Abstract
Pseudomonas Exotoxin A (PE) is the most toxic virulence factor of the pathogenic bacterium Pseudomonas aeruginosa. This review describes current knowledge about the intoxication pathways of PE. Moreover, PE represents a remarkable example for pathoadaptive evolution, how bacterial molecules have been structurally and functionally optimized under evolutionary pressure to effectively impair and kill their host cells.
Keywords: Pseudomonas aeruginosa, Pseudomonas Exotoxin A, virulence factor, ADP ribosylation, cytotoxic pathways, pathoadaptation
Introduction
Pseudomonas aeruginosa is a common Gram-negative, rod-shaped bacterium, which is optimally adapted in various environmental conditions. As an obligate respirer, it can use aerobic respiration as its optimal metabolism; however, it can also respire anaerobically on nitrate or other alternative electron acceptors (Su and Hassett, 2012). This is one reason, why P. aeruginosa is ubiquitously present in soil, water or sewage as well as in human, animal or plant hosts and why it is widespread around the world (Wiehlmann et al., 2007; Pirnay et al., 2009). Infection of healthy individuals by P. aeruginosa is very rare, but as an opportunistic bacterium it often colonizes immunocompromised patients with cystic fibrosis, burns, or AIDS (Gellatly and Hancock, 2013). The infections range from endophtalmitis, endocarditis, meningitis, and septicemia to chronic lung infections (Driscoll et al., 2007; Gomez and Prince, 2007; Gellatly and Hancock, 2013). Due to its inherent resistance to different antibiotics or chemotherapeutic agents, P. aeruginosa can only be eliminated with difficulty and leads to a high mortality rate (Maschmeyer and Braveny, 2000; Rowe et al., 2005).
A number of virulence factors enables P. aeruginosa to adhere to tissue surfaces, to damage tissue for dissemination and nutrition supply and to increase its survival rate (Coggan and Wolfgang, 2012; Jimenez et al., 2012; Balasubramanian et al., 2013). One of them is Pseudomonas Exotoxin A (PE), which has enzymatic activity and belongs to the mono-ADP-ribosyltransferase family (Liu, 1974). With regard to its function it is specified as NAD+-diphthamide-ADP-ribosyltransferase (EC 2.4.2.36) (Domenighini and Rappuoli, 1996). In the last years, the cytotoxic pathways of PE in eukaryotic host cells were investigated. Much relevant knowledge was obtained from studies with immunotoxins, in which the enzymatic active part of the toxin, coupled to antibodies, antibody fragments or ligands, was used for targeted therapeutic approaches against different cancers. Preclinical and clinical trials with PE-based immunotoxins were reviewed elsewhere (Wolf and Elsasser-Beile, 2009; Weidle et al., 2014). In the present article, we describe the cytotoxic pathways of PE (Figure 1) and how this molecule was structurally and functionally optimized under evolutionary pressure to effectively impair and finally kill its host cells.
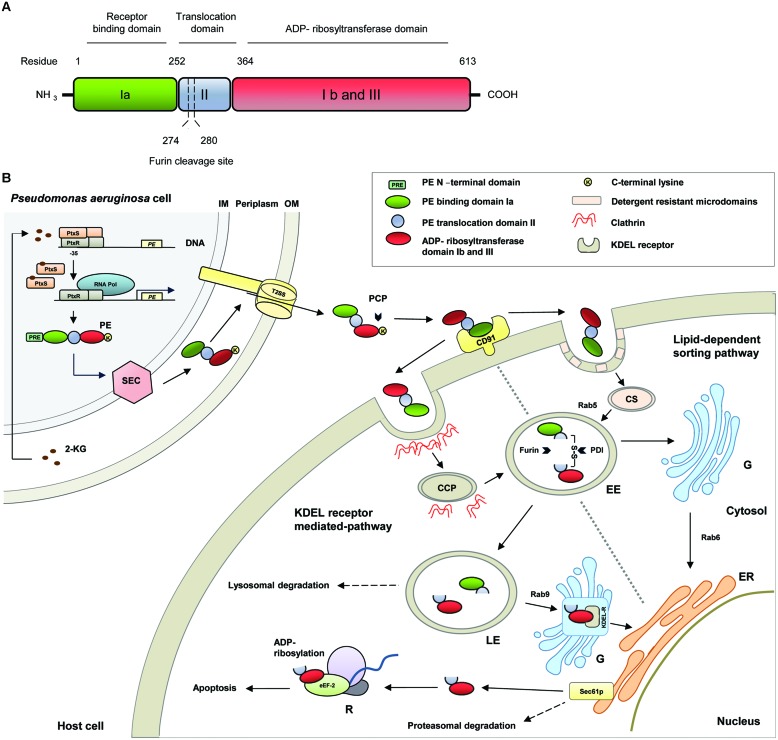
FIGURE 1.
(A) Schematic representation of the structural and functional domains of Pseudomonas Exotoxin A (PE). (B) Molecular pathways of PE. 2-KG, 2-ketogluconate; CCP, clathrin coated pit; CD91, CD91 receptor; CS, caveosome; EE, early endosome; eEF-2, eukaryotic elongation factor-2; ER, endoplasmatic reticulum; G, Golgi apparatus; KDEL-R, KDEL-receptor; PCP, plasma carboxypeptidases; PDI, protein disulfide isomerase; PtxR, PtxS, transcription regulators; R, ribosome; Rab, Rab-GTPase; RNA Pol, RNA polymerase; Sec61p, Sec61p translocon; T2SS, type II secretion system.
Pseudomonas Exotoxin A
Structure and Function
The PE gene was originally cloned from the P. aeruginosa strain PA 103 and analysis of the 5′ and 3′ flanking regions evidenced that the PE gene is translated from a monocystronic message (Gray et al., 1984). PE is expressed as a protein with a length of 638 amino acids (aa) and can be divided into several structural and functional domains (Wedekind et al., 2001; Figure 1A). Generally, PE belongs to the two-component AB toxin family, composed of an A domain with enzymatic activity and a B domain as cell binding subunit (Odumosu et al., 2010). In detail, PE contains a highly hydrophobic leader peptide of 25 aa at its N-terminus, which is removed during secretion. The leader sequence is followed by the receptor binding domain Ia (aa 1–252), which is composed of antiparallel ß-sheets. Domain II (aa 253–364) with six consecutive α-helices, enables the toxin to translocate across cell membranes. The last four residues (aa 400–404) of domain Ib (aa 365–404) together with domain III (aa 405–613) form the catalytic subunit of the toxin with ADP-ribosyltransferase activity (Siegall et al., 1989).
Molecular Pathways of Intoxication
The regulation of PE expression is complex and not fully understood to date. Different studies established a relation between PE expression and iron metabolism. The efficient uptake of iron is one important factor for P. aeruginosa allowing the colonization of the host. For this, the bacterium produces siderophores, such as pyoverdine, low-molecular weight excreted molecules that specifically chelate iron ions with high affinity. Interestingly, in the presence of iron ions, pyoverdine was found to activate a signaling pathway for the up-regulation of PE expression (Hunt et al., 2002; Lamont et al., 2002; Cornelis and Dingemans, 2013).
Recent data also suggest that there is a link to the bacterial glucose metabolism (Daddaoua et al., 2012, 2014). As a facultative aerobic organism, P. aeruginosa prefers respiration as metabolism. It gains energy by transferring electrons from glucose, a reduced substrate, to oxygen, the final electron acceptor. The initial step of glucose metabolism takes place in the periplasm and includes the oxidation of glucose to 2-ketogluconate, which enters the cytoplasm to be further metabolized. 2-ketogluconate is able to bind to the transcriptional repressor protein PtxS. In the absence of 2-ketogluconate, two PtxS molecules are bound to a dimer of the regulator PtxR, which again binds to the – 35 region to the PE promotor and inhibits the transcription of PE. After 2-ketogluconate binding, PtxS dissociates from the PtxR/DNA complex and PtxR can recruit RNA polymerase to facilitate the transcription of the toxin (Daddaoua et al., 2012, 2014; Figure 1B).
Pseudomonas Exotoxin A is secreted into the extracellular medium via the general secretory pathway, a two-step mechanism, which is highly conserved in Gram-negative bacteria (Voulhoux et al., 2000; Gerard-Vincent et al., 2002). After cytoplasmatic expression as an unfolded precursor protein, PE is initially transported to the periplasm using the Sec machinery (Douzi et al., 2012). During translocation through the inner membrane, the N-terminal signal peptide is cleaved off and PE is released into the periplasmatic space. In the hydrophilic environment of the periplasm, PE is folded to a mature conformational protein in a manner that can be recognized by the type II secretion system (T2SS), specifically called Xcp in P. aeruginosa, for secretion into the extracellular space (Voulhoux et al., 2000; Gerard-Vincent et al., 2002). Mutagenesis experiments gave evidence that two N-terminal glutamic acid residues at the +2 and +3 positions of domain Ia as well as domain II of PE are important for folding and extracellular secretion (Lu et al., 1993). It is therefore speculated that the corresponding residues are part of a still unknown conformational secretion signal of PE for recognition by T2SS or that they are important for the appropriate presentation of such a signal (Lu et al., 1993; Voulhoux et al., 2000).
Once secreted, the terminal lysine (aa 613) of PE can be cleaved from the toxin in the extracellular environment, presumably by plasma carboxypeptidases of the host. This leads to a formation of a C-terminal motif from REDLK (aa 609–613) to REDL (aa 609–612), which enables the toxin to bind to KDEL receptors at the Golgi apparatus during subsequent intracellular trafficking (Hessler and Kreitman, 1997). On the host cell surface, PE specifically binds via domain Ia to CD91, which is also known as alpha2-macroglobulin receptor/low-density lipoprotein receptor-related protein (α2MR/LRP; Kounnas et al., 1992). Then, there are two pathways open for PE to reach the Endoplasmatic Reticulum: the KDEL-receptor mediated pathway and the lipid-dependent sorting pathway.
KDEL-Receptor Mediated Pathway
CD91 bound PE molecules can be internalized via clathrin-coated pits. In the acidic early endosomal environment, PE dissociates from the CD91 receptor. Moreover, it undergoes a conformational change, which makes the furin-cleavable motif within domain II (aa 274–280, RHRQPRG) accessible. The protease furin cleaves PE between the residues R-279 and G-280, in two PE fragments. The first fragment (aa 1–279) of about 28 kDa in weight consists of domain I and parts of domain II. The second one (aa 280–613) of about 37 kDa contains parts of domain II, domains Ib, and domain III and holds the ADP-ribosylation activity (Ogata et al., 1992; Wedekind et al., 2001). After furin cleavage both fragments are still connected by a disulfide bond between C-265 and C-287, which encompasses the furin cleavage site. There is evidence that there is an unfolding event, possibly under the influence of chaperones, which leads to a surface exposure of the disulfide bond. The disulfide bond is then reduced, presumably by protein-disulfide-isomerases, and the 37 kDa fragment is detached (McKee and FitzGerald, 1999). After cleavage and transport into late endosomes, the 37 kDa PE fragment exploits a Rab9-regulated pathway to reach the trans Golgi network (TGN). Rab proteins are highly compartmentalized GTPases in organelle membranes. They coordinate consecutive stages of intracellular transport, such as vesicle formation and motility, or tethering of vesicles to their target membranes (Zerial and McBride, 2001). On the TGN, the C-terminal REDL motif of PE (aa 609–612) binds to the KDEL receptor and the toxin is transported to the ER in a retrograde manner (Kreitman and Pastan, 1995; Jackson et al., 1999). The KDEL-receptor cycles between the TGN and the ER via Golgi cisternae and is originally responsible for the recycling of cellular proteins bearing KDEL or KDEL-like sequences (Cancino et al., 2013).
Lipid-Dependent Sorting Pathway
Pseudomonas Exotoxin A can also use the lipid-dependent sorting pathway to reach the ER. In this pathway, CD91 bound PE associates with detergent-resistant microdomains (DRM), which facilitates the cellular uptake of the toxin-receptor complex via caveolin-mediated internalization. The receptor-toxin complex is then transported via caveosomes into early endosomes (EE) in a Rab5-dependent manner (Smith et al., 2006). After cleavage in the EE, the 37 kDa PE fragment can reach the TGN by a pathway, which was shown to be independent from Rab9. Then a Rab6-controlled lipid-dependent sorting pathway is used for trafficking to the ER (White et al., 1999; Smith et al., 2006).
ER-Associated Protein Degradation Pathway
Pseudomonas Exotoxin A uses the cellular ER-associated protein degradation pathway (ERAD) to get from the ER into the cytosol (Ogata et al., 1990; Theuer et al., 1993). Sequences inside the PE-domain II induce the translocation of the 37 kDa fragment via the Sec61p translocon, which normally serves as a channel to dislocate unfolded or misfolded proteins for subsequent proteasomal degradation (Hazes and Read, 1997; Koopmann et al., 2000).
ADP-Ribosylation of eEF-2
In the cytosol the 37 kDa PE fragment exerts its enzymatic activity and ADP-ribosylates the eukaryotic elongation factor-2 (eEF-2) on the ribosomes (Iglewski et al., 1977). eEF-2 belongs to the GTP-binding translation elongation factor family and promotes the GTP-dependent translocation of mRNA from the ribosomal A-site to the P-site (Proud, 1994).
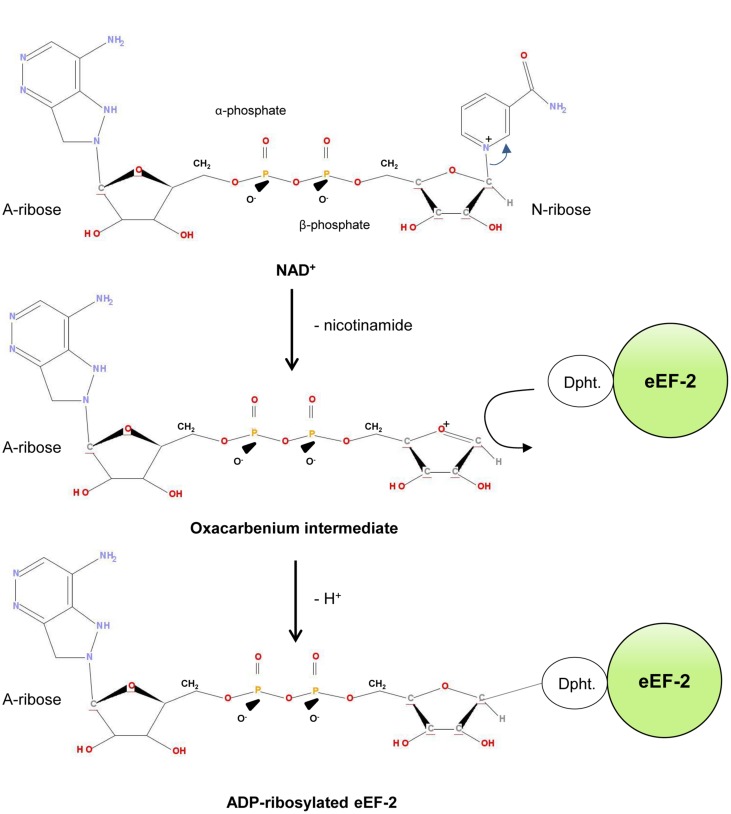
The ADP-ribosylation mechanism of PE was studied in detail and it turned out that it follows an SN1 nucleophilic substitution mechanism (Beattie et al., 1996; Armstrong et al., 2002; Jorgensen et al., 2005; Figure 2). Initially, the PE-fragment binds to NAD+ and interacts via the so-called “active-site loop L4” (aa 483–490 of domain III) with eEF-2 (Yates and Merrill, 2004). Afterward it facilicates the cleavage of the glycosidic bond (C-N) between the nicotinamide and N-ribose of NAD+. This results in a reactive oxacarbenium intermediate, which in turn is stabilized by residue E-553 of the PE-fragment (Li et al., 1996; Jorgensen et al., 2005). This step is followed by a nucleophilic attack of eEF-2, based on its nucleophilic residue diphthamide, a post-translationally modified histidine residue (2-(3-carboxyamido-3-[trimethylammonio]propyl) histidine) (Ortiz and Kinzy, 2005). The ADP-ribose group is subsequently transferred to the N3 atom of the diphthamide imidazole ring, which results in the ADP-ribosylated eEF-2 protein (Armstrong et al., 2002; Jorgensen et al., 2005).
FIGURE 2.
ADP-ribosylation of eEF-2. Dipth, diphthamide.
The ADP-ribosylation inactivates eEF-2 and the protein biosynthesis of the host cell comes to a standstill. As a consequence, apoptosis is induced and the host cell irreversibly dies. In cervix carcinoma cells, a decrease of cdc2 and cyclin B expression as well as an increase of the regulator protein 14-3-3 delta was observed. This suggests that PE induces cell cycle arrest, which is followed by apoptosis (Chang and Kwon, 2007). In mouse embryo fibroblasts, the regulation of pro- and anti-apoptotic proteins after PE intoxication was examined. In these cells, a rapid degradation of Mcl-1 was observed, which unleashed Bak to activate apoptosis (Du et al., 2010). In human mast cells, PE provoked the activation of caspase-8 and the down-regluation of FLIPs (Fas-associated death domain protein (FADD)-like interleukin-1β-converting enzyme (FLICE) (Caspase-8) inhibitory protein), giving evidence that PE can also activate the extrinsic apoptotic pathway (Jenkins et al., 2004).
The intoxication of PE only takes a short time. In studies, which examined PE uptake into rat liver, a rapid association of PE with plasma membranes after 5–30 min, an internalization within endosomes after 15–60 min, and a translocation into the cytosolic compartment after 30–90 min was measured (El Hage et al., 2010).
The intoxication pathways of PE are not fully elucidated yet. There is evidence that there is a further processing pathway for internalized PE, involving endosomal cathepsins B and D, resulting in a production of PE fragments that may contribute to cytotoxicity (El Hage et al., 2010). Moreover, genome-wide genetic screening identified hitherto unknown host factors for intracellular trafficking. A prime example is GPR107, an orphan G-protein coupled receptor, which, like the KDEL receptor, is located to the TGN and facilitates the retrograde transport of PE (Tafesse et al., 2014). Since there are differences of PE trafficking in different cell lines, it is presumed that the choice between the pathways seems to be dependent from the expression of host factors that are present in the cells.
Evolutionary Aspects of Intoxication
Bacteria and their hosts have coexisted for several millions of years. Over this time, bacteria developed a wide spectrum of adaptation to optimize infection and survival. One important mechanism P. aeruginosa developed, is the quorum sensing (QS) for intercellular communication. QS allows the bacteria to recognize the population density by sensing and measuring the accumulation of specific small signaling molecules that are secreted by the members of the colony. The bacteria now act as a community to perform tasks, which would be impossible for individual cells, e.g., cooperative activation of bacterial gene expression, biofilm formation, influence on the behavior of host cells, or the adequate production of virulence factors (Nguyen and Singh, 2006; Holm and Vikstrom, 2014).
Moreover, genome sequencing of bacterial pathogens and molecular analyses of intoxication pathways have shown how bacteria evolved via mutational changes, a mechanism, which is known as pathoadaptation.
Interestingly, the pathoadaptation of P. aeruginosa is exemplarily reflected in its virulence factor PE, which was structurally and functionally optimized especially in view of binding, processing, routing and toxicity (Marvig et al., 2015).
Both termini of the PE protein were formed for effective binding of target molecules. The N-terminus is able to specifically bind to the abundantly expressed CD91 antigen, which enables PE to reach many different host cells (Kounnas et al., 1992). The-C-terminus, containing the KDEL-like sequence, facilitates the retrograde transport of PE to the ER by binding to the KDEL-receptor (Kreitman and Pastan, 1995; Jackson et al., 1999).
Pseudomonas Exotoxin A developed specific aa motifs to be effectively processed by components of the host cell. For example, the C-terminus can be cleaved by plasma carboxypeptidases to form the KDEL-like sequence for subsequent intracellular trafficking (Hessler and Kreitman, 1997). Moreover, the molecule can be cleaved by furin, presumably to facilitate subsequent trafficking. Interestingly, the unfolding step in the EE for furin cleavage is also discussed to lead to a masquerade of the PE molecule as an unfolded/misfolded protein to be successfully transported to the cytosol (Pelham et al., 1992). PE is also able to exploit different intracellular routes controlled by both protein- and lipid-sorting signals. Especially, routing via caveasomes and the lipid-dependent pathway may contribute to protect the PE molecules against lysosomal degradation (Smith et al., 2006).
The toxicity of PE is marked by an induction of apoptosis in the host cells by specifically ADP-ribosylating the residue diphthamide. Diphthamide is named on the basis of the fact that it is also the target of Diphteria toxin produced by Corynebacterium diphteriae (Abdel-Fattah et al., 2013). Diphthamide is highly conserved among archaea and eukaryotes and was exclusively described in eEF-2 (Ortiz and Kinzy, 2005; Su et al., 2013). It represents an “Achilles heel” of the host cell, since its modification (ADP-ribosylation) can lead to the complete inhibition of protein biosynthesis and induction of programmed death.
Even the ADP-ribosylation mechanism of PE represents a good example for the evolutionary adaption of PE. X-ray structure analyses showed evidence that PE mimics the normal interaction between eEF-2 and the eukaryotic 80S ribosome, because a striking similarity was observed between the orientation of PE-bound ß-TAD (a non-hydrolysable NAD+ analog) and the phosphate backbone of two nucleotides in a conformational switch of 18S rRNA, with respect to the interaction with eEF-2. By optimizing this mimicry during evolution, PE minimizes the probability that the target organism could evolve resistance toward the invading toxin, because coordinated mutations in regions of eEF2 and the ribosome would be required that are crucial for function (Jorgensen et al., 2005).
Pseudomonas Exotoxin A evolved into a highly specific and toxic molecule; however, the optimization process seems to go on. Structural data suggest that there is no high-valency binding of PE to its receptor (Wedekind et al., 2001). Furthermore, lack of basic residues at the -2 aa position of the furin cleavage site could lead to less rapid cleavage by furin than proteins having the typical RX (K/R) R sequence (Gordon and Leppla, 1994). Moreover, many PE molecules are degraded in lysosomes and therefore it is necessary for the toxin to be in a sufficient concentration in the extracellular space for effective killing (Hessler and Kreitman, 1997).
Taken together, PE represents a remarkable molecule, which provides deep insight into pathoadaptive processes. Knowledge about its intoxication pathways should not only be used for the construction of PE-based immunotoxins for targeted therapy (Wolf and Elsasser-Beile, 2009; Weidle et al., 2014), but also for the development of strategies to alleviate P. aeruginosa infections (Jiang et al., 2014; Farajnia et al., 2015).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Abdel-Fattah W., Scheidt V., Uthman S., Stark M. J., Schaffrath R. (2013). Insights into diphthamide, key diphtheria toxin effector. Toxins (Basel) 5 958–968. 10.3390/toxins5050958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong S., Li J. H., Zhang J., Merrill A. R. (2002). Characterization of competitive inhibitors for the transferase activity of Pseudomonas aeruginosa exotoxin A. J. Enzyme Inhib. Med. Chem. 17 235–246. 10.1080/1475636021000010914 [DOI] [PubMed] [Google Scholar]
- Balasubramanian D., Schneper L., Kumari H., Mathee K. (2013). A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res. 41 1–20. 10.1093/nar/gks1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie B. K., Prentice G. A., Merrill A. R. (1996). Investigation into the catalytic role for the tryptophan residues within domain III of Pseudomonas aeruginosa exotoxin A. Biochemistry 35 15134–15142. 10.1021/bi961985t [DOI] [PubMed] [Google Scholar]
- Cancino J., Jung J. E., Luini A. (2013). Regulation of Golgi signaling and trafficking by the KDEL receptor. Histochem. Cell Biol. 140 395–405. 10.1007/s00418-013-1130-9 [DOI] [PubMed] [Google Scholar]
- Chang J. H., Kwon H. Y. (2007). Expression of 14-3-3delta, cdc2 and cyclin B proteins related to exotoxin A-induced apoptosis in HeLa S3 cells. Int. Immunopharmacol. 7 1185–1191. 10.1016/j.intimp.2007.05.001 [DOI] [PubMed] [Google Scholar]
- Coggan K. A., Wolfgang M. C. (2012). Global regulatory pathways and cross-talk control pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr. Issues Mol. Biol. 14 47–70. [PMC free article] [PubMed] [Google Scholar]
- Cornelis P., Dingemans J. (2013). Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front. Cell. Infect. Microbiol. 3:75 10.3389/fcimb.2013.00075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daddaoua A., Fillet S., Fernandez M., Udaondo Z., Krell T., Ramos J. L. (2012). Genes for carbon metabolism and the ToxA virulence factor in Pseudomonas aeruginosa are regulated through molecular interactions of PtxR and PtxS. PLoS ONE 7:e39390 10.1371/journal.pone.0039390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daddaoua A., Molina-Santiago C., de la Torre J., Krell T., Ramos J. L. (2014). GtrS and GltR form a two-component system: the central role of 2-ketogluconate in the expression of exotoxin A and glucose catabolic enzymes in Pseudomonas aeruginosa. Nucleic Acids Res. 42 7654–7663. 10.1093/nar/gku496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenighini M., Rappuoli R. (1996). Three conserved consensus sequences identify the NAD-binding site of ADP-ribosylating enzymes, expressed by eukaryotes, bacteria and T-even bacteriophages. Mol. Microbiol. 21 667–674. 10.1046/j.1365-2958.1996.321396.x [DOI] [PubMed] [Google Scholar]
- Douzi B., Filloux A., Voulhoux R. (2012). On the path to uncover the bacterial type II secretion system. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367 1059–1072. 10.1098/rstb.2011.0204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll J. A., Brody S. L., Kollef M. H. (2007). The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67 351–368. 10.2165/00003495-200767030-00003 [DOI] [PubMed] [Google Scholar]
- Du X., Youle R. J., FitzGerald D. J., Pastan I. (2010). Pseudomonas exotoxin A-mediated apoptosis is Bak dependent and preceded by the degradation of Mcl-1. Mol. Cell. Biol. 30 3444–3452. 10.1128/MCB.00813-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage T., Lorin S., Decottignies P., Djavaheri-Mergny M., Authier F. (2010). Proteolysis of Pseudomonas exotoxin A within hepatic endosomes by cathepsins B and D produces fragments displaying in vitro ADP-ribosylating and apoptotic effects. FEBS J. 277 3735–3749. 10.1111/j.1742-4658.2010.07775.x [DOI] [PubMed] [Google Scholar]
- Farajnia S., Peerayeh S. N., Tanomand A., Majidi J., Goudarzi G., Naghili B., et al. (2015). Protective efficacy of recombinant exotoxin A – flagellin fusion protein against Pseudomonas aeruginosa infection. Can. J. Microbiol. 61 60–64. 10.1139/cjm-2014-0501 [DOI] [PubMed] [Google Scholar]
- Gellatly S. L., Hancock R. E. (2013). Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog. Dis. 67 159–173. 10.1111/2049-632X.12033 [DOI] [PubMed] [Google Scholar]
- Gerard-Vincent M., Robert V., Ball G., Bleves S., Michel G. P., Lazdunski A., et al. (2002). Identification of XcpP domains that confer functionality and specificity to the Pseudomonas aeruginosa type II secretion apparatus. Mol. Microbiol. 44 1651–1665. 10.1046/j.1365-2958.2002.02991.x [DOI] [PubMed] [Google Scholar]
- Gomez M. I., Prince A. (2007). Opportunistic infections in lung disease: Pseudomonas infections in cystic fibrosis. Curr. Opin. Pharmacol. 7 244–251. 10.1016/j.coph.2006.12.005 [DOI] [PubMed] [Google Scholar]
- Gordon V. M., Leppla S. H. (1994). Proteolytic activation of bacterial toxins: role of bacterial and host cell proteases. Infect. Immun. 62 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray G. L., Smith D. H., Baldridge J. S., Harkins R. N., Vasil M. L., Chen E. Y., et al. (1984). Cloning, nucleotide sequence, and expression in Escherichia coli of the exotoxin A structural gene of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 81 2645–2649. 10.1073/pnas.81.9.2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazes B., Read R. J. (1997). Accumulating evidence suggests that several AB-toxins subvert the endoplasmic reticulum-associated protein degradation pathway to enter target cells. Biochemistry 36 11051–11054. 10.1021/bi971383p [DOI] [PubMed] [Google Scholar]
- Hessler J. L., Kreitman R. J. (1997). An early step in Pseudomonas exotoxin action is removal of the terminal lysine residue, which allows binding to the KDEL receptor. Biochemistry 36 14577–14582. 10.1021/bi971447w [DOI] [PubMed] [Google Scholar]
- Holm A., Vikstrom E. (2014). Quorum sensing communication between bacteria and human cells: signals, targets, and functions. Front. Plant Sci. 5:309 10.3389/fpls.2014.00309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt T. A., Peng W. T., Loubens I., Storey D. G. (2002). The Pseudomonas aeruginosa alternative sigma factor PvdS controls exotoxin A expression and is expressed in lung infections associated with cystic fibrosis. Microbiology 148 3183–3193. 10.1099/00221287-148-10-3183 [DOI] [PubMed] [Google Scholar]
- Iglewski B. H., Liu P. V., Kabat D. (1977). Mechanism of action of Pseudomonas aeruginosa exotoxin Aiadenosine diphosphate-ribosylation of mammalian elongation factor 2 in vitro and in vivo. Infect. Immun. 15 138–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M. E., Simpson J. C., Girod A., Pepperkok R., Roberts L. M., Lord J. M. (1999). The KDEL retrieval system is exploited by Pseudomonas exotoxin A, but not by Shiga-like toxin-1, during retrograde transport from the Golgi complex to the endoplasmic reticulum. J. Cell Sci. 112(Pt 4), 467–475. [DOI] [PubMed] [Google Scholar]
- Jenkins C. E., Swiatoniowski A., Issekutz A. C., Lin T. J. (2004). Pseudomonas aeruginosa exotoxin A induces human mast cell apoptosis by a caspase-8 and -3-dependent mechanism. J. Biol. Chem. 279 37201–37207. 10.1074/jbc.M405594200 [DOI] [PubMed] [Google Scholar]
- Jiang M., Yao J., Feng G. (2014). Protective effect of DNA vaccine encoding pseudomonas exotoxin A and PcrV against acute pulmonary P. aeruginosa Infection. PLoS ONE 9:e96609 10.1371/journal.pone.0096609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez P. N., Koch G., Thompson J. A., Xavier K. B., Cool R. H., Quax W. J. (2012). The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 76 46–65. 10.1128/MMBR.05007-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R., Merrill A. R., Yates S. P., Marquez V. E., Schwan A. L., Boesen T., et al. (2005). Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry. Nature 436 979–984. 10.1038/nature03871 [DOI] [PubMed] [Google Scholar]
- Koopmann J. O., Albring J., Huter E., Bulbuc N., Spee P., Neefjes J., et al. (2000). Export of antigenic peptides from the endoplasmic reticulum intersects with retrograde protein translocation through the Sec61p channel. Immunity 13 117–127. 10.1016/S1074-7613(00)00013-3 [DOI] [PubMed] [Google Scholar]
- Kounnas M. Z., Morris R. E., Thompson M. R., FitzGerald D. J., Strickland D. K., Saelinger C. B. (1992). The alpha 2-macroglobulin receptor/low density lipoprotein receptor-related protein binds and internalizes Pseudomonas exotoxin A. J. Biol. Chem. 267 12420–12423. [PubMed] [Google Scholar]
- Kreitman R. J., Pastan I. (1995). Importance of the glutamate residue of KDEL in increasing the cytotoxicity of Pseudomonas exotoxin derivatives and for increased binding to the KDEL receptor. Biochem. J. 307(Pt 1), 29–37. 10.1042/bj3070029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont I. L., Beare P. A., Ochsner U., Vasil A. I., Vasil M. L. (2002). Siderophore-mediated signaling regulates virulence factor production in Pseudomonasaeruginosa. Proc. Natl. Acad. Sci. U.S.A. 99 7072–7077. 10.1073/pnas.092016999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Dyda F., Benhar I., Pastan I., Davies D. R. (1996). Crystal structure of the catalytic domain of Pseudomonas exotoxin A complexed with a nicotinamide adenine dinucleotide analog: implications for the activation process and for ADP ribosylation. Proc. Natl. Acad. Sci. U.S.A. 93 6902–6906. 10.1073/pnas.93.14.6902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P. V. (1974). Extracellular toxins of Pseudomonas aeruginosa. J. Infect. Dis. 130(Suppl.), S94–S99. 10.1093/infdis/130.Supplement.S94 [DOI] [PubMed] [Google Scholar]
- Lu H. M., Mizushima S., Lory S. (1993). A periplasmic intermediate in the extracellular secretion pathway of Pseudomonas aeruginosa exotoxin A. J. Bacteriol. 175 7463–7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvig R. L., Sommer L. M., Molin S., Johansen H. K. (2015). Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat. Genet. 47 57–64. 10.1038/ng.3148 [DOI] [PubMed] [Google Scholar]
- Maschmeyer G., Braveny I. (2000). Review of the incidence and prognosis of Pseudomonas aeruginosa infections in cancer patients in the 1990s. Eur. J. Clin. Microbiol. Infect. Dis. 19 915–925. 10.1007/s100960000410 [DOI] [PubMed] [Google Scholar]
- McKee M. L., FitzGerald D. J. (1999). Reduction of furin-nicked Pseudomonas exotoxin A: an unfolding story. Biochemistry 38 16507–16513. 10.1021/bi991308+ [DOI] [PubMed] [Google Scholar]
- Nguyen D., Singh P. K. (2006). Evolving stealth: genetic adaptation of Pseudomonas aeruginosa during cystic fibrosis infections. Proc. Natl. Acad. Sci. U.S.A. 103 8305–8306. 10.1073/pnas.0602526103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odumosu O., Nicholas D., Yano H., Langridge W. (2010). AB toxins: a paradigm switch from deadly to desirable. Toxins (Basel) 2 1612–1645. 10.3390/toxins2071612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M., Chaudhary V. K., Pastan I., FitzGerald D. J. (1990). Processing of Pseudomonas exotoxin by a cellular protease results in the generation of a 37,000-Da toxin fragment that is translocated to the cytosol. J. Biol. Chem. 265 20678–20685. [PubMed] [Google Scholar]
- Ogata M., Fryling C. M., Pastan I., FitzGerald D. J. (1992). Cell-mediated cleavage of Pseudomonas exotoxin between Arg279 and Gly280 generates the enzymatically active fragment which translocates to the cytosol. J. Biol. Chem. 267 25396–25401. [PubMed] [Google Scholar]
- Ortiz P. A., Kinzy T. G. (2005). Dominant-negative mutant phenotypes and the regulation of translation elongation factor 2 levels in yeast. Nucleic Acids Res. 33 5740–5748. 10.1093/nar/gki882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H. R., Roberts L. M., Lord J. M. (1992). Toxin entry: how reversible is the secretory pathway? Trends Cell Biol. 2 183–185. 10.1016/0962-8924(92)90230-K [DOI] [PubMed] [Google Scholar]
- Pirnay J. P., Bilocq F., Pot B., Cornelis P., Zizi M., Van Eldere J., et al. (2009). Pseudomonas aeruginosa population structure revisited. PLoS ONE 4:e7740 10.1371/journal.pone.0007740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud C. G. (1994). Peptide-chain elongation in eukaryotes. Mol. Biol. Rep. 19 161–170. 10.1007/BF00986958 [DOI] [PubMed] [Google Scholar]
- Rowe S. M., Miller S., Sorscher E. J. (2005). Cystic fibrosis. N. Engl. J. Med. 352 1992–2001. 10.1056/NEJMra043184 [DOI] [PubMed] [Google Scholar]
- Siegall C. B., Chaudhary V. K., FitzGerald D. J., Pastan I. (1989). Functional analysis of domains II, Ib, and III of Pseudomonas exotoxin. J. Biol. Chem. 264 14256–14261. [PubMed] [Google Scholar]
- Smith D. C., Spooner R. A., Watson P. D., Murray J. L., Hodge T. W., Amessou M., et al. (2006). Internalized Pseudomonas exotoxin A can exploit multiple pathways to reach the endoplasmic reticulum. Traffic 7 379–393. 10.1111/j.1600-0854.2006.00391.x [DOI] [PubMed] [Google Scholar]
- Su S., Hassett D. J. (2012). Anaerobic Pseudomonas aeruginosa and other obligately anaerobic bacterial biofilms growing in the thick airway mucus of chronically infected cystic fibrosis patients: an emerging paradigm or “Old Hat?” Expert Opin. Ther. Targets 16 859–873. 10.1517/14728222.2012.708025 [DOI] [PubMed] [Google Scholar]
- Su X., Lin Z., Lin H. (2013). The biosynthesis and biological function of diphthamide. Crit. Rev. Biochem. Mol. Biol. 48 515–521. 10.3109/10409238.2013.831023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafesse F. G., Guimaraes C. P., Maruyama T., Carette J. E., Lory S., Brummelkamp T. R., et al. (2014). GPR107, a G-protein-coupled receptor essential for intoxication by Pseudomonas aeruginosa exotoxin A, localizes to the Golgi and is cleaved by furin. J. Biol. Chem. 289 24005–24018. 10.1074/jbc.M114.589275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theuer C. P., Buchner J., FitzGerald D., Pastan I. (1993). The N-terminal region of the 37-kDa translocated fragment of Pseudomonas exotoxin A aborts translocation by promoting its own export after microsomal membrane insertion. Proc. Natl. Acad. Sci. U.S.A. 90 7774–7778. 10.1073/pnas.90.16.7774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voulhoux R., Taupiac M. P., Czjzek M., Beaumelle B., Filloux A. (2000). Influence of deletions within domain II of exotoxin A on its extracellular secretion from Pseudomonas aeruginosa. J. Bacteriol. 182 4051–4058. 10.1128/JB.182.14.4051-4058.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedekind J. E., Trame C. B., Dorywalska M., Koehl P., Raschke T. M., McKee M., et al. (2001). Refined crystallographic structure of Pseudomonas aeruginosa exotoxin A and its implications for the molecular mechanism of toxicity. J. Mol. Biol. 314 823–837. 10.1006/jmbi.2001.5195 [DOI] [PubMed] [Google Scholar]
- Weidle U. H., Tiefenthaler G., Schiller C., Weiss E. H., Georges G., Brinkmann U. (2014). Prospects of bacterial and plant protein-based immunotoxins for treatment of cancer. Cancer Genomics Proteomics 11 25–38. [PubMed] [Google Scholar]
- White J., Johannes L., Mallard F., Girod A., Grill S., Reinsch S., et al. (1999). Rab6 coordinates a novel Golgi to ER retrograde transport pathway in live cells. J. Cell Biol. 147 743–760. 10.1083/jcb.147.4.743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiehlmann L., Wagner G., Cramer N., Siebert B., Gudowius P., Morales G., et al. (2007). Population structure of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 104 8101–8106. 10.1073/pnas.0609213104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf P., Elsasser-Beile U. (2009). Pseudomonas exotoxin A: from virulence factor to anti-cancer agent. Int. J. Med. Microbiol. 299 161–176. 10.1016/j.ijmm.2008.08.003 [DOI] [PubMed] [Google Scholar]
- Yates S. P., Merrill A. R. (2004). Elucidation of eukaryotic elongation factor-2 contact sites within the catalytic domain of Pseudomonas aeruginosa exotoxin A. Biochem. J. 379 563–572. 10.1042/BJ20031731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M., McBride H. (2001). Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2 107–117. 10.1038/35052055 [DOI] [PubMed] [Google Scholar]