inferred that Sis1 also has a role in translation initiation.
HSP70 enhances translation: nascent peptide folding
One way that Hsp70 proteins affect translation is by enhancing the folding of nascent peptides. This is suggested by the observations that Ssb1/2 (a ribosome-associated Hsp70 family in yeast) co-sediments with polysomes, interacts directly with nascent peptides [21,22], and that loss of Ssb function results in accumulation of aggregated nascent peptides [22]. Importantly, in higher eukaryotes the constitutive Hsp70 protein, referred to as Hsc70, is likely to perform this co-translational nascent peptide folding [23].
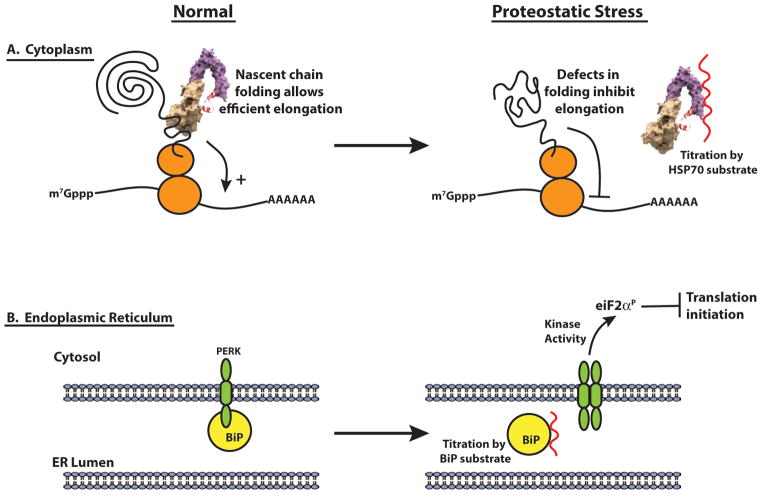
Two new studies demonstrate that the role of Hsp70 proteins in nascent chain folding is important for normal translation elongation [18,19]. The key result of these studies was that when either Hsp70 function was inhibited, or during proteotoxic stress, there was an increase in ribosomal pausing near in the first ~50 codons of ORFs [18,19]. The implication from these studies is that Hsp70 interacts with nascent peptides at the ribosome exit tunnel to promote translation elongation. During proteotoxic stress, Hsp70 would be titrated to other unfolded proteins, thus translation elongation is inhibited (Figure 1). The simplest model is that Hsp70 normally promotes elongation by binding to the nascent peptide and promoting its extrusion from the ribosome in a manner that promotes continued elongation. Consistent with this idea of protein:nascent peptide binding facilitating translation, an engineered heterologous protein interaction system enhanced translation only when the protein interacted with the nascent chain [19]. This supports a model of Hsp70-mediated nascent peptide ‘pulling’ from the ribosome exit tunnel to facilitate translation.
Figure 1.
Hsp70 effects on translation changes with stress. In the cytoplasm (A), Hsp70 proteins bind nascent polypeptides, thus enhancing translation. However, when cells are under proteostatic stress, Hsp70 no longer interacts with nascent peptides, which decreases translation elongation. In the endoplasmic reticulum (ER) (B), the Hsp70 family member Binding Immunoglobulin protein (BiP) normally binds PRKR-like endoplasmic reticulum kinase (PERK) in the ER. Misfolded/unfolded proteins in the ER titrate BIP away from PERK, allowing PERK activation, phosphorylation of eukaryotic initiation factor 2α, eIF2α and global downregulation of translation initiation.
An unresolved issue is why ribosomes only accumulate near the beginning of the ORF when Hsp70 is inhibited. In principle, Hsp70 interaction with the nascent peptide could be important throughout the ORF. One possibility is that the initial N terminus of the protein is highly sensitive to Hsp70 loss since later in the ORF a more complete nascent peptide would be formed, which could have already initiated folding and thereby have interactions in cis that contribute to “pulling” the nascent peptide from the ribosome exit tunnel.
It should be noted that this model implies a hierarchical order of Hsp70 client proteins. That is, Hsp70 proteins preferentially act on misfolded or aggregated proteins that are present during stress, as opposed to nascent peptides associated with the ribosome. Whether this would be accomplished by extrinsic factors (e.g., a change in Hsp70 subcellular localization) or intrinsic factors (e.g., differential substrate specificity of constitutive vs. induced Hsp70 isoforms) is unknown.
HSP70 enhances translation: strengthening translation initiation mRNP
Some evidence suggests that Hsp70 function may also affect the function of specific translation factors, such as Pab1 and eIF4F. A connection of Hsp70 to general translation was implied since a lack of Hsp70 function in yeast resulted in a decrease in polysomes and 35S incorporation [16]. A connection to Pab1 was proposed since Hsp70 defects led to a loss of the co-immunoprecipitation between Hsp70 proteins and Pab1 in heavy translation complexes, which were interpreted to be polysomes [16]. This raises the possibility that yeast Hsp70 proteins also promote translation through maintaining Pab1 activity, which is known to enhance translation initiation.
Hsp70 has also been suggested to affect translation initiation through eIF4F, a translation initiation complex consisting of eIF4A, eIF4E, and eIF4G that binds the 5′ cap of mRNAs to recruit ribosomes in mammalian cells via an eIF4G:eIF3:40S interaction [26,27]. In addition, translation is further stimulated by a direct interaction between eIF4G and polyA binding protein (PABP) [28]. During prolonged heat stress in human cell culture, eIF4G becomes insoluble and associates less with eIF4E, which limits the availability of the critical eIF4F complex. However, this effect is reversed with simultaneous overexpression of Hsc70 [29]. One interpretation of these observations is that eIF4G is a relatively aggregation prone protein, and interactions with Hsp27, Hsc70 and other Hsps promotes its function by maintaining its solubility [29]. The greater implication is that Hsp70 proteins may be necessary for cellular processes that require components of differing solubilities (or aggregation propensities). Since there are multiple connections of Hsp70 proteins to translation, determining the impact of these Hsp70 interactions with translation initiation factors will require additional work.
Collectively, these results suggest multiple roles for Hsp70 and Hsp40 proteins in coupling translation to the proteostatic state of the cell, presumably as defined by competition for Hsp70s due to the pool of unfolded/misfolded nascent or mature polypeptides. One clear role is that Hsp70/Hsp40 function at the ribosome exit tunnel to fold/prevent aggregation of nascent peptides. A second, and more speculative, role is that Hsp70/40 proteins may regulate the folded state of key translation initiation factors and/or their interactions with each other.
Coupling of the transcriptome to ER stress: the Unfolded Protein Response
The population and function of the transcriptome is also directly coupled to proteostatic stress in the endoplasmic reticulum (ER) through the unfolded protein response (UPR), which is initiated in part by a HSP70 family member, referred to as BiP, localized to the lumen of the ER. When misfolded proteins accumulate in the ER, they titrate BiP away from sites on stress sensors, which triggers sensor activation [30,31]. For example, normally the protein kinase PERK dimerizes with BiP and is in an inactive confromation, but titration of BiP to unfolded ER proteins allows PERK to self-activate by dimerization and then downregulate translation in the cytosol by an inhibitory phosphorylation of eIF2α (Figure 1).
The proteostatic stress triggering the UPR response affects three key aspects of the transcriptome. First, in organisms from yeast to man, the Hac1/Xbp1 transcription factor is produced, which induces the transcription of genes to modulate the ER stress [32,33]. Second, at least in metazoans, PERK is activated by ER stress, and this leads to a broad and general downregulation of translation [34,35]. Finally, in Drosophila and mammals, ER stress activates the preferential degradation of mRNAs encoding proteins targeted to the ER, thereby downregulating the pool of newly incoming substrates for ER folding [36]. It is striking how the sensing of misfolded proteins in the ER triggers such a broad response in mRNA metabolism to compensate for this perturbation of proteostasis.
Effects of cytoplasmic Hsp70/Hsp40 complexes on mRNA decay
Hsp70 proteins also have been suggested to play a role in mRNA decay pathways. Decay in eukaryotes is initiated by deadenylases, which remove 3′ adenosines from the polyA tracts of mRNAs. Once these polyA tails are at a critical length, 5′ decapping of mRNAs becomes kinetically favored, closely followed by 5′ to 3′ degradation by exonucleases, primarily XRN1. Alternatively, mRNAs can also be degraded via 3′ to 5′ decay either by the exosome, or (in mammalian cells) by a second 3′ to 5′ exonuclease called Dis3L [37–39].
One possible connection of Hsp proteins to mRNA decay is that stress can inhibit mRNA degradation in both yeast and mammals, mostly through inhibiting deadenylation [38]. Interestingly, it appears that this effect on mRNA decay rates in yeast requires a very extreme heat shock, since stabilization of mRNAs in Saccharomyces cerevisiae was observed at 46°C [3] but not 36°C [40]. This stabilization was attributed to an inhibition of the two major deadenylase complexes, the CCR4-NOT complex consisting of Ccr4, Pop2 and the Not proteins and the PAN2/3, which consisits of the catalytic Pan2 subunit and Pan3 [2]. Whether the effects of stress on mRNA decay involves Hsp70 function has not been directly examined, although there is no global change in mRNA decay rates in Hsp70 defective yeast strains at 37°C [41], which suggests that Hsp70 function may not be critical to this effect on mRNA stability.
Inhibition of ARE-mediated decay: possibly through direct Hsp70:RNA interactions
Several observations have suggested that Hsp70 proteins can directly modulate the decay rates of a subset of ARE-containing mRNAs. AREs are AU-rich sequence elements in the 3′ UTR of many mRNAs that typically promote rapid deadenylation and subsequent decay [42]. Evidence that Hsp70 proteins modulate mRNA decay stimulated by AREs first came from studies showing that heat shock stabilized reporter mRNAs with an ARE [43]. This effect appears to be due to Hsp70 proteins interacting with AUF1, a key RNA binding protein that binds ARE elements and promotes their mRNA degradation. Defects in Hsp70 function in yeast were also observed to lead to selective stabilization of the MFA2 mRNA in a manner dependent on an AU-rich mRNA decay element considered functionally similar to AREs [41,44]. In the case of the yeast MFA2 mRNA, Hsp70 function was required for normal deadenylation of the mRNA [41]. Both of these examples argue that Hsp70 can have a selective effect on differential mRNA decay rates, but the mechanism of this effect is not understood.
Some studies raise the possibility that Hsp70 may impact ARE-mediated decay by directly binding RNA. In vitro, Hsp70 binds RNA and prefers AU-rich sequences [45,46], U-rich sequences [47], polyA stretches [46], specific ARE sequences from individual mRNAs (TNFα; [49]), or an entire 3′UTR containing multiple AU rich elements (Bim mRNA/Hsc70; [49]). Hsp70 binding to RNA is inhibited by ATP binding and is influenced by its peptide binding domain[45,6], arguing that RNA binding will be influenced by both substrate availability and the ATP state of Hsp70, which is influenced by Hsp40 and nucleotide exchange factors. In addition to these studies, several Hsp70 proteins were independently identified as efficiently cross-linking to mRNA in vivo [46]. Interestingly, in this same study several mammalian Hsp40 proteins were also identified as possible RNA binding proteins, consistent with both Hsp70 and Hsp40 proteins interacting with mRNAs in vivo. A key experiment to determine if Hsp70 binding to mRNAs directly modulates specific mRNA function will be to map the Hsp70-mRNA interactions on a genome wide scale and determine the functional impact of those specific interactions.
Inhibition of ARE-mediated decay: alteration of effector mRNP
Some evidence suggests that Hsp70 proteins modulate the availability and/or composition of key protein complexes that affect mRNA turnover rates. ARE-containing mRNAs are stabilized during heat shock [43], where Hsp70 expression is induced. It is thought that AUF1 binds to PABP, either removing it from the polyA tail, and/or recruiting deadenylase components, thus initiating decay [48]. Hsp70 may inhibit ARE-mediated decay as it disrupts the AUF1:PABP interaction in vitro [44], and is found in a complex with AUF1 and PABP with heat stress in vivo [43]. However, in vitro, it is principally the addition of an ARE-containing RNA that inhibits the AUF1:PABP interaction [48]. The presence of ARE-containing mRNAs in Hsp70:AUF1 complexes was not examined in vivo [43].
There is an additional example of Hsp70 proteins changing mRNP complexes in mammalian cells. During steady state conditions, a pro-apoptotic ARE-containing mRNA, Bim, is bound by Hsc70 via its 3′UTR [49]. With interleukin-3 treatment, the Bim mRNA is destabilized, concurrent with loss of Hsc70 binding to the mRNA and the formation of an Hsc70, Bag4 (a nucleotide exchange factor), and CHIP (an E3 ubiquitin ligase) complex [49]. In this manner, Hsc70 plays a role in stabilizing the mRNA and its removal may allow for the binding of other proteins that stimulate mRNA turnover.
Taken together, it is now clear that Hsp70 function can affect mRNP composition and the degradation of at least some mRNAs.
Hsp70/Hsp40, translational recovery from stress and clearance of stress granules
Several pieces of data argue that Hsp70/Hsp40 complexes play a role in helping translation recover from stress. First, knockdown of Hsp72 in mammalian cells prevented translation recovery after prolonged proteasomal inhibition [51]. Second, in Drosophila S2 cells, adding a pan-Hsp70 inhibitor during recovery from heat stress limited the reformation of polysomes [52]. Third, in human cell culture, Hsp70 overexpression rescues this translational repression caused by overexpression of the prion related domain (PRD) of the stress granule component TIA-1, a sequence specific RNA binding proteins [6]. Finally, yeast strains lacking the Ydj1 Hsp40 protein were shown to be compromised in translation recovery after NaN3 stress [14].
Additional evidence suggests that yeast Hsp70/Hsp40 complexes play roles in the disassembly and/or clearance of stress granules. Not only do stress granules contain Hsp70 and Hsp40 proteins [14], but stress granule clearance is reduced during stress recovery in both Hsp70 and Hsp40 deficient yeast strains [14].
An unresolved question is how stress granule dissipation directly relates to translation recovery. It appears clear that stress granule formation and translation are inversely correlated [4]. Moreover, stress granules contain mRNPs that resemble stalled translation complexes [53,54], and often loss of stress granules coincides with resumption of translation [14,51,55]. However, there are at least two situations where cells resume translation while maintaining stress granules [14,56]. One explanation is that there may be subclasses of stress granules, or components within individual stress granules, that have distinct fates: some will remodel to re-enter translation, and some will be trafficked to the vacuole for degradation [14].
Some evidence now argues that either different stress granule components, or different classes of stress granules, have different fates during stress granule recovery. First, during recovery from paraquat-induced stress granules in mammalian cells, HuR exits stress granules efficiently while TDP-43 remains present in granules for prolonged periods [58]. Second, while deletion of the yeast Hsp40 family members Ydj1 and Sis1 similarly caused stress granule persistence after stress, translation recovered in Sis1-depleted but not Ydj1-depleted cells after stress [14]. These results have been interpreted to suggest that Ydj1 affects stress granule disassembly in a manner that promotes mRNAs re-entering translation, while Sis1 targets them for degradation in the vacuole (Figure 2, Key Figure). Thus, subsets of stress granules, or their components, have alternative fates based on the function of specific Hsp40 proteins.
Figure 2. Key Figure.
Distinct Hsp70:Hsp40 pairings elicit different stress granule outcomes. Hsp40 member Ydj1 may resolve stress granules to facilitate translation recovery following stress (left). Conversely, Hsp40 Sis1 traffics stress granules to the vacuole for degradation.
Collectively, these observations suggest that, at least in some circumstances, Hsp70s are necessary for translation recovery and stress granule dissipation.
Concluding remarks
The observations discussed in this review highlight the multiple mechanisms by which cells couple the proteostatic state of the cell to modulate mRNA metabolism through Hsp70 family members. Hsp70 proteins can directly affect translation by regulating nascent polypeptide chain folding, which can feedback on translation elongation. At least in the lumen of the ER, an Hsp70 family member functions to limit a proteostatic stress signal transduction pathway. This example illustrates the principle that Hsp70 family members may play a broader role in controlling cell signaling in response to proteostatic stress. Finally, Hsp70 proteins have been described to affect both individual mRNP composition, as well as macromolecular mRNP stress granules. Important areas of future work will be to determine the specific mechanisms of these effects, and if other regulatory systems coupling proteostasis and ribostasis exist.
OUTSTANDING QUESTIONS.
Several significant questions about how proteostasis and ribostasis are coupled to allow optimal cellular function.
How do changes in proteostatic stress perturb Hsp70 action on specific client proteins and do cells use this mechanism to modulate key aspects of mRNA biogenesis and function during stress conditions?
What is the global nature and scope of Hsp70 RNA binding? A global analysis of Hsp70 RNA binding may yield important insights about the specificity and function of direct Hsp70:RNA interactions.
Are the functions of Hsp70/40 in translation recovery and stress granule clearance directly linked? A related issue is what the targets of Hsp70 activity within stress granules are. One intriguing possibility is that the high degree of intrinsically disordered protein domains in these assemblies creates a high local context of Hsp70/Hsp40 clients, which is important to disassemble to allow recovery from stress.
Given the importance of cellular homeostasis, are there other regulatory pathways that ensure that proteostasis and ribostasis are properly balanced?
TRENDS BOX.
The proper control of the proteome (proteostasis) and transcriptome (ribostasis) is critical to cell function.
By multiple mechanisms, Hsp70 family members sense perturbation of proteostasis and then modulate aspects of mRNA metabolism.
Hsp70 family members promote nascent protein folding and when defective this leads to an inhibition of translation elongation.
Hsp70 family members can be titrated by unfolded proteins leading to the activation of stress responsive signal transduction systems that modulate the transcriptome.
Hsp70 family members can modulate the protein composition of individual mRNPs, thereby affecting their function.
Hsp70 proteins promote disassembly of stress granules and are important for recovery of translation after stress.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–27. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 2.Hilgers V, et al. Translation-independent inhibition of mRNA deadenylation during stress in Saccharomyces cerevisiae. RNA. 2006;12:1835–45. doi: 10.1261/rna.241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gowrishankar G, et al. Inhibition of mRNA deadenylation and degradation by different types of cell stress. Biol Chem. 2006;387:323–7. doi: 10.1515/BC.2006.043. [DOI] [PubMed] [Google Scholar]
- 4.Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheth U, Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilks N, et al. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–98. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Decker CJ, et al. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007;179:437–49. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reijns MA, et al. A role for Q/N-rich aggregation-prone regions in P-body localization. J Cell Sci. 2008;121:2463–72. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato M, et al. Cell-free Formation of RNA Granules: Low Complexity Sequence Domains Form Dynamic Fibers within Hydrogels. Cell. 2012;149:753–67. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chappell TG, et al. Uncoating ATPase is a member of the 70 kilodalton family of stress proteins. Cell. 1986;45:3–13. doi: 10.1016/0092-8674(86)90532-5. [DOI] [PubMed] [Google Scholar]
- 11.Liao T, Tang L. The critical roles of HSC70 in physiological and pathological processes. Curr Pharm Des. 2014;20:101–7. doi: 10.2174/13816128113199990585. [DOI] [PubMed] [Google Scholar]
- 12.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–92. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ungewickell E, et al. Role of auxilin in uncoating clathrin-coated vesicles. Nature. 1995;378:632–5. doi: 10.1038/378632a0. [DOI] [PubMed] [Google Scholar]
- 14.Walters RW, et al. Differential effects of Ydj1 and Sis1 on Hsp70 mediated clearance of stress granules in Saccharomyces cerevisiae. RNA. 2015 doi: 10.1261/rna.053116.115. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minami Y, et al. Regulation of the heat-shock protein 70 reaction cycle by the mammalian DnaJ homolog Hsp40. J Biol Chem. 1996;271:19617–24. doi: 10.1074/jbc.271.32.19617. [DOI] [PubMed] [Google Scholar]
- 16.Horton LE, et al. The yeast hsp70 homologue Ssa is required for translation and interacts with Sis1 and Pab1 on translating ribosomes. J Biol Chem. 2001;276:14426–33. doi: 10.1074/jbc.M100266200. [DOI] [PubMed] [Google Scholar]
- 17.Zhong T, Arndt KT. The yeast SIS1 protein, a DnaJ homolog, is required for the initiation of translation. Cell. 1993;73:1175–86. doi: 10.1016/0092-8674(93)90646-8. [DOI] [PubMed] [Google Scholar]
- 18.Shalgi R, et al. Widespread Regulation of Translation by Elongation Pausing in Heat Shock. Mol Cell. 2013;49:439–52. doi: 10.1016/j.molcel.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B, et al. Cotranslational Response to Proteotoxic Stress by Elongation Pausing of Ribosomes. Mol Cell. 2013;49:453–63. doi: 10.1016/j.molcel.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson RJ, et al. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- 21.Pfund C, et al. The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome–nascent chain complex. EMBO J. 1998;17:3981–9. doi: 10.1093/emboj/17.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willmund F, et al. The Cotranslational Function of Ribosome-Associated Hsp70 in Eukaryotic Protein Homeostasis. Cell. 2007;152:196–209. doi: 10.1016/j.cell.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfund C, et al. Divergent functional properties of the ribosome-associated molecular chaperone Ssb compared with other Hsp70s. Mol Biol Cell. 2001;12:3773–82. doi: 10.1091/mbc.12.12.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frydman J, et al. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 1994;370:111–7. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- 25.Frydman J, Hartl FU. Principles of chaperone-assisted protein folding: differences between in vitro and in vivo mechanisms. Science. 1996;272:1497–502. doi: 10.1126/science.272.5267.1497. [DOI] [PubMed] [Google Scholar]
- 26.Villa N, et al. Human eukaryotic initiation factor 4G (eIF4G) protein binds to eIF3c,-d, and-e to promote mRNA recruitment to the ribosome. J Biol Chem. 2013;288:32932–40. doi: 10.1074/jbc.M113.517011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pestova TV, Kolupaeva VG. Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci USA. 2001;13:7029–36. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kahvejian A, et al. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–13. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuesta R, et al. Chaperone hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap-initiation complexes. Genes Dev. 2000;14:1460–70. [PMC free article] [PubMed] [Google Scholar]
- 30.Bertolotti A, et al. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–32. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 31.Ma K, et al. Dimerization and release of molecular chaperone inhibition facilitate activation of eukaryotic initiation factor-2 kinase in response to endoplasmic reticulum stress. J Biol Chem. 2002;277:18728–35. doi: 10.1074/jbc.M200903200. [DOI] [PubMed] [Google Scholar]
- 32.Aragon T, et al. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature. 2009;457:736–40. doi: 10.1038/nature07641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshida H, et al. XPB1 mRNA is induced by ATF6 and spliced by IRE1 in respone to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–91. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 34.Harding HP, et al. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–4. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 35.Wek RC, Cavener DR. Translational control and the unfolded protein response. Antioxid Redox Signal. 2007;12:2357–71. doi: 10.1089/ars.2007.1764. [DOI] [PubMed] [Google Scholar]
- 36.Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;5783:104–7. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 37.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–7. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 38.Garneau NL, et al. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–26. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 39.Malecki M, et al. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J. 2013;32:1842–54. doi: 10.1038/emboj.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herrick D, et al. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2269–84. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duttagupta R, et al. A yeast homologue of Hsp70, Ssa1p, regulates turnover of the MFA2 transcript through its AU-rich 3′ untranslated region. Mol Cell Biol. 2003;23:2623–32. doi: 10.1128/MCB.23.8.2623-2632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gratacós FM, Brewer G. The role of AUF1 in regulated mRNA decay. Wiley Interdiscip Rev RNA. 2011;1:457–73. doi: 10.1002/wrna.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laroia G, et al. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 44.Muhlrad D, Parker R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 1992;6:2100–11. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- 45.Henics T, et al. Mammalian Hsp70 and Hsp110 proteins bind to RNA motifs involved in mRNA stability. J Biol Chem. 1999;274:17318–24. doi: 10.1074/jbc.274.24.17318. [DOI] [PubMed] [Google Scholar]
- 46.Zimmer C, et al. Analysis of sequence-specific binding of RNA to Hsp70 and its various homologs indicates the involvement of N- and C-terminal interactions. RNA. 2001;7:1628–37. [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson GM, et al. Thermodynamics and kinetics of Hsp70 association with A + U-rich mRNA-destabilizing sequences. J Biol Chem. 2001;276:44450–6. doi: 10.1074/jbc.M108521200. [DOI] [PubMed] [Google Scholar]
- 48.Lu J, et al. Assembly of AUF1 with eIF4G-poly(A) binding protein complex suggests a translation function in AU-rich mRNA decay. RNA. 2006;12:883–93. doi: 10.1261/rna.2308106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsui H, et al. Cytokines direct the regulation of Bim mRNA stability by heat-shock cognate protein 70. Mol Cell. 2007;25:99–112. doi: 10.1016/j.molcel.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 50.Castello A, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 51.Mazroui R, et al. Inhibition of the ubiquitin-proteasome system induces stress granule formation. Mol Biol Cell. 2007;18:2603–18. doi: 10.1091/mbc.E06-12-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cherkasov V, et al. Coordination of translational control and protein homeostasis during severe heat stress. Curr Biol. 2013;23:2452–62. doi: 10.1016/j.cub.2013.09.058. [DOI] [PubMed] [Google Scholar]
- 53.Buchan JR, et al. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183:441–55. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buchan JR, et al. Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J Cell Sci. 2011;124:228–39. doi: 10.1242/jcs.078444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gallouzi I. Could stress granules be involved in age-related diseases? Aging (Albany NY) 2009;1:753–7. doi: 10.18632/aging.100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loschi M, et al. Dynein and kinesin regulate stress-granule and P-body dynamics. J Cell Sci. 2009;122:3973–82. doi: 10.1242/jcs.051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buchan JR, et al. Eukaryotic Stress Granules Are Cleared by Autophagy and Cdc48/VCP Function. Cell. 2013;153:1461–74. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parker SJ, et al. Endogenous TDP-43 localized to stress granules can subsequently form protein aggregates. Neurochem Int. 2012;60:415–24. doi: 10.1016/j.neuint.2012.01.019. [DOI] [PubMed] [Google Scholar]