Abstract
In this study we characterized and sequenced the genome of Arcobacter anaerophilus strain IR-1 isolated from enrichment cultures used in nitrate-amended corrosion experiments. A. anaerophilus IR-1 could grow lithoautotrophically on hydrogen and hydrogen sulfide and lithoheterothrophically on thiosulfate and elemental sulfur. In addition, the strain grew organoheterotrophically on yeast extract, peptone, and various organic acids. We show for the first time that Arcobacter could grow on the complex organic substrate tryptone and oxidize acetate with elemental sulfur as electron acceptor. Electron acceptors utilized by most Epsilonproteobacteria, such as oxygen, nitrate, and sulfur, were also used by A. anaerophilus IR-1. Strain IR-1 was also uniquely able to use iron citrate as electron acceptor. Comparative genomics of the Arcobacter strains A. butzleri RM4018, A. nitrofigilis CI and A. anaerophilus IR-1 revealed that the free-living strains had a wider metabolic range and more genes in common compared to the pathogen strain. The presence of genes for NAD+-reducing hydrogenase (hox) and dissimilatory iron reduction (fre) were unique for A. anaerophilus IR-1 among Epsilonproteobacteria. Finally, the new strain had an incomplete denitrification pathway where the end product was nitrite, which is different from other Arcobacter strains where the end product is ammonia. Altogether, our study shows that traditional characterization in combination with a modern genomics approach can expand our knowledge on free-living Arcobacter, and that this complementary approach could also provide invaluable knowledge about the physiology and metabolic pathways in other Epsilonproteobacteria from various environments.
Keywords: Arcobacter, Epsilonproteobacteria, genomics, metabolism, hox hydrogenase, ferric citrate reduction
Introduction
Free-living, environmental Epsilonproteobacteria are found to be ubiquitous in marine and terrestrial habitats, such as hydrothermal systems, marine sediments, pelagic seawater, acid mine drainage, lakes, springs, sulfidic caves and hydrocarbon-rich ground water (Campbell et al., 2006; Nakagawa and Takaki, 2009). They are in general associated with sulfide rich environments where they play a key role in the cycling of carbon, nitrogen, and sulfur (Campbell et al., 2006). Epsilonproteobacteria may also facilitate the colonization of other microbial groups due to detoxification of sulfur species, such as hydrogen sulfide (Campbell et al., 2006). Modern meta-omics technologies have revealed a wide distribution of free-living Epsilonproteobacteria (Huber et al., 2010; Akerman et al., 2013; Headd and Engel, 2014; Urich et al., 2014; Pop Ristova et al., 2015; Suh et al., 2015) and provided knowledge about their in situ metabolism (Dahle et al., 2013; Urich et al., 2014), however, there are relatively few cultivated representatives within this group. Isolates are obtained within the genera Sulfurovum (Inagaki et al., 2004; Mino et al., 2014), Sulfurospirillum (Finster et al., 1997; Stolz et al., 1999; Luijten et al., 2003; Kodama et al., 2007), Nautiliales (Alain et al., 2002; Takai et al., 2005; Grosche et al., 2015), Nitratifractor (Nakagawa et al., 2005b), Sulfuricurvum (Kodama and Watanabe, 2004), Sulfurimonas (Inagaki et al., 2003; Takai et al., 2006; Labrenz et al., 2013), Thiomicrospira (Brinkhoff et al., 1999; Knittel et al., 2005; Sorokin et al., 2006), Thioreductor (Nakagawa et al., 2005a) and Arcobacter (Donachie et al., 2005; Collado et al., 2009; Kim et al., 2010). Many of the free-living Epsilonproteobacteria associated with hydrothermal vents can perform oxidation of hydrogen or sulfur species (e.g., elemental sulfur, thiosulfate and hydrogen sulfide) with reduction of elemental sulfur, sulfite, thiosulfate, nitrate or low concentrations of oxygen (Alain et al., 2002; Miroshnichenko et al., 2002; Inagaki et al., 2003, 2004; Kodama and Watanabe, 2004; Nakagawa et al., 2005a, 2007; Mino et al., 2014). However, organoheterotrophic species within Arcobacter (McClung and Patriquin, 1980; Gevertz et al., 2000), Thiomicrospira (Takai et al., 2004) and Sulfurospirillum (Finster et al., 1997; Stolz et al., 1999; Luijten et al., 2003) have also been isolated from various environments. Of these, Arcobacter is the only genus with both pathogenic and free-living, non-pathogenic taxa. The first cultivated representatives of the genus Arcobacter were isolated from aborted bovine fetuses nearly 40 years ago (Ellis et al., 1977) and classified within the genus Campylobacter. As this genus includes many pathogens, the main focus of the characterization of first Arcobacter isolates was to distinguish between different species and survey the strains for resistance to antibiotics (Neill et al., 1985; Kiehlbauch et al., 1991; Vandamme et al., 1992). The genus Arcobacter was resolved by Vandamme et al. (1991), and since then several new species have been described, both pathogenic and free-living strains. The new species have been isolated from a remarkably broad range of habitats, such as humans and animals (Houf et al., 2005, 2009), marine environments (Wirsen et al., 2002; Kim et al., 2010), roots of estuarine salt march plant (McClung and Patriquin, 1980), sewage (Collado et al., 2011; Levican et al., 2013), shellfish (Collado et al., 2009; Figueras et al., 2011a,b; Levican et al., 2012), hypersaline environments (Teske et al., 1996; Donachie et al., 2005), estuarine sediments (Sasi Jyothsna et al., 2013), and even an oil field brine (Gevertz et al., 2000). The metabolic potential of most environmental free-living Arcobacter species has not been examined in detail. A lithoautotrophic lifestyle has only been described for the free-living Arcobacter strains FWKO B and CAB (Gevertz et al., 2000; Carlström et al., 2013), while the remaining strains are cultivated as organoheterotrophs. Furthermore, genomic information is available for a few Arcobacter species, and only one of the four available genomes originate from a free-living species; i.e., A. nitrofigilis CI (Pati et al., 2010).
Here we have isolated and characterized the novel strain IR-1 affiliated with Arcobacter anaerophilus originating from nitrate-amended corrosion experiments. The thorough growth experiments and genomic information revealed that the strain has several unique features relative to Epsilonproteobacteria in general and to Arcobacter in particular. The characterization showed that the IR-1 strain has a lithoautotrophic, lithoheterotrophic, or organoheterotrophic lifestyle coupled to a wide selection of electron acceptors. Genomic information from the IR-1 strain, the pathogenic A. butzleri RM4018 and free-living A. nitrofigilis CI showed that the free-living strains had more genes in common and in general a wider metabolic range than the pathogenic strain. We also extend the metabolic properties of Epsilonproteobaceteria by showing an ability to use iron-citrate as an electron acceptor, utilize tryptone, oxidize acetate with elemental sulfur and involve a hox hydrogenase in the central metabolism. Unique metabolic traits for Arcobacter were also revealed by strain IR-1, such as genes encoding nitrogen-fixation and an incomplete denitrification pathway, where nitrite is the end product from the reduction of nitrate.
Materials and Methods
Sampling Site and Isolation
The IR-1 strain was isolated from an enrichment culture grown on water from the Utsira Aquifer (UA) added 1:150 injection water (i.e., production water from Oilfield A and aquifer water mixed 1:1), a slice of sterile iron foil (20–50 mg, 0.1 mm, Alfa Aesar) and 12 mM So (Merck), as previously described (2014). The UA water is anaerobic and fully saturated with CH4 and CO2, saline (40%) and has a naturally low concentration of sulfate (0–5 mM; Drønen et al., 2014). The bottle was shaken at 100 rpm for 8 months at 25°C, before the remaining iron chips were harvested and stored at -80°C in enrichment medium added 15% glycerol. From here fresh enrichment cultures were initiated on Marine Broth 2216 added 5 mM NaNO3. The culture was transferred to agar plates of Marine Broth 2216 added 5 mM NaNO3 in anaerobic atmosphere (10% H2, 20% CO2, and 70% N2), and single colonies were transferred to new plates twice. Cells with a single morphology remained in the culture, and sequencing the 16S rRNA gene of the isolate using the primers 8f (5′-AGAGTTTGATCCTGGCTCAG-3′) (Edwards et al., 1989) and 1392r (5′-ACGGGCGGTGTGTRC-3′) (Lane et al., 1985) confirmed that the strain was 99% identical to A. anaerophilus strain JC83T (Sasi Jyothsna et al., 2013), hence we named the new isolate A. anaerophilus IR-1.
Cultivation
Growth medium for the isolate was based on an anaerobic mineral medium for nitrate reducers (NRB-medium) buffered with bicarbonate and supplied with trace element solution SL-10 and vitamins, as described by Myhr and Torsvik (2000). During experiments to determine optimal growth temperature, NaCl concentration and pH range; cultures were supplied with 30 mM acetate and 0.05% yeast extract as energy source and 8.5 mM nitrate as electron acceptor. For the pH range experiments, the medium was buffered with either 30 mM bicarbonate (Sigma–Aldrich) or 10 mM HEPES (Sigma). A. anaerophilus IR-1 was transferred to fresh medium twice prior to the growth curve experiments. Optical density (OD) measurements were generated every 10–15 min at 600 nm, measured by the Cary 100 Bio UV/VIS-spectrophotometer (Varian) in 10 mm quartz glass cuvettes (Hellma). Instead of Gram staining, a KOH string test was performed as described by Ryu (1938). For a metabolic characterization of the isolate, NRB medium with 2% NaCl and pH of 7.2–7.3 was used. Substrates and e-acceptors were added according to Table 2. The closest relative, A. anaerophilus JC83T (=DSM-24636), was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) and included in the characterization as a reference strain.
Table 2.
Characteristics and substrate range of A. anaerophilus IR-1 and the closest related species A. anaerophilus JC83T.
| Characteristic | A. anaerophilus IR-1 | A. anaerophilus JC83T |
|---|---|---|
| Cell size | 1.5–2 μm long, 0.4–0.5 μm wide | 1–2 μm long, 0.1–0.3μm widea |
| Motility | + | + |
| DNA G + C content | 30.2% mol | 24.6% mola |
| Genome size | 3.26 Mbp | ND |
| Substrate utilization | ||
| Acetate | + | + |
| Caproate | - | - |
| Cellobiose | - | - |
| Citrate | - | - |
| D-Fructose | - | - |
| D-Galactose | - | - |
| D-Glucose | - | - |
| DL-Lactate | + | - |
| Elemental sulfur | +b | +b |
| Ferrous (Fe2+) iron | - | - |
| Formate | - | - |
| H2 | + | + |
| H2S | + | + |
| L-Arabinose | - | - |
| Peptone | + | + |
| Pyruvate | + | + |
| Sucrose | - | - |
| Thiosulfate | +b | +b |
| Tryptone | + | + |
| Yeast extract | + | + |
| Electron acceptors | ||
| Nitrate | + | + |
| Elemental sulfurc | + | + |
| Sulfate | - | - |
| Thiosulfate | - | - |
| Ferric (Fe3+) citrate | + | + |
| 3% O2 | + | + |
| 5% O2 | + | + |
| 10% O2 | + | + |
| Air | - | - |
| Enzymatic reactions | ||
| Urease | + | + |
| Indoxyl acetate hydrolysis | - | + |
| Oxidase | + | + |
| Catalase | - | - |
a(Sasi Jyothsna et al., 2013).
bCulture supplied with organic carbon source.
cSulfide in the medium might have formed polysulfide from sulfur.
Presence of enzymatic activity, such as urease, indoxyl acetate hydrolysis and oxidase, was investigated using Diatabs (Rosco Diagnostica) according to the manufacturer’s protocol. In addition, a catalase test was performed using 30% (w/w) hydrogen peroxide solution (Sigma) and colonies of Escherichia coli as positive control.
Nitrite formation was quantified using colorimetry, where the coloration generated by a nitrite test kit (Sera) was quantified in a Cary 100 Bio UV/VIS-spectrophotometer (Varian) at 550 nm. Here, A. anaerophilus IR-1 was cultivated on 10 mM acetate and 5 mM nitrate for 48 h at 35°C. The nitrite standard curve, negative control and parallel cultures of A. anaerophilus IR-1 were measured in duplicates.
DNA Extraction and Sequencing
DNA was extracted from cells in parallel cultures of 30 ml NRB medium added 30 mM acetate and 8.5 mM nitrate. Cells were harvested by centrifugation at 5000 × g for 25 min, and the DNA extraction was based on the protocol described by Marmur (1961). In short: The pellet was dissolved in TE buffer (pH = 8) and 1% SDS, and the mix was incubated at 65°C for 5 min. Then perchlorate was added to a final concentration of 1 M and shaken well. The solution was added equal amounts of chloroform:isoamyl alcohol (24:1), followed by centrifugation at 5000 × g for 10 min. This step was repeated twice using only the aqueous phase. The nucleic acids were mixed 1:2 with 96% ethanol, incubated for 30 min on ice, and finally centrifuged at 13000 × g for 20 min at 4°C. The pellet was washed twice in 70% ethanol and dissolved in TE buffer (pH = 8). For the RNase treatment, the parallel DNA extractions were pooled. Ribonuclease A (Sigma) was added to a final concentration of 50 ng/μl, and the sample was incubated at 37°C for 30 min. One step of deproteinization with chloroform:isoamylalcohol (24:1) was performed as described above. The aqueous phase was added sodium acetate (pH = 5.2) to a final concentration of 0.3 M, and the DNA was precipitated and purified with ethanol. The DNA pellet was dissolved in 10 mM Tris buffer (pH = 8.0). A total of 35 μg high quality DNA (RatioA260/280 = 1.98) was obtained and the DNA was sequenced at the Norwegian Sequencing Center for sequencing. A library was prepared using the Pacific Biosciences 10 kb library protocol, and size selection was done using BluePippin. The library was sequenced on a Pacific Biosciences RS II instrument using P4-C2 chemistry, where three SMRT cells were used in total.
In addition, DNA:DNA hybridization analysis of high quality DNA of A. anaerophilus IR-1 and A. anaerophilus JC83T was performed by DSMZ by renaturation rate measurements (De Ley et al., 1970), under consideration of the modifications described by Huss et al. (1983) using a model Cary 100 Bio UV/VIS-spectrophotometer equipped with a peltier-thermostatted 6 × 6 multicell changer and a temperature controller (Varian).
Bioinformatics
Reads were assembled into contigs using the software Hierarchical Genome Assembly Process (HGAP) v2 (Chin et al., 2013) from Pacific Biosciences. The Prokka software (Seemann, 2014) and the RAST-server (Aziz et al., 2008; Overbeek et al., 2014) were used for automatic annotation of the genome. Predicted genes were also aligned to the NCBI database using a standalone BlastP search (Altschul et al., 1990). For identification of peptidases and adhesion-associated proteins encoded in the genome, the MEROPS peptidase database (Rawlings and Morton, 2008) and Pfam protein families database (Finn et al., 2014) were used. PacBio sequencing raw-data have been submitted to the Sequencing Read Archive (SRA) under BioProject PRJNA273926 and BioSample SAMN03316823.
Contigs of the assembled draft genome of A. anaerophilus IR-1 have been deposited as a WGS project in GenBank under the accession number JXXG01000000.
Results
Characterization of Isolate
The A. anaerophilus IR-1 strain was isolated from injection water in an enrichment culture added UA water, zerovalent iron, and elemental sulfur, which was used in corrosion monitoring (Drønen et al., 2014). The 16S rRNA gene of A. anaerophilus IR-1 was 99% identical to A. anaerophilus strain JC83T (Sasi Jyothsna et al., 2013), and the DNA:DNA hybridization analysis confirmed that A. anaerophilus IR-1 is a new strain within A. anaerophilus with a 76.3–80.2% DNA:DNA hybridization value in comparison with A. anaerophilus strain JC83T.
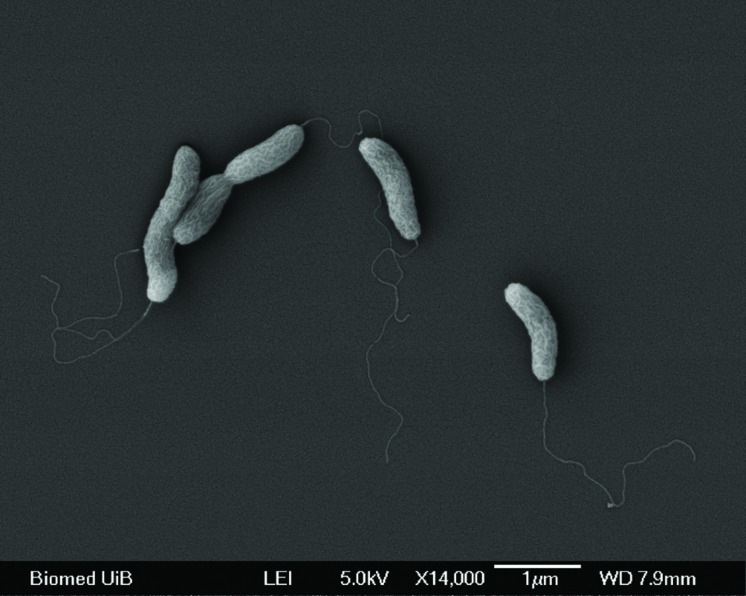
The cells of the new strain were gram negative, curved rods, 1.5–2 μm long and 0.4–0.5 μm wide (Figure 1), and were observed as single cells or in chains of 2–6 cells during active growth. Cells were motile by a single polar flagellum (Figure 1), and were particularly active at early stages of growth. Cells were non-spore forming. The physiological characteristics of strain IR-1 were compared to published descriptions of other Arcobacter species (Table 1) using a selection of parameters devised to distinguish members of the family Campylobacteraceae, as resolved by the International Committee on Systematic Bacteriology (Ursing et al., 1994). Strain IR-1 grew under microaerophilic conditions at 37°C, but not at 42°C or under aerobic conditions. Growth was observed at both 0.5 and 4% NaCl. The strain did not utilize 1% glycine, and no hemolysis was observed. The strain was oxidase and urease positive, but did not have enzyme activity for catalase or indoxyl acetate hydrolysis. The new isolate showed a unique profile of characteristics among the chosen representatives of Arcobacter, and could be distinguished from other Arcobacter species based on three or more parameters (Table 1).
FIGURE 1.
Scanning electron micrographs of Arcobacter anaerophilus IR-1. Cells are curved rods with a single polar flagellum.
Table 1.
Characteristics that differentiate Arcobacter anaerophilus IR-1 from other strains within the genus Arcobacter.
| Characteristics | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Growth in/on | |||||||||||
| Air at 37°C | - | - | - | + | + | + | ND | ND | + | ND | + |
| CO2 at 37°C∗ | + | - | - | + | + | + | + | + | + | + | + |
| CO2 at 42°C∗ | - | - | - | - | - | (+)a | + | - | (+)a | - | (+)a |
| 0.5% (w/v) NaCl | + | + | + | - | - | + | + | + | + | + | ND |
| 4% (w/v) NaCl | + | + | + | + | + | + | +b | - | - | -b | + |
| 1% (w/v) glycine | - | + | - | + | - | ND | + | - | - | - | - |
| Hemolysis | - | - | - | - | - | - | V | + | - | - | - |
| Enzyme activity | |||||||||||
| Oxidase | + | + | + | + | + | + | + | + | + | + | + |
| Catalase | - | - | + | - | - | (+) | (+) | + | (+) | + | + |
| Urease | + | - | + | - | - | - | - | - | + | - | - |
| Nitrate reduction | + | + | + | + | + | - | + | + | + | - | + |
| Indoxyl acetate hydrolysis | - | + | + | + | + | - | + | + | + | + | - |
1, Arcobacter anaerophilus IR-1 (This study); 2, A. anaerophilus JC83 DSM-24636T (Sasi Jyothsna et al., 2013); 3, A. nitrofigilis DSM-7299T (McClung and Patriquin, 1980; Sasi Jyothsna et al., 2013); 4, A. marinus DSM-24769T (Kim et al., 2010; Sasi Jyothsna et al., 2013); 5, A. halophilus DSM-18005T (Donachie et al., 2005); 6, A. mytilii DSM-23625T (Collado et al., 2009); 7, A. butzleri DSM-8739T (Vandamme et al., 1992); 8, A. skirrowii DSM-7302T (Vandamme et al., 1992); 9, A. defluvii DSM-25359T (Collado et al., 2011); 10, A. cryaerophilus DSM-7289T (Vandamme et al., 1992); 11, A. molluscorum (Figueras et al., 2011a).
+, positive; –, negative; (+), weakly positive; V, variable among strains; ND, not determined.
∗Growth at microaerophilic conditions.
aAt aerobic atmosphere.
bAt 3.5% (w/v) NaCl.
A more comprehensive characterization showed that the isolate could be cultivated under anaerobic or microaerophilic conditions. The temperature range was 15–40°C, with an optimum at 37°C. The salinity range was 0.5–6% NaCl, with an optimum at 2% NaCl. No growth was observed using 0% NaCl in the medium. The pH optimum was 7.5 in bicarbonate buffered medium, and pH 7.2 in the HEPES buffered medium. Carbon sources and rich complexes, such as acetate, lactate, peptone, pyruvate, tryptone, and yeast extract were utilized with oxygen or nitrate as electron acceptor (Table 2). Yeast extract, peptone and pyruvate were also fermented without an external electron acceptor. Inorganic compounds, such as hydrogen (H2), elemental sulfur, hydrogen sulfide, and thiosulfate could also be utilized if CO2 or small amounts of acetate (1 mM) was provided as carbon source (Table 2). Sugars were not utilized. In addition to respiration with oxygen at microaerophilic levels (3–10%); nitrate, elemental sulfur, and ferric citrate were also identified as electron acceptors. Ferric hydroxide and sulfate were not used as terminal acceptors. The A. anaerophilus IR-1 could be distinguished from its closest relative, A. anaerophilus JC83T, on the basis of lactic acid utilization and the capacity to hydrolyse indoxyl acetate (Table 2).
Genomic Information
Sequencing of the A. anaerophilus IR-1 genome generated 92 845 reads with an average read length of 6073 bp. The reads were assembled into seven contigs, comprising 3.257 Mbp in total. The size-distribution and sequencing coverage of assembled contigs suggested that three of the contigs constitute the chromosomal genome, while the remaining four could be extra chromosomal elements, such as plasmids. By using the Prokka annotation tool, 3596 protein coding genes were identified (Supplementary Table S1). The genome also contained 60 non-coding genes comprising four rDNA operons (Supplementary Table S2) localized on two different contigs, in addition to 48 tRNAs. The GC-ratio was 30.2% mol.
Central Metabolism
From analyses of the A. anaerophilus IR-1 genome we identified a genotype that was in agreement with the phenotype revealed by the physiological characterization (Table 3). Presence of genes encoding energy converting Ni/Fe hydrogenase (hydABC), a Ni/Fe uptake hydrogenase (hupSL) and a cytoplasmic NAD+-reducing hydrogenase (hoxEFHUY) were congruent with the use of H2 as an electron donor. A complete SOX system (soxABCDXYZ) and sulfide:quinone oxidoreductase (sqr) was most likely involved in oxidation of hydrogen sulfide, thiosulfate, and elemental sulfur. The SOX system may also be involved in sulfite oxidation; however, sulfite was not tested as a substrate during the physiological characterization. Regarding central carbon metabolism, the genome encoded a complete TCA cycle, pentose phosphate pathway, Entner-Doudoroff pathway and glycolysis and gluconeogenesis, with the exception of genes encoding hexokinase/glucose 6-phosphatase. Also, genes encoding a reductive TCA cycle were the only identified genes associated with a CO2 fixation pathway. Key genes for utilization of lactate and pyruvate were found, including L-lactate dehydrogenase (ldh), pyruvate synthase (por) and phosphoenol pyruvate synthase (pps). Two pathways for acetate oxidation were encoded: a two-step reaction involving acetate kinase (ack) and phosphate acetyltransferase (pta) or a single step reaction involving acetyl coenzyme A synthetase [both the ADP dependent (acd) and AMP forming (asc) genes were found]. The acetyl-CoA formed could enter the TCA cycle by citrate synthase (glt) or malate synthase (glc) or be included in various anabolic pathways. The presence of formate dehydrogenase (fdh) genes indicates a potential for growth by formate oxidation, however, neither A. anaerophilus IR-1 nor A. anaerophilus JC83T could grow on this substrate (Table 2).
Table 3.
Comparison of genes involved in central metabolisms within selected species of Arcobacter, based on RAST annotations.
| Pathway | A. anaerophilus IR-1 | A. nitrofigilis CI | A. butzleri RM4018 |
|---|---|---|---|
| Genome size (Mbp) | 3.26 | 3.22 | 2.33 |
| Hydrogenase | |||
| NAD+-reducing | HoxEFHUY | - | - |
| Ni/Fe hydrogenase | HydABC | HydABC | HydABC |
| Ni/Fe uptake hydrogenase | HupSL | HupSL | HupSL |
| Sulfur oxidation | |||
| Sox | SoxABCDXYZ | SoxABCDHXYZ | SoxABCDXYZ |
| Sulfide:quinone oxidoreductase | Sqr∗ | Sqr∗∗ | Sqr∗∗ |
| Central carbon metabolism | |||
| Glycolysis/Gluconeogenesis | + | + | + |
| Entner-Duorodorf pathway | + | + | - |
| Pentose phosphate way | + | + | + |
| Lactate dehydrogenase | + | + | - |
| Pyruvate:ferredoxin oxidoreductase | + | + | - |
| Phosphoenol pyruvate synthase (pps) | + | - | - |
| Acetyl coenzyme A synthetase | + | + | + |
| Malate synthase | + | - | - |
| TCA | + | + | + |
| rTCA | + | + | + |
| Nitrogen fixation | |||
| Nitrogenase molybdenum-iron protein | NifDHK | NifDHK | - |
| Oxygen reduction | |||
| NADH ubiquinone oxidoreductase | NuoABCDEFGHIJKLMN | NuoABCDEFGHIJKLMN | NuoABDEFGIJKLMN |
| Succinate degydrogenase | FrdABC | FrdABC | FrdABC |
| Cytochrome bc1 | PetABC | PetABC | PetABC |
| Cytochrome c oxidoreductase | CcoNOP | CcoNOP | CcoNOP |
| Cytochrome d ubiquinol oxidase | CydAB | CydAB | CydAB |
| Nitrate reduction | |||
| Nitrate reductase | NapABCDFGHL | NapABCDFGHL | NapABDFGHL |
| Nitrite reductase | - | NirBD | NrfAH |
| Nitric oxide reductase (nor) | - | - | NorB |
| Nitrous oxide reductase (nos) | NosZ | - | - |
| Sulfur reduction | |||
| Polysulfide reductase | PsrAB | NrfD∗∗ | - |
| Tetrathionate reductase | TtrABC | TtrABC | - |
| Anaerobic dimethyl sulfoxide reductase | DsmABC | - | - |
| Iron reduction | |||
| Ferric reductase | Fre | - | - |
| Formate-dependent nitrite reductase | NrfD | - | - |
∗Present in dataset from Prokka.
∗∗Present in dataset from NCBI.
As the new strain could utilize organic rich substrates like yeast extract, peptone and tryptone, a search in the MEROPS peptide database was performed in order to survey the genome for membrane associated peptidases that could initiate the degradation of complex protein molecules. In total, 80 different peptidases were identified, which were classified within the families: aspartic (4), cycteine (19), metallo (26), asparagine (2), serine (21), threonine (4) and unknown (4). Indications of protein-rich substrate as a preferable carbon source in A. anaerophilus IR-1 was strengthen by identification of genes encoding dipeptide chemoreceptor protein (tap) and dipeptide transport ATP-binding protein (ddp) involved in chemotaxis toward peptides, and membrane transport proteins, such as inner membrane amino-acid ABC transporter permease protein (yecS, yhdGY), oligopeptide transport system permease protein (oppC) and transporter permeases for leucine, isoleucine, valine, glutamine, arginine, glycine, and proline.
Two gene clusters encoding nitrogenase (nifDHK) were identified in the genome of strain IR-1 (Table 3), where one gene cluster is also present in A. nitrofigilis. These genes have previously not been described for members of the genus Arcobacter, however, our results suggest that free-living strains within this genus can sustain themselves with a nitrogen source by converting dinitrogen to ammonia.
Respiration
As expected, genes for aerobic respiration were found in the genome of the microaerophilic A. anaerophilus IR-1. Genes encoding complex I–IV of the respiratory chain was present, including NADH ubiquinone oxidoreductase (nuoABCDEFGHIJKLMN; complex I), succinate dehydrogenase (frdABC; complex II), cytochrome bc1 complex (petABC; complex III) and cytochrome c oxidoreductase (ccoNOP; complexIV; Table 3). In addition, genes for cytochrome d ubiquinol oxidase (cydAB) were found (complex IV). The respiratory chain is linked to a F0F1 ATPase that generates ATP, and genes (atpABCDEFGH) encoding both domains were found. The Arcobacter strain IR-1 could also respire with nitrate to form nitrite, and accordingly, nitrate reductase (napABCDFGHL) genes were present in the genome (Table 3). However, the denitrification pathway was incomplete, where genes encoding nitrite reductase (nir/nrf) and nitric oxide reductase (nor) were lacking, while genes encoding nitrous oxide reductase (nos) were identified (Table 3). Congruent with the genomic information, a colorimetric method revealed an average nitrite concentration of 0.22 mM after 48 h in cultures grown on acetate and nitrate. Furthermore, cultures of A. anaerophilus IR-1 grew well in medium supplemented with elemental sulfur as terminal electron acceptor. However, the strain may also have benefitted from polysulfide that might have formed chemically in the medium since hydrogen sulfide was used as reducing agent. Polysulfide reductase genes (psrAB) were identified in the genome, suggesting a role for reduction of polysulfide to sulfide. Genes for tetrathionate reductase (ttrABC) were also found, which could form thiosulfate from tetrathionate, but genes for thiosulfate reductase were missing. Observation of anaerobic dimethyl sulfoxide (DMSO) reductase genes (dsmABC) in the genome indicated that DMSO could be a possible electron acceptor, but his was never tested in vitro. Genes for dissimilatory sulfate reduction were not found. The genomic survey also revealed a putative ferric iron reductase (fre), which could support the observed respiration with ferric citrate by the new isolate.
Comparative Genomics
The genome of A. anaerophilus IR-1 was compared to the free-living A. nitrofigilis CI (DSM 7299; Pati et al., 2010) and the pathogen A. butzleri RM4018 (DSM 8739; Miller et al., 2007). Genomes of A. nitrofigilis (NC_014166) and A. butzleri (NC_009850) were obtained from NCBI and uploaded in RAST for comparison with A. anaerophilus IR-1. The genome analyses of A. anaerophilus (3.26 Mbp) resulted in 1432 annotated genes in RAST, while 1542, and 1244 annotated genes, respectively, were identified in A. nitrofigilis (3.22 Mbp) and A. butzleri (2.33 Mbp). Duplicate genes in each genome were removed from the dataset; leaving A. anaerophilus, A. nitrofigilis, and A. butzleri with 1008, 1069, and 837 unique genes, respectively; of which 633 genes were common for all three genomes (Figure 2). The genomes of free-living A. anaerophilus and A. nitrofigilis showed highest similarity, with 221 shared genes, while the pathogen A. butzleri had around 60 genes in common with the two free-living species (Figure 2). The genomes also comprised genes that were unique for each specimen: A. anaerophilus (93), A. nitrofigilis (157), and A. butzleri (85).
FIGURE 2.
Comparative genomics. Unique genes from A. anaerophilus IR-1 (1008), A. nitrofigilis CI (1069), and A. butzleri RM4018 (837) annotated in RAST was extracted and compared.
The function based comparison tool in RAST and searches in the Prokka dataset (A. anaerophilus) or the genome dataset from NCBI (A. nitrofigilis and A. butzleri) were used for identifying key genes in central carbon metabolism, sulfur oxidation, hydrogen oxidation, and respiration with oxygen, nitrate, sulfur, and iron; thereby providing genomic comparisons of the metabolic capacity based on selected pathways (Table 3). This approach revealed that A. anaerophilus was the most metabolically versatile species, with unique genes involved in hydrogen oxidation (hox), nitrate reduction (nosZ), reduction of sulfur species (psr, dsm), iron reduction (fre), in addition to central carbon metabolism, such as phosphoenol pyruvate synthase and malate synthase (Table 3). Furthermore, the free-living strains were sharing some metabolic properties that seemed to be absent in A. butzleri, such as genes for nitrogen fixation (nif), the napC subunit in the nitrate reductase complex, tetrathionate reduction (ttr) and several pathways in central carbon metabolism. Interestingly, A. anaerophilus IR-1 was lacking genes encoding a nitrite reductase, while nitrite reductase (nir/nrf) genes were found in the other two Arcobacter species. This indicates that both A. nitrofigilis and A. butzleri could have a mechanism for nitrite detoxification, which is not found in A. anaerophilus. The A. butzleri genome also comprised nitric oxide reductase (norB), which was not present in the other species.
Discussion
New Metabolic Properties of Free-Living Arcobacter
Today there are 20 acknowledged species within the genus Arcobacter, including free-living strains and pathogens isolated from mammal and human sources. The genus is dominated by free-living strains isolated from various environments; however, knowledge about the metabolic capacity of these remains incomplete. In this study, extensive cultivation experiments with A. anaerophilus IR-1 revealed a versatile metabolism, including lithoautotrophy, lithoheterotrophy, and organoheterotrophy (Table 2) that was confirmed by genome analysis. A lithoautotrophic lifestyle has previously only been described for the free-living Arcobacter strains FWKO B and CAB (Gevertz et al., 2000; Carlström et al., 2013), however, this capability might be more widespread among the members of Arcobacter than anticipated, as most free-living species were never grown lithoautotrophically when characterized. During lithoautotrophic and lithoheterotrophic growth of A. anaerophilus IR-1, elemental sulfur, hydrogen sulfide, thiosulfate or H2 were used as electron donors in combination with organic molecules or CO2 as carbon source. This metabolic profile is similar to those of the free-living Epsilonproteobacteria of the genera Nautiliales, Sulfurovum, and Sulfurimonas isolated from hydrothermal vents and marine sediments (Alain et al., 2002; Miroshnichenko et al., 2002; Inagaki et al., 2003, 2004; Takai et al., 2006; Cai et al., 2014; Mino et al., 2014). The hydrogen sulfide may be oxidized autotrophically in A. anaerophilus IR-1 by sqr (Table 3). The presence of sqr was unique for this Arcobacter strain, but transcriptomic studies of microbial mats from hydrothermal vents and sediments have shown that the gene is highly expressed in free-living Epsilonproteobacteria (Dahle et al., 2013; Urich et al., 2014). Oxidation of elemental sulfur and thiosulfate, which required an organic carbon source, may be catalyzed by the SOX system (soxABCDXYZ) using the previously described mechanism (Friedrich et al., 2001; Bagchi and Ghosh, 2005; Dahl et al., 2008). The genes encoding the SOX system was found in all three Arcobacter genomes, which fits well with the metabolic traits of Epsilonproteobacteria in general. The Ni/Fe hydrogenase (hyd) used for hydrogen uptake was found in the Arcobacter genomes investigated in this study and is also found in other epsilonproteobacterial genera, such as Campylobacter, Helicobacter, Sulfurimonas, Wolinella (Dross et al., 1992; Benoit and Maier, 2008; Cordwell et al., 2008; Sievert et al., 2008). On the contrary, the NAD+-reducing hydrogenase (hox) seemed unique for A. anaerophilus IR-1 among Arcobacter and even among Epsilonproteobacteria (Table 3). The NAD+-reducing hydrogenase is a cytoplasmic, oxygen tolerant, bidirectional hydrogenase that has previously been identified in Cyanobacteria (Papen et al., 1986; Tamagnini et al., 2002), Gammaproteobacteria (Coppi, 2005), Betaproteobacteria (Burgdorf et al., 2005), Actinobacteria (Grzeszik et al., 1997) and phototrophic bacteria (Rákhely et al., 2004; Long et al., 2007). The exact function of this hydrogenase is not well understood, however, it has been suggested to play a role in removing excess electrons from fermentation or photosynthesis in Cyanobacteria and phototrophic bacteria, or supply complex I with reducing compounds (NADH) in aerobic bacteria in order to maintain a proton motive force (Horch et al., 2012). The NADH could also be converted to NADPH in the cytoplasm, hence the NAD+-reducing hydrogenase could provide electron donors for CO2 fixation in some bacteria. In A. anaerophilus IR-1, this hydrogenase may thus be involved in regulation of the NADH levels in the cells or possibly in NADPH generation for CO2 fixation. An organoheterotrophic lifestyle was also verified for the A. anaerophilus strains. To our knowledge, this is the first time a free-living Arcobacter has been shown to degrade tryptone with nitrate as electron acceptor, while degradation of yeast extract and peptone is also described for Arcobacter sp. CAB (Carlström et al., 2013). Utilization of smaller organic molecules, such as acetate, lactate, and pyruvate, seems to be a common trait within Arcobacter, and has been described for several species (McClung and Patriquin, 1980; Teske et al., 1996; Carlström et al., 2013; Sasi Jyothsna et al., 2013), including strain IR-1. Interestingly, A. anaerophilus JC83T was unable to utilize lactate. The observed growth upon acetate with sulfur as the terminal electron acceptor by A. anaerophilus IR-1 has previously not been observed among Epsilonproteobacteria, and is so far only described for strains of Desulfuromonas and Desulfurella within Deltaproteobacteria (Pfennig and Biebl, 1976; Bonch-Osmolovskaya et al., 1990). Formate did not support growth of A. anaerophilus IR-1 or A. anaerophilus JC83T in our growth experiments, although growth on formate is previously observed in Arcobacter sp. (Teske et al., 1996; Gevertz et al., 2000). The fdh in E. coli is oxygen sensitive, and the fdhF gene is induced by increasing formate concentrations and repressed by nitrate (Pecher et al., 1983; Wang and Gunsalus, 2003). The lack of growth of A. anaerophilus IR-1 on formate when oxygen or nitrate was provided as electron acceptor may be explained by a similar mechanism as in E. coli.
With a few exceptions, free-living Arcobacter are described as microaerophilic and nitrate reducing bacteria (Gevertz et al., 2000; Donachie et al., 2005; Collado et al., 2009, 2011; Kim et al., 2010; Figueras et al., 2011a,b; Levican et al., 2012, 2013; Sasi Jyothsna et al., 2013); and even elemental sulfur has been reported as electron acceptor (Gevertz et al., 2000). We observed that the A. anaerophilus strain IR-1 could use all of these during growth on organic or inorganic compounds (Table 2). The genome analysis suggests that complex IV in the electron transport chain comprise cytochrome c oxidoreductase or cytochrome d ubiquinol oxidase when oxygen is provided as terminal electron acceptor. In the betaproteobacterial Azoarcus sp. BH72, a cytochrome c oxidase and a quinol oxidase are expressed, however, they are upregulated according to oxygen concentrations where the former dominate during aerobic conditions and the latter during microaerophilic conditions (Reinhold-Hurek and Zhulin, 1997). In E. coli, the cytochrome d containing enzyme complex has a higher affinity for oxygen (Miller and Gennis, 1983; Kita et al., 1984), and this can thus indicate that this terminal oxidase may operate at low oxygen concentrations in A. anaerophilus IR-1. The cytochrome c oxidoreductase may operate at higher oxygen concentration, but still within the range of microaerophilic conditions.
All three Arcobacter strains included in the genome comparison have the capability of nitrate reduction (McClung and Patriquin, 1980; Miller et al., 2007), which is also recognized as a common property among other epsilonproteobacterial taxa (Vetriani et al., 2014). Genome sequencing of free-living Campylobacter, Nitratiruptor, Sulfurimonas, Sulfurovum, and Wolinella has revealed a complete denitrification pathway or nitrite ammonification pathway, meaning that nitrate can be reduced to dinitrogen or ammonium (Voordeckers et al., 2005; Takai et al., 2006; Nakagawa et al., 2007; Kern and Simon, 2009; Sikorski et al., 2010; Grote et al., 2012). The periplasmic enzyme nitrite reductase (Nap) is catalyzing the nitrate reduction to nitrite, and the nap operon (napAGHBFLD) in A. anaerophilus IR-1, A. nitrofigilis, and A. butzleri had the same gene orientation as commonly found in epsilonproteobacterial species (Sparacino-Watkins et al., 2014). In these species, electrons are transferred from the menaquinone pool to the terminal reductase (NapAB) via a ferredoxin containing integral membrane protein (NapGH; Kern and Simon, 2009; Sparacino-Watkins et al., 2014). However, the free-living strains within Arcobacter might also have the transmembrane protein NapC as an alternative electron transition pathway to NapAB, as genes encoding NapC were found downstream the nap operon in both A. anaerophilus and A. nitrofigilis (Table 3). This could provide the Arcobacter strains with a more advanced nitrate reductase complex that has previously only been seen in Gammaproteobacteria (Sparacino-Watkins et al., 2014). However, further studies are needed to confirm a function in nitrate reduction by NapC in free-living Arcobacter strains. The remaining denitrification pathway in A. anaerophilus IR-1 was incomplete, where only the genes encoding nos were identified (Table 3), indicating that nitrite produced by nitrate reductase may represent a metabolic end product. These findings were also supported by the detection of nitrite (0.22 mM) in cultures supplied with nitrate as electron acceptor. In comparison, A. nitrofigilis and A. butzleri encode nitrite reductase [NAD(P)H] (nirBD) and cytochrome C nitrite reductase (nrfAH), respectively (Table 3), which catalyzes the reduction of nitrite to ammonia. The periplasmic nitrite reductase in A. butzleri has probably direct supply of nitrite produced by nitrate reductase; however, the nitrite reductase in A. nitrofigilis is a cytoplasmic enzyme. Here, expression of the nitrite transporter gene found in the same gene cluster as the nitrite reductase might be involved in shuttling the nitrite over the membrane to the cytoplasmic enzyme complex. In the epsilonproteobacterial Nautilia profundicola, a different mechanism of nitrite removal is known, where nitrite is used in an assimilatory nitrogen pathways via hydroxylamine (Campbell et al., 2009). Genes encoding a similar pathway were not found in the genome of A. anaerophilus IR-1. Instead, the new Arcobacter strain seems to tolerate increasing nitrite concentrations (up to 0.3 mM in batch cultures).
In A. anaerophilus IR-1 a gene cluster comprising nosZDFLY and napGH was found, which is conserved in Wolinella succinogenes (Simon et al., 2004), while the gene cluster was lacking in A. nitrofigilis and A. butzleri. This gene cluster in the IR-1 strain also included two hypothetical genes that are annotated as cytochrome c in W. succinogenes. This gene cluster is also widely conserved among the Epsilonproteobacteria, and seems to be a unique characteristic of this bacterial class (Kern and Simon, 2009). The encoded proteins provide a link between the reduction of nitrous oxide to N2, catalyzed by NosZ, and electron transfer from the menaquinol pool by NapGH and cytochrome c. However, the metabolic importance of nitrous oxide reduction is not well studied for most Epsilonproteobacteria and the role in A. anaerophilus IR-1 metabolism remains to be identified.
The A. anaerophilus IR-1 strain was also found to reduce ferric iron with acetate as electron donor. The genome analysis suggests that a NrfD protein coupled to ferric reductase takes part in this reaction; a mechanism equivalent to the one proposed for Melioribacter roseus, where putative ferric reductase (fre) in the outer membrane is coupled to a NrfCD protein located in the inner membrane via a c-type cytochrome shuttle (Kadnikov et al., 2013). In A. anaerophilus IR-1, the NrfD protein was identified as a membrane bound formate-dependent nitrite reductase (Acr_01494) in the original Prokka annotation list. However, a standalone BlastP search using a 420 aa query sequence resulted in best hit against polysulfide reductase (Sulfurospirillum arcachonese) with a bit-score of 593 and 72% identity. Although the identity of the protein was difficult to determine, the protein was classified within the NrfD superfamily, which is involved in electron transfer from the quinone pool to a periplasmic receptor. Growth experiments showed that A. anaerophilus IR-1 could only reduce the soluble ferric citrate, which indicates that the strain requires chelated iron compounds for iron reduction. The insoluble ferric hydroxide was not utilized, and genes encoding conductive pili homologous to the nanowires in Geobacter (Reguera et al., 2005) were not found. Ferric iron minerals are abundant in anaerobic aquifers and sediments (Nealson, 1997; Orcutt et al., 2011), and humic substances have been shown to stimulate iron reduction in these environments where the humic substances serve as an electron shuttle between microorganisms and minerals (Lovley et al., 1996; Nevin and Lovley, 2000). However, this was not tested in vitro for strain IR-1. Overall, A. anaerophilus IR-1 could possibly reduce chelated iron minerals in a natural environment, and this ability expands the ecological niche of the free-living Epsilonproteobacteria to take part in cycling of iron.
Conclusion
Through physiological characterization and genomics of A. anaerophilus IR-1, new knowledge about the metabolic properties of free-living Epsilonproteobacteria has been achieved, such as utilization of the complex organic substrate tryptone, reduction of ferric iron citrate and presence of genes encoding a NAD+-reducing hydrogenase (Hox). Comparative genomics of three Arcobacter species showed that the free-living strains had more in common compared to the pathogen strain. Their metabolic range of lithoautotrophy and organoheterotrophy is comparable to many free-living Epsilonproteobacteria isolated from hydrothermal vents and marine sediments. All the Arcobacter strains had an incomplete denitrification pathway; hence we suggest that members of the genus Arcobacter are nitrate reducers rather than denitrifiers. A. anaerophilus IR-1 did not have a mechanism for nitrite removal, and therefore seems to tolerate high concentrations of nitrite, while the other Arcobacter species encodes genes for nitrite reduction to ammonia.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by VISTA (Norwegian Academy of Science and Letters, and Statoil, project 6507). The work was also supported by the Norwegian Research Council (project 179560). The authors wish to thank Anne Nyhaug and Endy Spriet at the Molecular Imaging Center for their helpful assistance during SEM imaging.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00987
References
- Akerman N. H., Butterfield D. A., Huber J. A. (2013). Phylogenetic diversity and functional gene patterns of sulfur-oxidizing subseafloor Epsilonproteobacteria in diffuse hydrothermal vent fluids. Front. Microbiol. 4:185 10.3389/fmicb.2013.00185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alain K., Querellou J., Lesongeur F., Pignet P., Crassous P., Raguenes G., et al. (2002). Caminibacter hydrogeniphilus gen. nov., sp. nov., a novel thermophilic, hydrogen-oxidizing bacterium isolated from an East Pacific Rise hydrothermal vent. Int. J. Syst. Evol. Microbiol. 52 1317–1323. 10.1099/ijs.0.02142-0 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi A., Ghosh T. C. (2005). A structural study towards the understanding of the interactions of SoxY. SoxZ, and SoxB, leading to the oxidation of sulfur anions via the novel global sulfur oxidizing (sox) operon. Biochem. Biophys. Res. Commun. 335 609–615. [DOI] [PubMed] [Google Scholar]
- Benoit S. L., Maier R. J. (2008). Hydrogen and nickel metabolism in Helicobacter species. Ann. N. Y. Acad. Sci. 1125 242–251. 10.1196/annals.1419.014 [DOI] [PubMed] [Google Scholar]
- Bonch-Osmolovskaya E. A., Sokolova T. G., Kostrikina N. A., Zavarzin G. A. (1990). Desulfurella acetivorans gen. nov. and sp. nov. —a new thermophilic sulfur-reducing Eubacterium. Arch. Microbiol. 153 151–155. 10.1007/BF00247813 [DOI] [Google Scholar]
- Brinkhoff T., Muyzer G., Wirsen C. O., Kuever J. (1999). Thiomicrospira kuenenii sp. nov. and Thiomicrospira frisia sp. nov., two mesophilic obligately chemolithoautotrophic sulfur-oxidizing bacteria isolated from an intertidal mud flat. Int. J. Syst. Bacteriol. 2 385–392. 10.1099/00207713-49-2-385 [DOI] [PubMed] [Google Scholar]
- Burgdorf T., Lenz O., Buhrke T., van der Linden E., Jones A. K., Albracht S. P. J., et al. (2005). [NiFe]-hydrogenases of Ralstonia eutropha H16: modular enzymes for oxygen-tolerant biological hydrogen oxidation. J. Mol. Microbiol. Biotechnol. 10 181–196. 10.1159/000091564 [DOI] [PubMed] [Google Scholar]
- Cai L., Shao M.-F., Zhang T. (2014). Non-contiguous finished genome sequence and description of Sulfurimonas hongkongensis sp. nov., a strictly anaerobic denitrifying, hydrogen- and sulfur-oxidizing chemolithoautotroph isolated from marine sediment. Stand Genomic Sci. 9 1302–1310. 10.4056/sigs.4948668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell B. J., Engel A. S., Porter M. L., Takai K. (2006). The versatile Epsilonproteobacteria: key players in sulphidic habitats. Nat. Rev. Microbiol. 4 458–468. 10.1038/nrmicro1414 [DOI] [PubMed] [Google Scholar]
- Campbell B. J., Smith J. L., Hanson T. E., Klotz M. G., Stein L. Y., Lee C. K., et al. (2009). Adaptations to submarine hydrothermal environments exemplified by the genome of Nautilia profundicola. PLoS Genet. 5:e1000362 10.1371/journal.pgen.1000362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlström C. I., Wang O., Melnyk R. A., Bauer S., Lee J., Engelbrektson A., et al. (2013). Physiological and genetic description of dissimilatory perchlorate reduction by the novel marine bacterium Arcobacter sp. strain CAB. mBio 4:e217–13 10.1128/mBio.00217-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C. S., Alexander D. H., Marks P., Klammer A. A., Drake J., Heiner C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10 563–569. 10.1038/nmeth.2474 [DOI] [PubMed] [Google Scholar]
- Collado L., Cleenwerck I., Van Trappen S., De Vos P., Figueras M. J. (2009). Arcobacter mytili sp. nov., an indoxyl acetate-hydrolysis-negative bacterium isolated from mussels. Int. J. Syst. Evol. Microbiol. 59 1391–1396. 10.1099/ijs.0.003749-0 [DOI] [PubMed] [Google Scholar]
- Collado L., Levican A., Perez J., Figueras M. J. (2011). Arcobacter defluvii sp. nov., isolated from sewage samples. Int. J. Syst. Evol. Microbiol. 61 2155–2161. 10.1099/ijs.0.025668-0 [DOI] [PubMed] [Google Scholar]
- Coppi M. V. (2005). The hydrogenases of Geobacter sulfurreducens: a comparative genomic perspective. Microbiology 151 1239–1254. 10.1099/mic.0.27535-0 [DOI] [PubMed] [Google Scholar]
- Cordwell S. J., Len A. C. L., Touma R. G., Scott N. E., Falconer L., Jones D., et al. (2008). Identification of membrane-associated proteins from Campylobacter jejuni strains using complementary proteomics technologies. Proteomics 8 122–139. 10.1002/pmic.200700561 [DOI] [PubMed] [Google Scholar]
- Dahl C., Friedrich C., Kletzin A. (2008). “Sulfur oxidation in prokaryotes,” in Encyclopedia of Life Sciences (ELS) (Chichester: John Wiley & Sons, Ltd.). [Google Scholar]
- Dahle H., Roalkvam I., Thorseth I. H., Pedersen R. B., Steen I. H. (2013). The versatile in situ gene expression of an Epsilonproteobacteria-dominated biofilm from a hydrothermal chimney. Environ. Microbiol. Rep. 5 282–290. 10.1111/1758-2229.12016 [DOI] [PubMed] [Google Scholar]
- De Ley J., Cattoir H., Reynaerts A. (1970). The quantitative measurement of DNA hybridization from renaturation rates. Eur. J. Biochem. 12 133–142. [DOI] [PubMed] [Google Scholar]
- Donachie S. P., Bowman J. P., On S. L., Alam M. (2005). Arcobacter halophilus sp. nov., the first obligate halophile in the genus Arcobacter. Int. J. Syst. Evol. Microbiol. 55 1271–1277. 10.1099/ijs.0.63581-0 [DOI] [PubMed] [Google Scholar]
- Drønen K., Roalkvam I., Beeder J., Torsvik T., Steen I. H., Skauge A., et al. (2014). Modeling of heavy nitrate corrosion in anaerobe aquifer injection water biofilm: a case study in a flow rig. Environ. Sci. Technol. 48 8627–8635. 10.1021/es500839u [DOI] [PubMed] [Google Scholar]
- Dross F., Geisler V., Lenger R., Theis F., Krafft T., Fahrenholz F., et al. (1992). The quinone-reactive Ni/Fe-hydrogenase of Wolinella succinogenes. Eur. J. Biochem. 206 93–102. 10.1111/j.1432-1033.1992.tb16905.x [DOI] [PubMed] [Google Scholar]
- Edwards U., Rogall T., Blocker H., Emde M., Bottger E. C. (1989). Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17 7843–7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis W. A., Neill S. D., O’Brien J. J., Ferguson H. W., Hanna J. (1977). Isolation of Spirillum/Vibrio-like organisms from bovine fetuses. Vet. Rec. 100 451–452. 10.1136/vr.100.21.451 [DOI] [PubMed] [Google Scholar]
- Figueras M. J., Collado L., Levican A., Perez J., Solsona M. J., Yustes C. (2011a). Arcobacter molluscorum sp. nov., a new species isolated from shellfish. Syst. Appl. Microbiol. 34 105–109. 10.1016/j.syapm.2010.10.001 [DOI] [PubMed] [Google Scholar]
- Figueras M. J., Levican A., Collado L., Inza M. I., Yustes C. (2011b). Arcobacter ellisii sp. nov., isolated from mussels. Syst. Appl. Microbiol. 34 414–418. 10.1016/j.syapm.2011.04.004 [DOI] [PubMed] [Google Scholar]
- Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., Eddy S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42 D222–D230. 10.1093/nar/gkt1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finster K., Liesack W., Tindall B. J. (1997). Sulfurospirillum arcachonense sp. nov., a new microaerophilic sulfur-reducing bacterium. Int. J. Syst. Bacteriol. 47 1212–1217. 10.1099/00207713-47-4-1212 [DOI] [PubMed] [Google Scholar]
- Friedrich C. G., Rother D., Bardischewsky F., Quentmeier A., Fischer J. (2001). Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism? Appl. Environ. Microbiol. 67 2873–2882. 10.1128/AEM.67.7.2873-2882.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevertz D., Telang A. J., Voordouw G., Jenneman G. E. (2000). Isolation and characterization of strains CVO and FWKO B, two novel nitrate-reducing, sulfide-oxidizing bacteria isolated from oil field brine. Appl. Environ. Microbiol. 66 2491–2501. 10.1128/AEM.66.6.2491-2501.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosche A., Sekaran H., Pérez-Rodríguez I., Starovoytov V., Vetriani C. (2015). Cetia pacifica gen. nov., sp. nov., a chemolithoautotrophic, thermophilic, nitrate-ammonifying bacterium from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 65 1144–1150. 10.1099/ijs.0.000070 [DOI] [PubMed] [Google Scholar]
- Grote J., Schott T., Bruckner C. G., Glöckner F. O., Jost G., Teeling H., et al. (2012). Genome and physiology of a model Epsilonproteobacterium responsible for sulfide detoxification in marine oxygen depletion zones. Proc. Natl. Acad. Sci. U.S.A. 109 506–510. 10.1073/pnas.1111262109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeszik C., Lübbers M., Reh M., Schlegel H. G. (1997). Genes encoding the NAD-reducing hydrogenase of Rhodococcus opacus MR11. Microbiology 143 1271–1286. 10.1099/00221287-143-4-1271 [DOI] [PubMed] [Google Scholar]
- Headd B., Engel A. S. (2014). Biogeographic congruency among bacterial communities from terrestrial sulfidic springs. Front. Microbiol. 5:473 10.3389/fmicb.2014.00473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horch M., Lauterbach L., Lenz O., Hildebrandt P., Zebger I. (2012). NAD(H)-coupled hydrogen cycling – structure–function relationships of bidirectional [NiFe] hydrogenases. FEBS Lett. 586 545–556. 10.1016/j.febslet.2011.10.010 [DOI] [PubMed] [Google Scholar]
- Houf K., On S. L., Coenye T., Debruyne L., De Smet S., Vandamme P. (2009). Arcobacter thereius sp. nov., isolated from pigs and ducks. Int. J. Syst. Evol. Microbiol. 59 2599–2604. 10.1099/ijs.0.006650-0 [DOI] [PubMed] [Google Scholar]
- Houf K., On S. L., Coenye T., Mast J., Van Hoof J., Vandamme P. (2005). Arcobacter cibarius sp. nov., isolated from broiler carcasses. Int. J. Syst. Evol. Microbiol. 55 713–717. 10.1099/ijs.0.63103-0 [DOI] [PubMed] [Google Scholar]
- Huber J. A., Cantin H. V., Huse S. M., Welch D. B., Sogin M. L., Butterfield D. A. (2010). Isolated communities of Epsilonproteobacteria in hydrothermal vent fluids of the Mariana Arc seamounts. FEMS Microbiol. Ecol. 73 538–549. 10.1111/j.1574-6941.2010.00910.x [DOI] [PubMed] [Google Scholar]
- Huss V. A., Festl H., Schleifer K. H. (1983). Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst. Appl. Microbiol. 4 184–192. 10.1016/S0723-2020(83)80048-4 [DOI] [PubMed] [Google Scholar]
- Inagaki F., Takai K., Kobayashi H., Nealson K. H., Horikoshi K. (2003). Sulfurimonas autotrophica gen. nov., sp. nov., a novel sulfur-oxidizing Epsilonproteobacterium isolated from hydrothermal sediments in the Mid-Okinawa Trough. Int. J. Syst. Evol. Microbiol. 53 1801–1805. 10.1099/ijs.0.02682-0 [DOI] [PubMed] [Google Scholar]
- Inagaki F., Takai K., Nealson K. H., Horikoshi K. (2004). Sulfurovum lithotrophicum gen. nov., sp. nov., a novel sulfur-oxidizing chemolithoautotroph within the Epsilonproteobacteria isolated from Okinawa Trough hydrothermal sediments. Int. J. Syst. Evol. Microbiol. 54 1477–1482. 10.1099/ijs.0.03042-0 [DOI] [PubMed] [Google Scholar]
- Kadnikov V. V., Mardanov A. V., Podosokorskaya O. A., Gavrilov S. N., Kublanov I. V., Beletsky A. V., et al. (2013). Genomic analysis of Melioribacter roseus, facultatively anaerobic organotrophic bacterium representing a novel deep lineage within Bacteriodetes/Chlorobi group. PLoS ONE 8:e53047 10.1371/journal.pone.0053047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern M., Simon J. (2009). Electron transport chains and bioenergetics of respiratory nitrogen metabolism in Wolinella succinogenes and other Epsilonproteobacteria. Biochim. Biophys. Acta 1787 646–656. 10.1016/j.bbabio.2008.12.010 [DOI] [PubMed] [Google Scholar]
- Kiehlbauch J. A., Brenner D. J., Nicholson M. A., Baker C. N., Patton C. M., Steigerwalt A. G., et al. (1991). Campylobacter butzleri sp. nov. isolated from humans and animals with diarrheal illness. J. Clin. Microbiol. 29 376–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. M., Hwang C. Y., Cho B. C. (2010). Arcobacter marinus sp. nov. Int. J. Syst. Evol. Microbiol. 60 531–536. 10.1099/ijs.0.007740-0 [DOI] [PubMed] [Google Scholar]
- Kita K., Konishi K., Anraku Y. (1984). Terminal oxidases of Escherichia coli aerobic respiratory chain. II. Purification and properties of cytochrome b558-d complex from cells grown with limited oxygen and evidence of branched electron-carrying systems. J. Biol. Chem. 259 3375–3381. [PubMed] [Google Scholar]
- Knittel K., Kuever J., Meyerdierks A., Meinke R., Amann R., Brinkhoff T. (2005). Thiomicrospira arctica sp. nov. and Thiomicrospira psychrophila sp. nov., psychrophilic, obligately chemolithoautotrophic, sulfur-oxidizing bacteria isolated from marine Arctic sediments. Int. J. Syst. Evol. Microbiol. 55 781–786. 10.1099/ijs.0.63362-0 [DOI] [PubMed] [Google Scholar]
- Kodama Y., Ha L. T., Watanabe K. (2007). Sulfurospirillum cavolei sp. nov., a facultatively anaerobic sulfur-reducing bacterium isolated from an underground crude oil storage cavity. Int. J. Syst. Evol. Microbiol. 57 827–831. 10.1099/ijs.0.64823-0 [DOI] [PubMed] [Google Scholar]
- Kodama Y., Watanabe K. (2004). Sulfuricurvum kujiense gen. nov., sp. nov., a facultatively anaerobic, chemolithoautotrophic, sulfur-oxidizing bacterium isolated from an underground crude-oil storage cavity. Int. J. Syst. Evol. Microbiol. 54 2297–2300. 10.1099/ijs.0.63243-0 [DOI] [PubMed] [Google Scholar]
- Labrenz M., Grote J., Mammitzsch K., Boschker H. T., Laue M., Jost G., et al. (2013). Sulfurimonas gotlandica sp. nov., a chemoautotrophic and psychrotolerant epsilonproteobacterium isolated from a pelagic redoxcline, and an emended description of the genus Sulfurimonas. Int. J. Syst. Evol. Microbiol. 63 4141–4148. 10.1099/ijs.0.048827-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. J., Pace B., Olsen G. J., Stahl D. A., Sogin M. L., Pace N. R. (1985). Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. U.S.A. 82 6955–6959. 10.1073/pnas.82.20.6955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levican A., Collado L., Aguilar C., Yustes C., Dieguez A. L., Romalde J. L., et al. (2012). Arcobacter bivalviorum sp. nov. and Arcobacter venerupis sp. nov., new species isolated from shellfish. Syst. Appl. Microbiol. 35 133–138. 10.1016/j.syapm.2012.01.002 [DOI] [PubMed] [Google Scholar]
- Levican A., Collado L., Figueras M. J. (2013). Arcobacter cloacae sp. nov. and Arcobacter suis sp. nov., two new species isolated from food and sewage. Syst. Appl. Microbiol. 36 22–27. 10.1016/j.syapm.2012.11.003 [DOI] [PubMed] [Google Scholar]
- Long M., Liu J., Chen Z., Bleijlevens B., Roseboom W., Albracht S. J. (2007). Characterization of a HoxEFUYH type of [NiFe] hydrogenase from Allochromatium vinosum and some EPR and IR properties of the hydrogenase module. J. Biol. Inorg. Chem. 12 62–78. 10.1007/s00775-006-0162-1 [DOI] [PubMed] [Google Scholar]
- Lovley D. R., Coates J. D., BluntHarris E. L., Phillips E. J. P., Woodward J. C. (1996). Humic substances as electron acceptors for microbial respiration. Nature 382 445–448. 10.1038/382445a0 [DOI] [Google Scholar]
- Luijten M. L., de Weert J., Smidt H., Boschker H. T., de Vos W. M., Schraa G., et al. (2003). Description of Sulfurospirillum halorespirans sp. nov., an anaerobic, tetrachloroethene-respiring bacterium, and transfer of Dehalospirillum multivorans to the genus Sulfurospirillum as Sulfurospirillum multivorans comb. nov. Int. J. Syst. Evol. Microbiol. 53 787–793. 10.1099/ijs.0.02417-0 [DOI] [PubMed] [Google Scholar]
- Marmur J. (1961). A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 3 208–218. 10.1016/S0022-2836(61)80047-8 [DOI] [Google Scholar]
- McClung C. R., Patriquin D. G. (1980). Isolation of a nitrogen-fixing Campylobacter species from the roots of Spartina alterniflora Loisel. Can. J. Microbiol. 26 881–886. 10.1139/m80-153 [DOI] [PubMed] [Google Scholar]
- Miller M. J., Gennis R. B. (1983). The purification and characterization of the cytochrome d terminal oxidase complex of the Escherichia coli aerobic respiratory chain. J. Biol. Chem. 258 9159–9165. [PubMed] [Google Scholar]
- Miller W. G., Parker C. T., Rubenfield M., Mendz G. L., Wösten M. M., Ussery D. W., et al. (2007). The complete genome sequence and analysis of the Epsilonproteobacterium Arcobacter butzleri. PLoS ONE 2:e1358 10.1371/journal.pone.0001358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mino S., Kudo H., Arai T., Sawabe T., Takai K., Nakagawa S. (2014). Sulfurovum aggregans sp. nov., a hydrogen-oxidizing, thiosulfate-reducing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent chimney, and an emended description of the genus Sulfurovum. Int. J. Syst. Evol. Microbiol. 64 3195–3201. 10.1099/ijs.0.065094-0 [DOI] [PubMed] [Google Scholar]
- Miroshnichenko M. L., Kostrikina N. A., L’Haridon S., Jeanthon C., Hippe H., Stackebrandt E., et al. (2002). Nautilia lithotrophica gen. nov., sp. nov., a thermophilic sulfur-reducing Epsilonproteobacterium isolated from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 52 1299–1304. 10.1099/ijs.0.02139-0 [DOI] [PubMed] [Google Scholar]
- Myhr S., Torsvik T. (2000). Denitrovibrio acetiphilus, a novel genus and species of dissimilatory nitrate-reducing bacterium isolated from an oil reservoir model column. Int. J. Syst. Evol. Microbiol. 50 1611–1619. 10.1099/00207713-50-4-1611 [DOI] [PubMed] [Google Scholar]
- Nakagawa S., Inagaki F., Takai K., Horikoshi K., Sako Y. (2005a). Thioreductor micantisoli gen. nov., sp. nov., a novel mesophilic, sulfur-reducing chemolithoautotroph within the Epsilonproteobacteria isolated from hydrothermal sediments in the Mid-Okinawa Trough. Int. J. Syst. Evol. Microbiol. 55 599–605. 10.1099/ijs.0.63480-0 [DOI] [PubMed] [Google Scholar]
- Nakagawa S., Takai K., Inagaki F., Horikoshi K., Sako Y. (2005b). Nitratiruptor tergarcus gen. nov., sp. nov. and Nitratifractor salsuginis gen. nov., sp. nov., nitrate-reducing chemolithoautotrophs of the Epsilonproteobacteria isolated from a deep-sea hydrothermal system in the Mid-Okinawa Trough. Int. J. Syst. Evol. Microbiol. 55 925–933. 10.1099/ijs.0.63480-0 [DOI] [PubMed] [Google Scholar]
- Nakagawa S., Takaki Y. (2009). “Nonpathogenic Epsilonproteobacteria,” in Encyclopedia of Life Sciences (ELS) (Chichester: John Wiley & Sons, Ltd.). [Google Scholar]
- Nakagawa S., Takaki Y., Shimamura S., Reysenbach A.-L., Takai K., Horikoshi K. (2007). Deep-sea vent epsilonproteobacterial genomes provide insights into emergence of pathogens. Proc. Natl. Acad. Sci. U.S.A. 104 12146–12150. 10.1073/pnas.0700687104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nealson K. H. (1997). Sediment bacteria: who’s there, what are they doing, and what’s new? Annu. Rev. Earth Planetary Sci. 25 403–434. 10.1146/annurev.earth.25.1.403 [DOI] [PubMed] [Google Scholar]
- Neill S. D., Campbell J. N., Obrien J. J., Weatherup S. T. C., Ellis W. A. (1985). Taxonomic position of Campylobacter cryaerophila sp. nov. Int. J. Syst. Bacteriol. 35 342–356. 10.1099/00207713-35-3-342 [DOI] [Google Scholar]
- Nevin K. P., Lovley D. R. (2000). Potential for nonenzymatic reduction of Fe(III) via electron shuttling in subsurface sediments. Environ. Sci. Technol. 34 2472–2478. 10.1021/es991181b [DOI] [Google Scholar]
- Orcutt B. N., Sylvan J. B., Knab N. J., Edwards K. J. (2011). Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol. Mol. Biol. Rev. 75 361–422. 10.1128/MMBR.00039-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R., Olson R., Pusch G. D., Olsen G. J., Davis J. J., Disz T., et al. (2014). The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42 D206–D214. 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papen H., Kentemich T., Schmulling T., Bothe H. (1986). Hydrogenase activities in cyanobacteria. Biochimie 68 121–132. 10.1016/S0300-9084(86)81077-X [DOI] [PubMed] [Google Scholar]
- Pati A., Gronow S., Lapidus A., Copeland A., Glavina Del Rio T., Nolan M., et al. (2010). Complete genome sequence of Arcobacter nitrofigilis type strain (CIT). Stand. Genomic Sci. 2 300–308. 10.4056/sigs.912121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecher A., Zinoni F., Jatisatienr C., Wirth R., Hennecke H., Bock A. (1983). On the redox control of synthesis of anaerobically induced enzymes in Enterobacteriaceae. Arch. Microbiol. 136 131–136. 10.1007/BF00404787 [DOI] [PubMed] [Google Scholar]
- Pfennig N., Biebl H. (1976). Desulfuromonas acetoxidans gen. nov. and sp. nov., a new anaerobic, sulfur-reducing, acetate-oxidizing bacterium. Arch. Microbiol. 110 3–12. 10.1007/BF00416962 [DOI] [PubMed] [Google Scholar]
- Pop Ristova P., Wenzhöfer F., Ramette A., Felden J., Boetius A. (2015). Spatial scales of bacterial community diversity at cold seeps (Eastern Mediterranean Sea). ISME J. 9 1306–1318. 10.1038/ismej.2014.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rákhely G., Kovács Á. T., Maróti G., Fodor B. D., Csanádi G., Latinovics D., et al. (2004). Cyanobacterial-type, heteropentameric. NAD(+)-reducing NiFe hydrogenase in the purple sulfur photosynthetic bacterium Thiocapsa roseopersicina. Appl. Environ. Microbiol. 70 722–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings N. D., Morton F. R. (2008). The MEROPS batch BLAST: a tool to detect peptidases and their non-peptidase homologues in a genome. Biochimie 90 243–259. 10.1016/j.biochi.2007.09.014 [DOI] [PubMed] [Google Scholar]
- Reguera G., McCarthy K. D., Mehta T., Nicoll J. S., Tuominen M. T., Lovley D. R. (2005). Extracellular electron transfer via microbial nanowires. Nature 435 1098–1101. 10.1038/nature03661 [DOI] [PubMed] [Google Scholar]
- Reinhold-Hurek B., Zhulin I. B. (1997). Terminal oxidases of Azoarcus sp. BH72, a strictly respiratory diazotroph. FEBS Lett. 404 143–147. 10.1016/S0014-5793(97)00111-7 [DOI] [PubMed] [Google Scholar]
- Ryu E. (1938). On the Gram-differentiation of bacteria by the simplest method. J. Vet. Med. Sci. 17 31. [Google Scholar]
- Sasi Jyothsna T. S., Rahul K., Ramaprasad E. V., Sasikala C., Ramana C. V. (2013). Arcobacter anaerophilus sp. nov., isolated from an estuarine sediment and emended description of the genus Arcobacter. Int. J. Syst. Evol. Microbiol. 63 4619–4625. 10.1099/ijs.0.054155-0 [DOI] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Sievert S. M., Scott K. M., Klotz M. G., Chain P. S., Hauser L. J., Hemp J., et al. (2008). Genome of the epsilonproteobacterial chemolithoautotroph Sulfurimonas denitrificans. Appl. Environ. Microbiol. 74 1145–1156. 10.1128/AEM.01844-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski J., Munk C., Lapidus A., Ngatchou D., Lucas S., Glavina T. D., et al. (2010). Complete genome sequence of Sulfurimonas autotrophica type strain (OK10T). Stand. Genomic Sci. 3 194–202. 10.4056/sigs.1173118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J., Einsle O., Kroneck P. M., Zumft W. G. (2004). The unprecedented nos gene cluster of Wolinella succinogenes encodes a novel respiratory electron transfer pathway to cytochrome c nitrous oxide reductase. FEBS Lett. 569 7–12. 10.1016/j.febslet.2004.05.060 [DOI] [PubMed] [Google Scholar]
- Sorokin D. Y., Tourova T. P., Kolganova T. V., Spiridonova E. M., Berg I. A., Muyzer G. (2006). Thiomicrospira halophila sp. nov., a moderately halophilic, obligately chemolithoautotrophic, sulfur-oxidizing bacterium from hypersaline lakes. Int. J. Syst. Evol. Microbiol. 56 2375–2380. 10.1099/ijs.0.64445-0 [DOI] [PubMed] [Google Scholar]
- Sparacino-Watkins C., Stolz J. F., Basu P. (2014). Nitrate and periplasmic nitrate reductases. Chem. Soc. Rev. 43 676–706. 10.1039/c3cs60249d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz J. F., Ellis D. J., Blum J. S., Ahmann D., Lovley D. R., Oremland R. S. (1999). Sulfurospirillum barnesii sp. nov. and Sulfurospirillum arsenophilum sp. nov., new members of the Sulfurospirillum clade of the Epsilonproteobacteria. Int. J. Syst. Bacteriol. 49 1177–1180. 10.1099/00207713-49-3-1177 [DOI] [PubMed] [Google Scholar]
- Suh S.-S., Park M., Hwang J., Kil E.-J., Jung S. W., Lee S., et al. (2015). Seasonal dynamics of marine microbial community in the South Sea of Korea. PLoS ONE 10:e0131633 10.1371/journal.pone.0131633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K., Hirayama H., Nakagawa T., Suzuki Y., Nealson K. H., Horikoshi K. (2004). Thiomicrospira thermophila sp. nov., a novel microaerobic, thermotolerant, sulfur-oxidizing chemolithomixotroph isolated from a deep-sea hydrothermal fumarole in the TOTO caldera. Mariana Arc, Western Pacific. Int. J. Syst. Evol. Microbiol. 54 2325–2333. 10.1099/ijs.0.63284-0 [DOI] [PubMed] [Google Scholar]
- Takai K., Hirayama H., Nakagawa T., Suzuki Y., Nealson K. H., Horikoshi K. (2005). Lebetimonas acidiphila gen. nov., sp. nov., a novel thermophilic, acidophilic, hydrogen-oxidizing chemolithoautotroph within the Epsilonproteobacteria, isolated from a deep-sea hydrothermal fumarole in the Mariana Arc. Int. J. Syst. Evol. Microbiol. 55 183–189. 10.1099/ijs.0.63330-0 [DOI] [PubMed] [Google Scholar]
- Takai K., Suzuki M., Nakagawa S., Miyazaki M., Suzuki Y., Inagaki F., et al. (2006). Sulfurimonas paralvinellae sp. nov., a novel mesophilic, hydrogen- and sulfur-oxidizing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent polychaete nest, reclassification of Thiomicrospira denitrificans as Sulfurimonas denitrificans comb. nov. and emended description of the genus Sulfurimonas. Int. J. Syst. Evol. Microbiol. 56 1725–1733. [DOI] [PubMed] [Google Scholar]
- Tamagnini P., Axelsson R., Lindberg P., Oxelfelt F., Wunschiers R., Lindblad P. (2002). Hydrogenases and hydrogen metabolism of cyanobacteria. Microbiol. Mol. Biol. Rev. 66 1–20. 10.1128/MMBR.66.1.1-20.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske A., Sigalevich P., Cohen Y., Muyzer G. (1996). Molecular identification of bacteria from a coculture by denaturing gradient gel electrophoresis of 16S ribosomal DNA fragments as a tool for isolation in pure cultures. Appl. Environ. Microbiol. 62 4210–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urich T., Lanzen A., Stokke R., Pedersen R. B., Bayer C., Thorseth I. H., et al. (2014). Microbial community structure and functioning in marine sediments associated with diffuse hydrothermal venting assessed by integrated meta-omics. Environ. Microbiol. 16 2699–2710. 10.1111/1462-2920.12283 [DOI] [PubMed] [Google Scholar]
- Ursing J. B., Lior H., Owen R. J. (1994). Proposal of minimal standards for describing new species of the family Campylobacteraceae. Int. J. Syst. Bacteriol. 44 842–845. 10.1099/00207713-44-4-842 [DOI] [PubMed] [Google Scholar]
- Vandamme P., Falsen E., Rossau R., Hoste B., Segers P., Tytgat R., et al. (1991). Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: emendation of generic descriptions and proposal of Arcobacter gen. nov. Int. J. Syst. Evol. Microbiol. 41 88–103. [DOI] [PubMed] [Google Scholar]
- Vandamme P., Vancanneyt M., Pot B., Mels L., Hoste B., Dewettinck D., et al. (1992). Polyphasic taxonomic study of the emended genus Arcobacter with Arcobacter butzleri comb. nov. and Arcobacter skirrowii sp. nov., an aerotolerant bacterium isolated from veterinary specimens. Int. J. Syst. Bacteriol. 42 344–356. 10.1099/00207713-42-3-344 [DOI] [PubMed] [Google Scholar]
- Vetriani C., Voordeckers J. W., Crespo-Medina M., O’Brien C. E., Giovannelli D., Lutz R. A. (2014). Deep-sea hydrothermal vent Epsilonproteobacteria encode a conserved and widespread nitrate reduction pathway (Nap). ISME J. 8 1510–1521. 10.1038/ismej.2013.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voordeckers J. W., Starovoytov V., Vetriani C. (2005). Caminibacter mediatlanticus sp. nov., a thermophilic, chemolithoautotrophic, nitrate-ammonifying bacterium isolated from a deep-sea hydrothermal vent on the Mid-Atlantic Ridge. Int. J. Syst. Evol. Microbiol. 55 773–779. 10.1099/ijs.0.63430-0 [DOI] [PubMed] [Google Scholar]
- Wang H., Gunsalus R. P. (2003). Coordinate regulation of the Escherichia coli formate dehydrogenase fdnGHI and fdhF genes in response to nitrate, nitrite, and formate: roles for NarL and NarP. J. Bacteriol. 185 5076–5085. 10.1128/JB.185.17.5076-5085.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirsen C. O., Sievert S. M., Cavanaugh C. M., Molyneaux S. J., Ahmad A., Taylor L. T., et al. (2002). Characterization of an autotrophic sulfide-oxidizing marine Arcobacter sp. that produces filamentous sulfur. Appl. Environ. Microbiol. 68 316–325. 10.1128/AEM.68.1.316-325.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.