Abstract
Verticillium wilt of olive (VWO) is caused by the vascular pathogen Verticillium dahliae. One of the best VWO management measures is the use of tolerant cultivars; however, our knowledge on VWO tolerance/resistance genetics is very limited. A transcriptomic analysis was conducted to (i) identify systemic defense responses induced/repressed in aerial tissues of the tolerant cultivar Frantoio upon root colonization by V. dahliae, and (ii) determine the expression pattern of selected defense genes in olive cultivars showing differential susceptibility to VWO. Two suppression subtractive hybridization cDNA libraries, enriched in up-regulated (FU) and down-regulated (FD) genes respectively, were generated from “Frantoio” aerial tissues. Results showed that broad systemic transcriptomic changes are taking place during V. dahliae-“Frantoio” interaction. A total of 585 FU and 381 FD unigenes were identified, many of them involved in defense response to (a)biotic stresses. Selected genes were then used to validate libraries and evaluate their temporal expression pattern in “Frantoio.” Four defense genes were analyzed in cultivars Changlot Real (tolerant) and Picual (susceptible). An association between GRAS1 and DRR2 gene expression patterns and susceptibility to VWO was observed, suggesting that these transcripts could be further evaluated as markers of the tolerance level of olive cultivars to V. dahliae.
Keywords: verticillium wilt, Olea europaea, vascular pathogen, systemic defense responses, tolerance, susceptibility, defoliating pathotype
Introduction
Verticillium wilt of olive (Olea europaea L.) (VWO) is one of the most serious diseases affecting this relevant woody crop in many regions of the Mediterranean Basin. It is caused by the soil-borne fungus Verticillium dahliae Kleb. The disease is very difficult to control and the implementation of an integrated disease management strategy is therefore recommended. This framework must combine measures such as the use of tolerant cultivars, pathogen-free propagation material, appropriate cultural practices, and/or application of biological control agents (BCAs) (López-Escudero and Mercado-Blanco, 2011). Obviously, the use of olive varieties tolerant/resistant to VWO would be the most efficient and environmentally-friendly approach to control the disease (Tsror, 2011; Arias-Calderón et al., 2015). Up-to-date, however, no olive cultivar has been reported as fully resistant to VWO, although a number of studies have aimed to search and evaluate sources of genetic resistance to V. dahliae (López-Escudero and Mercado-Blanco, 2011, and references therein). We use the term tolerance (Robb, 2007) to refer to those olive cultivars able to cope with V. dahliae infections without developing severe symptoms of the disease (i.e., “Frantoio” and “Changlot Real”) in contrast to susceptible ones (i.e., “Picual”). “Frantoio” is considered one of the most tolerant cultivar to this harmful disease. A recent study has shown that “Frantoio” gave rise to a high number of tolerant seedlings, even when crossed with a very susceptible cultivar such as “Picual” (Trapero et al., 2015). However, not all VWO-tolerant genitors conferred tolerance to their offspring. For instance, “Changlot Real” and “Empeltre,” which are also considered very tolerant to V. dahliae (López-Escudero et al., 2004; Martos-Moreno et al., 2006; Trapero et al., 2013), mostly produced susceptible descendants (Trapero et al., 2015).
Understanding the mechanisms triggered in the host plant by the presence of the pathogen would be instrumental to design novel disease control strategies. Even though our knowledge on plant-pathogen interactions has been enhanced from studies based on powerful histological, histochemical, microscopy, molecular, and “omics” approaches, the information about the genetic bases underlying plant defense responses against vascular (Yadeta and Thomma, 2013) and/or root pathogens (Okubara and Paulitz, 2005; Larroque et al., 2013) is yet scant. Defense mechanisms deployed by the host plant can be the generation of structural barriers, i.e., tyloses (Dixon and Pegg, 1969), activation of metabolic responses, i.e., phytoalexins biosynthesis (Hammerschmidt, 1999), and/or mounting/triggering complex defense-related genetic cascades mediated by diverse signaling molecules, i.e., salicylate, jasmonate, etc. (Derksen et al., 2013). In the case of V. dahliae attacks, plant tissue responses so far reported can be structural, i.e., formation of tyloses in the xylem and/or biochemical, i.e., phenolic compounds accumulation (Baídez et al., 2007; Markakis et al., 2010). Moreover, these responses can be either constitutive (Mueller and Morgham, 1993) or induced in response to the pathogen infection (Daayf et al., 1997; Markakis et al., 2010). In olive, tolerance of cultivar Frantoio to V. dahliae has been proposed to be mediated by biochemical mechanisms activated in the root tissues rather than by plant structural characteristics such as vascular plugging (Bubici and Cirulli, 2012). These reactions are reported to be less noticeable in susceptible than in tolerant cultivars (Baídez et al., 2007; Markakis et al., 2010; Bubici and Cirulli, 2012). Interestingly, Daayf et al. (1997) observed similar responses (i.e., accumulation of paramural and cell wall coatings, phenolic compound deposits) in cotton (Gossypium hirsutum L.) plants infected by V. dahliae, these reactions being also more pronounced in tolerant than in susceptible cultivars. Up-to-date, no genetic and/or genomic information about systemic defense responses taking place in olive upon root colonization by V. dahliae is available, nor whether these responses could be related to VWO susceptibility level. Therefore, the objectives of this study were: (i) to elucidate whether early systemic responses can be differentially triggered in above-ground tissues of “Frantoio” plants upon root inoculation with a representative of the V. dahliae highly-virulent, defoliating (D) pathotype; and (ii) to determine whether specific systemic defense responses correlate with the differential VWO susceptibility level showed by diverse olive cultivars. From the wide range of differential responses found, seven genes identified in “Frantoio” cDNA libraries, namely transcription factor (TF) GRAS1, caffeoyl-O-methyltransferase (CO-MT), defensin protein 1 (DEF), disease resistance-responsive protein (DRR2), 1-aminocyclopropane-1-carboxylate oxidase (ACO), pathogenesis related protein 10 (PR10), and acetone cyanohydrin lyase (ACL) were selected for validation and to assess time-course gene expression pattern during V. dahliae-“Frantoio” interaction. We also analyzed the expression of ACL, ACO, DRR2, and GRAS1 in two additional olive cultivars differing in VWO susceptibility to assess their potential as markers associated with tolerance to V. dahliae in a woody host of commercial relevance.
Materials and methods
Plant material and olive root inoculation with Verticillium dahliae
Two different olive-V. dahliae experiments were performed. In the first one, olive plants (3-month-old) of the tolerant cultivar Frantoio were utilized to generate cDNA libraries (see below). A second experiment, aiming to evaluate gene expression pattern of selected genes (see below), was carried out with olive cultivars Picual (VWO susceptible, 3-month-old), Frantoio and Changlot Real, (VWO tolerant, 8-month-old) (López-Escudero et al., 2004; Martos-Moreno et al., 2006). All olive plants originated from a commercial nursery located in Córdoba (southern Spain). Prior to V. dahliae treatment, plants were acclimated for 3 weeks in a growth chamber under conditions described below. Olive plants manipulation and root dip inoculation in a conidial suspension (30 min, 1·107 conidia ml−1) of V. dahliae isolate V937I, representative of the highly-virulent D pathotype (Maldonado-González et al., 2015), were performed as previously described (Mercado-Blanco et al., 2002). Roots of control plants (non-inoculated) were dipped in water. Then, plants were individually transplanted into polypropylene pots containing an autoclaved sandy substrate (Prieto and Mercado-Blanco, 2008). Plants were maintained at controlled conditions (23 ± 1°C, 60–90% relative humidity, 14-h photoperiod of fluorescent light at 360 μE m−2) during 21 days. In order to ease plant stress after manipulation, inoculation and transplanting, the photoperiod was increased gradually (Gómez-Lama Cabanás et al., 2014). In the first experiment, aerial tissues (stems and leaves) of each olive plant were sampled at 8 h and 1, 2, 3, 4, 5, 6, 7, 10, 13, 15, and 21 (two plants/time point) DAI (days after inoculation) to obtain a broad range of differential (induced or repressed genes) responses. Therefore, aerial tissues of 48 plants (24 V. dahliae inoculated and 24 non-inoculated) were sampled, rapidly frozen in liquid nitrogen, and stored at −80°C until processing. In the second experiment, above-ground tissues (stems and leaves) of 24 plants (12 V. dahliae inoculated and 12 non-inoculated; two plants/time point/cultivar) were sampled at 2 and 10DAI. In addition, roots of three plants per cultivar were collected at 15DAI for assessment of V. dahliae infection. All samples were stored as described above.
mRNA purification
Isolation of total RNA from olive tissue samples of the two experiments was performed according to Asif et al. (2000). RNA was treated with DNaseI (Roche Applied Science, Mannheim, Germany) according to the manufacturer's instructions. In the case of the first experiment (cDNA libraries generation), all RNA samples corresponding to each treatment (V. dahliae-inoculated and non-inoculated plants) were pooled separately to obtain two independent RNA pools prior to mRNA purification. Poly A+ mRNA was purified from approximately 400 μg of total RNA of each pool using the Dynabeads® mRNA Purification Kit (Dynal Biotech, Oslo, Norway) according to the manufacturer's indications. Purity and quality of all RNA samples were assessed electrophoretically and by spectrophotometry using a ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE).
Generation of “suppression substractive hybridization” cDNA libraries
Two different cDNA libraries were constructed from aerial olive tissues originated from the first experiment (“Frantoio”-V. dahliae V937I). “Suppression Subtractive Hybridization” (SSH) technology (Diatchenko et al., 1996) was used in order to clone and identify systemically FU library and FD library genes during the V. dahliae- “Frantoio” roots interaction. The PCR-Select™ cDNA Subtraction Kit (BD Biosciences, Palo Alto, CA) was used to generate cDNA libraries, following the manufacturer's instructions and as previously described (Schilirò et al., 2012; Gómez-Lama Cabanás et al., 2014). The constitutively-expressed β-actin gene from olive (Accession number AF545569), whose expression was checked as not influenced by V. dahliae inoculation, was used to check subtraction efficiency. The fragment (308-bp) was amplified using the primer pair Act1-fw: 5′-GCTTGCTTATGTTGCTCTCGAC-3′/Act1-rv: 5′-TGATTTCCTTGCTCATACGGTC-3′.
Cloning and sequencing
cDNAs resulting from each SSH were ligated in the pGEM-T Easy Vector (Promega, Madison, WI) and cloned into Escherichia coli CH3-Blue competent cells (Bioline, London, UK) according to manufacturer's instructions. Positive (white) colonies were selected and picked in 96-well microtiter plates containing LB medium amended with 100 mg ml−1 ampicillin and then incubated at 37°C for 22 h. Finally, forward T7 universal primer was utilized to sequence 1344 bacterial clones from each SSH library. DNA sequencing was performed at a commercial service (Sistemas Genómicos S.L., Valencia, Spain).
Bioinformatics analysis of ESTs
Contaminating vector and adaptors sequences were removed from ESTs (Expressed Sequence Tags) by mass alignment using the “CLC Main Workbench 6.8.1” (CLC bio, Aarhus, Denmark) software. Sequences that showed low quality or length (<100 bp) were ruled out from the analysis. EST data set assembly was performed by using the “CLC Main Workbench 6.8.1,” aiming to find contiguous sequences and redundancy. Computational annotation of ESTs originating from V. dahliae-“Frantoio” interaction was carried out by using the software “Blast2GO PRO trial” (Conesa et al., 2005) available at https://www.blast2go.com/. To find homologies, the non-redundant (nr) GenBank protein database was searched, running the Blastx algorithm (Altschul et al., 1990) with the E-value set to 1.0E-3 and the High-scoring Segment Pairs (HSP) length cut off fixed to 33 (Schilirò et al., 2012). The ‘Blast2GO PRO trial software was used to perform Gene Ontology (GO) analysis from retrieved database matches. InterPro Scan (Zdobnov and Apweiler, 2001) and functional annotation was used to associate functional information and GO terms to the protein of interest, implementing the specific tool in the Blast2GO software with the default parameters. The “Augment Annotation by ANNEX” function was finally used to improve the annotation profiles information. The GOslim “goslim_plant.obo” was run to achieve specific plant GO terms by means of a plant-specific reduced version of the GO (available at http://www.arabidopsis.org/). Enzyme mapping of annotated sequences was retrieved by direct GO to Enzyme annotation and used to query the Kyoto Encyclopaedia of Genes and Genomes (KEGG - http://www.genome.jp/kegg/) to define the main metabolic pathways involved. The distribution of hits obtained against entries for other plants within the NCBI database was used to get a descriptive view of the newly generated dataset.
Accession numbers
ESTs reported in this study have been deposited in the dbESTs database of the National Center for Biotechnology Information (NCBI) under GenBank accession numbers JZ844237 (dbEST_Id79672628)-JZ844946 (dbEST_Id79673337) (FU library) and JZ822972 (dbEST_Id79640674)-JZ823534 (dbEST_Id79641236) (FD library).
Data validation and time-course gene expression profiles during Verticillium dahliae-olive “frantoio” interaction
Selected ESTs identified in FU and/or FD cDNA libraries by the Blast2GO tool were used for validation by quantitative real-time PCR (qRT-PCR) experiments. Seven transcripts from the whole dataset of nr sequences, two of them from FU (GRAS1 [Accession number JZ844283] and CO-MT [JZ844695]), four from FD (DEF [JZ823187], DRR2 [JZ823156], ACO [JZ823262] and PR10 [JZ823324]), and one identified in both FU and FD (salicylic acid-binding protein 2-like = acetone cyanohydrin lyase, ACL [JZ844792 and JZ823163]), all involved in plant defense responses, were chosen for data validation (Table 1). All these ESTs fulfilled the criteria of >100-bp long and E = 1.0E-3. Expression patterns of these genes in above-ground organs were assessed at 8 h, 2, 4, 10, and 15DAI after V. dahliae root inoculation. ACL and CO-MT primer pairs have been previously described (Schilirò et al., 2012). The “CLC Main Workbench 6.8.1” (CLC bio) software was used to design specific primer pairs for the remaining genes (Table 1). All primer pairs were empirically tested for their specificity in the temperature range 53°C–63°C by conventional PCR. To find the pertinent concentration range at which target cDNA amplified more efficiently, specific qRT-PCR experiments were implemented using cDNAs synthesized from 10-fold serially diluted (1 μg, 100 ng, 10 ng, 1 ng, 100 pg) RNA samples. To draw standard curves for each selected transcript reverse transcribed cDNA was used from serial dilutions (300, 30, 3, 0.3 ng) of remnant total RNA samples not used for SSH. Ct values and the log cDNA concentrations were linearly correlated for each of the examined genes, and PCR efficiencies were automatically calculated by iQ5 optical system software v.2.1 (BioRad, Hercules, CA). Synthesis of cDNA was performed from 100 ng of total RNA in each of five different times assayed using the “iScript cDNA Synthesis Kit” (BioRad, Hercules, CA) following the manufacturer's indications. qRT-PCR experiments and analyses were done in a thermal cycler iQ5 Real-Time PCR System (BioRad) provided with a 96-well sample block. Relative expression for each selected gene was repeated at least once in independent qRT-PCR assays, and three replicas per condition studied and per plate were routinely included. All qRT-PCR reactions, melting profiles and normalizations were performed as previously described (Gómez-Lama Cabanás et al., 2014). Relative expression levels at different times were calculated according to Livak and Schmittgen (2001). The average of each expression gene fold change was categorized as follows: “low” ≥ −1.0 to ≤ 1.0; “medium” ≥ −2.0 to < −1.0 or > 1.0 to ≤ 2.0; and “high,” < −2.0 or > 2.0 (Kim et al., 2008). All relative expression data in four different times for each gene were represented as means ± SD of at least two independent qRT-PCR experiments, each performed with triplicate samples.
Table 1.
List of selected transcripts induced in “Frantoio” aerial tissues upon Verticillium dahliae colonization used in qRT-PCR experiments.
| Clone ID | Putative gene | Process | Primer pair | Amplicon length (bp) | Linear equation | R2 | PCR efficiency |
|---|---|---|---|---|---|---|---|
| FD07-B04T7 | 1-Aminocyclopropane-1-carboxylate oxidase | Ethylene biosynthesis | Fw: CTCAAGTTGATCCCCAAT Rv: GCATTCCATGGCTCTAAA |
231 | y = −3.334x + 26.333 | 0.99 | 99.5 |
| FD04-H01T7 | Defensin protein 1 | Defense response | Fw:ACACCATGAGCAGGAAAA Rv:TGGCTATTGCAGGGGATT |
166 | y = −3.452x + 27.980 | 0,99 | 94.8 |
| FU08-D05T7 | Caffeoyl-o-methyltransferase | Phenylpropanoids biosynthesis | Fw:ACCAGAGGCCATGAAAGAAC Rv:ATTGCCAAAATCTTCCCATC |
204 | y = −3.399x + 26.905 | 1.00 | 96.9 |
| FD08-H06 T7 | Pathogenesis-related protein 10 | Defense response | Fw:GATGTGTGGAGAGGCTTT Rv: CGTCATTTTTCTTCCTAGGT |
153 | y = −3.351x + 23.514 | 0.99 | 98.8 |
| FD-C16 FU-C145 | Acetone-cyanohydrin lyase | Salicylic acid-binding protein 2 | Fw: GAAAGAGATGGAAGCGGAAA Rv: ACACAGGGAAATGCATCAAA |
246 | y = −3.390x + 28.218 | 0.99 | 97.2 |
| FU01-E11T7 | Transcription factor GRAS1 | Signal transduction defense response | Fw:TCAGTGGGTGTTCCTTATT Rv: GGTGCAGCATATAAGGAAA |
269 | y = −3.415x + 31.094 | 0.99 | 96.3 |
| FD-C65 | Disease resistance-responsive family protein | Defense response | Fw:CCAATGCCCGTAAAGTAA Rv: TACAGCGTTTCTTCCCAA |
311 | y = −3.774x + 26.080 | 1.00 | 84.1 |
| (1) AF545569 | Olea europea beta-actin (act1) | Citoskeletal integrity | Fw: GCTTGCTTATGTTGCTCTCGAC Rv: TGATTTCCTTGCTCATACGGTC |
308 | y = −3.465x + 27.632 | 0.99 | 94.4 |
For all transcripts, relative expression analysis was repeated at least two times in independent qRT-PCR experiments (see text and Figure 2). Clone ID, gene name, biological process, primers sequences, amplicon length, linear equations, correlation coefficients (R2), and PCR efficiencies are indicated. Fw, Forward; Rv, Reverse; FU, Frantoio aerial tissues induced gene; FD, Frantoio aerial tissues repressed gene; (1), olive act-1 gene used as reference to normalize relative expression (Gene Bank Accession Number).
Gene expression patterns in olive cultivars displaying differential Verticillium dahliae susceptibility level
Four genes were selected to assess their expression patterns at two different time points (2 and 10DAI) in olive cultivars “Picual” (susceptible) and “Frantoio” and “Changlot Real” (tolerant) (Trapero et al., 2013) from the second experiment. The expression of one up-regulated (GRAS1), two down-regulated (ACO and DRR2) and one found in both libraries (ACL) genes was studied following the approach previously described. Linear equations, correlation coefficients (R2) and PCR efficiencies were estimated for each transcript (Table 2). For each selected gene, expression was quantified at least two times in independent qRT-PCR experiments, and three replicas per time point studied and per plate were routinely included.
Table 2.
List of selected transcripts induced in olive aerial tissues upon Verticillium dahliae colonization used in qRT-PCR experiments with susceptible (Picual) and tolerant (Frantoio and Changlot Real) olive cultivars.
| Putative gene | Cultivars | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Picual | Frantoio | Changlot real | |||||||
| Linear equation | R2 | PCR efficiency | Linear equation | R2 | PCR efficiency | Linear equation | R2 | PCR efficiency | |
| 1-Aminocyclopropane-1-carboxylate oxidase | y = −3.580x + 26.556 | 0.99 | 90.2 | y = −3.305x + 27.123 | 0.98 | 100.7 | y = −3.542x + 29.629 | 0.99 | 91.6 |
| Acetone-cyanohydrin lyase | y = −3.311x + 28.941 | 0.99 | 100.5 | y = −3.447x + 28.292 | 0.99 | 95.0 | y = −3.536x + 31.503 | 0.98 | 91.8 |
| Transcription factor GRAS1 | y = −3.275x + 27.722 | 0.99 | 102.0 | y = −3.301x + 28.952 | 0.99 | 100.9 | y = −3.341x + 29.082 | 0.99 | 99.2 |
| Disease resistance-responsive family protein | y = −3.753x + 24.626 | 0.99 | 84.7 | y = −3.808x + 25.865 | 0.99 | 99.4 | y = −3.528x + 27.767 | 0.98 | 92.1 |
| Olea europea beta-actin (act1) | y = −3.491x + 28.287 | 0.99 | 93.4 | y = −3.475x + 27.558 | 0.98 | 94.0 | y = −3.718x + 28.072 | 0.99 | 85.8 |
For all transcripts, relative expression (RE) analysis was repeated at least two times in independent qRT-PCR experiments (see text and Figure 3). Gene name, linear equations, correlation coefficients (R2) and PCR efficiencies are indicated for each gene/cultivar combination.
Detection of V. dahliae in root tissues
Presence of V. dahliae in olive roots was checked by PCR. Reactions were performed containing 25 ng of DNA, 0.1 μM of V. dahliae specific primers (DB19 and DB22) (Carder et al., 1994), 2x iQ™ SYBR® Green Supermix (BioRad) and H2O up to a total volume of 20 μL. PCR protocol was: denaturation for 4 min at 95°C, followed by 50 cycles of 1 min at 94°C, 45 s at 54°C and 45 s at 72°C and a final extension step of 10 min at 72°C. Melting curves of products were assessed from 54°C to 100°C to confirm the amplification of single PCR bands. For all samples reaction protocol was as follows: 5 min at 95°C for initial denaturation, cooling to 54°C and melting from 54°C to 100°C with a 0.5°C transition rate every 10 s. For each plant root, presence of V. dahliae DNA was verified two times in independent experiments, and three replicas per point studied and per plate were routinely included.
Results
Construction of cDNA libraries enriched in differentially-expressed olive genes from aerial tissues during the colonization of “frantoio” roots by V. dahliae
Two different cDNA libraries enriched in up- and down-regulated transcripts from above-ground tissues of olive cv. Frantoio were generated. A total of 1344 EST sequences were sequenced and analyzed for each library. On the one hand, 585 unigenes were identified in the up-regulated cDNA library (FU), which were assembled into 57 contigs (average length 524 bp) each composed of 2–4 sequences and 528 singlets (average length 427 bp) (Table S1). On the other hand, ESTs from the down-regulated cDNA library (FD) were eventually assembled into 52 contigs (average length 479 bp), each composed of 2–13 sequences, and 329 singlets (average length 382 bp) (Table S2). Despite the fact that the number of ESTs sequenced in both libraries was the same, the number of up-regulated unigenes found in aerial tissues was considerably higher than that of down-regulated unigenes.
Querying (Blastx) the nr NCBI database allowed the attribution of homologous hits for nearly 77% of the ESTs in both libraries. Hits distribution of the complete EST set against sequences from different plant species revealed that 39% of the ESTs matched to coding sequences previously identified in genomes of woody plants such as robusta coffee (Coffea canephora L. Linden) (9.8% present in FU and 7.1% in FD) grapevine (Vitis vinifera L.,) (6.1 and 6.5%), and cacao (Theobroma cacao L.) (2.7 and 2%). E-values ranged from 1.00E-03 to 5.54E-169 for FU cDNA library and 1.66E-3 to 1.94E-162 for FD cDNA library. Only 3.5 and 5.9% of the unigenes found in FU and FD libraries, respectively, showed significant homology with olive genes [NCBIdbEST (http://www.ncbi.nlm.nih.gov/dbEST/)] (Tables 3, 4). This indicates an as yet important lack of genetic/genomic information for this relevant woody crop. Finally, 23% of induced and repressed transcripts found in both libraries were of unknown function.
Table 3.
List of induced EST sequences identified after Blastx analysis as homologous to olive genes previously indexed in databases.
| EST sequence name | Putative protein function | Accession number | E-Value |
|---|---|---|---|
| *FU02-D07T7 | Beta-glucosidase 12-like | AAL93619 | 1.89E-41 |
| FU02-E10T7 | Lipoxygenase family protein | ACD43483 | 1.81E-85 |
| FU03-C10T7 | Thaumatin-like protein | E3SU11 | 5.49E-28 |
| FU04-D05T7 | Alcohol dehydrogenase | AEQ04839 | 2.49E-91 |
| FU04-G10T7 | 24-methylenesterol c-methyltransferase 2 | AGR55393 | 1.81E-74 |
| FU05-E01T7 | Lipid transfer protein | ABS72013 | 6.81E-19 |
| FU06-C04T7 | Beta-glucosidase 12-like | AAL93619 | 1.49E-41 |
| FU08-D04T7 | Serine mitochondrial-like | ABS72016 | 7.01E-136 |
| FU11-A09T7 | b chain structures of alkaloid biosynthetic glucosidases decode substrate specificity | AAL93619 | 7.38E-44 |
| FU11-B02T7 | Beta-glucosidase 44-like | AAL93619 | 1.19E-18 |
| FU11-F10T7 | Ribulose- -bisphosphate carboxylase oxygenase activase | ABS72022 | 5.60E-90 |
| FU12-A03T7 | Cytochrome b5-like | CAA04702 | 5.53E-14 |
| FU12-F09T7 | Beta-glucosidase isozyme 2 precursor | AAL93619 | 8.19E-48 |
| FU13-D06T7 | Cytochrome p450 subunit cyp72a13 | AFS28694 | 1.38E-21 |
| FU14-H02T7 | Carbonic chloroplast precursor | CBL86547 | 2.77E-48 |
| FU-C17 | Hypothetical protein, partial | AFP49328 | 9.00E-13 |
| FU-C58 | Hypothetical protein, partial | AFP49328 | 1.01E-05 |
| FU- C224 | Linoleate 13s-lipoxygenase 2- chloroplastic-like | ACD43485 | 3.09E-116 |
Unigene found in both libraries with the same accession number.
Table 4.
List of repressed EST sequences identified after Blastx analysis as homologous to olive genes previously indexed in databases.
| EST sequence name | Putative protein function | Accession number | E-Value |
|---|---|---|---|
| FD01-E11T7 | Protein chloroplastic | AFP49328 | 1.69E-42 |
| FD01-H08T7 | Ole e 5 olive pollen allergen | ABX26138 | 1.13E-45 |
| FD02-E02T7 | Chloroplast ribulose—bisphosphate carboxylase oxygenase small subunit | ABS71998 | 3.00E-08 |
| FD03-C06T7 | Glycolate oxidase | ABS72011 | 1.40E-67 |
| FD03-E06T7 | Photosystem i reaction center subunit chloroplastic-like | ABU39903 | 3.59E-48 |
| FD03-F07T7 | Fatty acid hydroperoxide lyase | ACD43482 | 6.90E-82 |
| FD03-F09T7 | 4-hydroxy-3-methylbut-2-enyl diphosphate partial | AFS28680 | 3.21E-57 |
| FD04-C07T7 | Isopentenyl diphosphate isomerase | AFS28681 | 1.99E-68 |
| FD04-H01T7 | Defensin ec-amp-d2-like | ABS72000 | 1.66E-01 |
| FD05-D05T7 | Aquaporin tip1-3-like | ABB76813 | 2.30E-83 |
| FD06-E12T7 | Isopentenyl-diphosphate delta-isomerase i | ACF05532 | 4.17E-10 |
| FD07-B09T7 | Cytochrome p450 family protein | AFS28690 | 8.18E-66 |
| FD07-E06T7 | Hypothetical protein, partial | AFP49328 | 4.22E-06 |
| FD08-F04T7 | Ribulose-bisphosphate carboxylase oxygenase activase | ABS72022 | 2.25E-91 |
| FD09-A03T7 | Glyceraldehyde-3-phosphate dehydrogenase chloroplastic | ABS72003 | 3.44E-50 |
| FD14-E01T7 | Achain crystal structure of perakine founder member of a novel akr subfamily with unique conformational changes during nadph binding | ABS72001 | 4.39E-27 |
| FD-C129 | Thaumatin-like protein | E3SU11 | 9.31E-49 |
| FD-C156 | Metallothionein 1 | AFP49330 | 5.50E-15 |
| FD-C163 | Salt tolerance-related protein | ABS72020 | 4.80E-62 |
| *FD-C203 | Beta-glucosidase partial protein | AAL93619 | 1.78E-111 |
Unigene found in both libraries with the same accession number.
Functional characterization of EST data set
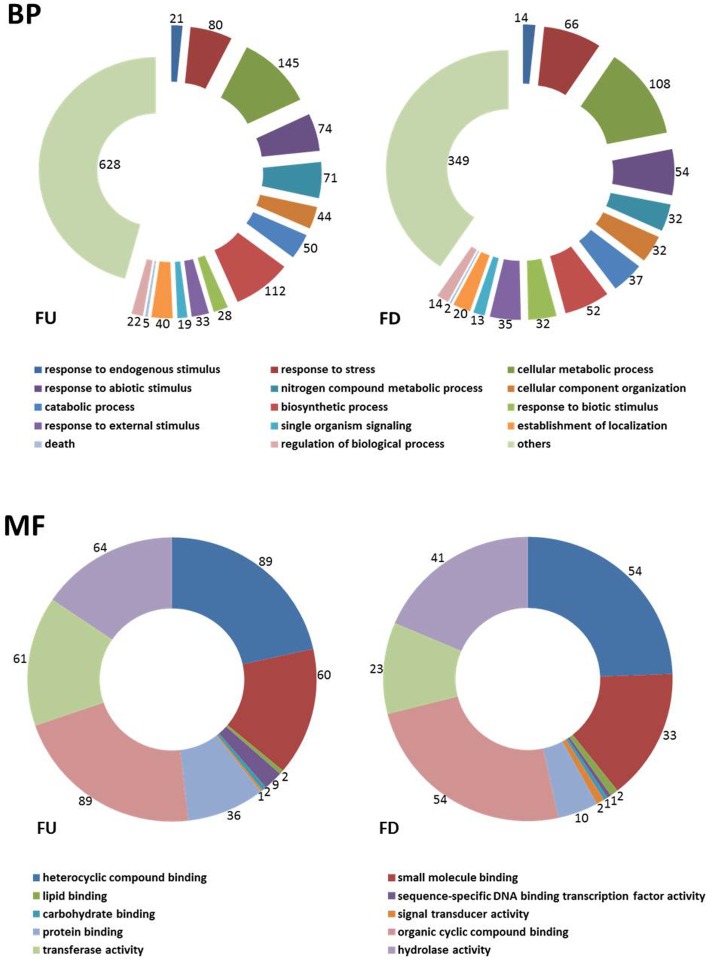
Blast2GO analysis of the EST set enabled annotation of expressed sequences according to the terms of the three main GO vocabularies, i.e., “Biological Process” (BP), “Molecular Function” (MF), and “Cellular Component” (CC). GO annotation was only feasible for 57.3% (up-regulated genes) and 68.3% (down-regulated genes) of the sequences, i.e., 141 and 89 ESTs, respectively. A total of 140 FD and 88 FU assigned to “unknown” category and 1 FU and 1 FD assigned to “predicted” category were automatically excluded from the analysis by the program. Since a number of transcripts were identified by different GO terms, the mapped ESTs distribution for BP and MF main categories shown in Figure 1 resulted in more than 585 (FU) or 381 (FD) sequences. The distribution of assignments into GO categories “level 3” was 336 (FU) and 226 (FD) (for BP), 311 (FU) and 202 (FD) (for MF), and 266 (FU) and 201 (FD) (for CC). Regarding to BP main GO vocabulary, transcripts representing GO terms categories non-related to plant defense processes (e.g., anatomical structure development, primary metabolic process, reproductive process, etc.), were grouped as “other” (Figure 1, BP). Concerning to plant defense-related categories, ESTs found to be induced or repressed in above-ground organs upon V. dahliae olive root colonization were assigned to processes such as “response to stress” (80 FU and 66 FD unigenes), “response to abiotic stimuli” (74 FU and 54 FD), “response to biotic stimuli” (28 FU and 32 FD), “response to external stimuli” (33 FU and 35 FD) or “response to endogenous stimuli” (21 FU and 11 FD). GO terms included catalases (CATs), proteins involved in the phenylpropanoid pathway, ET biosynthesis (ACO) or terpenoids biosynthesis, proteins related to SA (ACL), linolenic acid metabolism and PR protein. In addition, we identified several transcription factors (TF) such as WRKY's (WRKY20, WRKY44, WRKY33, WRKY40) or GRAS1, ET-responsive TF rap2-12-like, elongation factors (EF-1α, EF-1β, and EF-1Δ) and proteins directly related with stress responses (for a complete list of unigenes, see Tables S1, S2).
Figure 1.
“Level 3” Gene Ontology (GO) “Biological processes” (BP) and “Molecular function” (MF) terms distribution of 381 unigenes repressed (FD) and 585 induced (FU) in olive (Olea europaea L.) aerial tissues colonized by Verticillium dahliae. Unigenes were categorized using the “Blast2GO” software.
For the main GO vocabulary term MF, 10 different categories were identified for “level 3” (Figure 1, MF). The three main categories were “heterocyclic compound binding” (89 unigenes in FU like disease resistance protein and 54 unigenes in FD such as ribulose-bisphosphate carboxylase oxygenase activase), “organic cyclic compound binding” (89 unigenes in FU as adp-ribosylation factor 2-like and 54 unigenes in FD such as glycine dehydrogenase) and “hydrolase activity” (64 unigenes in FU such as thaumatin-like protein and 41 unigenes in FD like cysteine proteinase inhibitor) (Figure 1 MF and Tables S1, S2).
Finally, the same pattern of categories distribution for main GO term vocabulary CC was found in both libraries (data not shown). Thus, most of the unigenes identified were assigned to: “cell part” (242 unigenes in FU such as glutamine synthetase and 187 unigenes in FD like selenium-binding protein 2) and “membrane-bounded organelle” (180 unigenes in FU like ET-responsive transcription factor rap2-12-like and 149 unigenes in FD as protein thylakoid chloroplastic-like) (Tables S1, S2).
Identification of defense response genes in FU and FD libraries
Analysis of the 585 olive ESTs from the FU library showed that 19.8% of the induced genes in aerial olive tissues related with plant responses to different stimuli and stresses ([a]biotic, endogenous, extracellular, and/or external). For instance, genes potentially coding for a catalase or a tyrosyl-dna phosphodiesterase 1-like (response to stress), calcium-dependent protein kinase sk5-like or calmodulin-binding family protein (response to endogenous stimuli), auxin-responsive protein iaa14-like or cysteine proteinase 15a-like (response to external stimuli), ATP sulfurylase chloroplastic-like or 2-oxoisovalerate dehydrogenase subunit beta mitochondrial-like (response to extracellular stimuli), peptidyl-prolyl cis-trans isomerase fkbp65-like (response to wounding), gdt1-like protein 4, ET-responsive transcription factor rap2-12-like, or protein dehydration-induced 19 homolog 4-like (response to abiotic stimulus), ACL, pectin methylesterase, linoleate 13s LOX 2-chloroplastic-like, feroina like protein or thaumatin like-protein (response to biotic stimulus) and hva22-like protein a-like (response to abiotic and endogenous stimulus, response to stress), were found to be up-regulated. In addition, several TF related to plant defense responses (i.e., WRKY factors or GRAS1) were identified in the FU cDNA library as well (Table S1).
Similarly, analysis of the 381 olive ESTs from the FD library showed that 25.7% of the repressed genes in above-ground tissues were also related with plant responses to different stimuli and stresses. Thus, presence of the pathogen in the roots repressed genes related to response to biotic stimuli such as defensin, chloroplastic 6-phosphogluconolactonase and ribose-5-phosphate isomerase, oxoglutarate 3-dioxygenase-like or phosphatase 2c 25. Some unigenes were identified in both libraries coding for calcium (Ca2+)-binding proteins likely related with plant defense reaction, e.g., calmodulin, calmodulin binding, Ca2+ transporting ATPase, Ca2+ exchanger 4-like, and Ca2+ dependent protein kinase sk5-like, ACL, as well as β-amylases (chloroplast beta amylase isoform and inactive beta amylase 9-like) (Tables S1, S2). It is worth mentioning that some of these unigenes shared the putative protein function but had different sequences and accession numbers in the databases (Tables S1, S2), while others shared the putative protein function and accession number but sequences were different (e.g., glucosidases) (Tables 3, 4). We also detected 37 unigenes with the same sequence, putative protein function and accession number in both libraries, accounting for 7.2% (FU) and 9.7% (FD) of the sequences in the libraries. Finally, it is interesting to emphasize that GO term assignment revealed that 4% (FU library) and 19% (FD library) of the identified unigenes related with the photosynthesis process. The complete lists of ESTs induced and repressed in above-ground organs after inoculation of olive roots with V. dahliae are shown in Tables S1, S2, respectively. In addition, Tables S3, S4 display contigs identified in FU and FD cDNA libraries with their corresponding contiguous/overlapping ESTs. The 37 ESTs founded in both libraries and ESTs whose functions were unknown are not included in these tables.
Data validation and time-course of expression of selected defense response olive genes to Verticillium dahliae
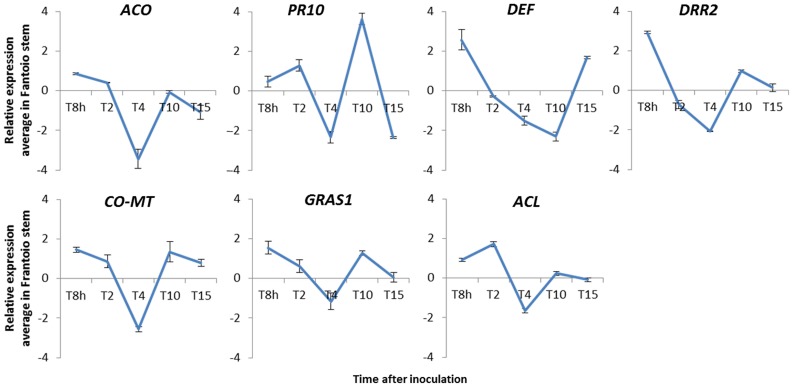
Seven genes identified in FU, FD or both libraries, CO-MT, GRAS1 (up-regulated genes), ACO, PR10, DEF, DRR2 (down-regulated genes), and ACL (found in both libraries) were selected to analyze short- and mid-term (8 h, 2, 4, 10, and 15 DAI) gene expression pattern. Overall, results showed a gene repression at 4DAI, followed by an increase at 10DAI. Most of the genes reached the maximal relative expression level within the first hours after pathogen inoculation (Figure 2). The two up-regulated genes identified in the FU library (CO-MT and GRAS1) were validated at all-time points except at 4DAI. Down regulation of two (ACO and DEF) out of four genes identified in the FD library was confirmed at three different times after pathogen inoculation. On the contrary, down-regulated expression of DRR2 and PR10 was validated only at two times (Figure 2). Finally, ACL gene, found in both libraries, displayed an ambiguous expression pattern along time. The vast majority of relative fold changes were assigned to medium (>+1.0 to ≤ +2.0 or < −1.0 to ≥ −2.0) or low (≥ −1.0 to ≤ +1.0 or ≤ 1.0 to ≥ −1.0) categories of differential expression, but for some cases such as DRR2 (8 h and 4DAI) and DEF (8 h and 10DAI), ACO and CO-MT (4DAI) or PR10 (4, 10, and 15DAI) which were assigned to the high category (>+2 or < −2) (Figure 2). Linear equations, correlation coefficients (R2) and PCR efficiencies for each gene are shown in Table 1.
Figure 2.
Relative expression (RE) average of seven genes identified in FD and/or FU cDNA libraries from aerial tissues of “Frantoio” olive plants at different time points after Verticillium dahliae inoculation in roots. ACO, 1-aminocyclopropane-1-carboxylate oxidase; DRR2, disease resistant response protein; ACL, acetone cyanohydrin lyase; GRAS1, transcription factor GRAS1; CO-MT, caffeoyl-o-methyltransferase; PR10, pathogenesis-related protein 10; and DEF, defensin protein 1. Error bars represent the SD from at least two independent qRT-PCR experiments. RE values (log2-fold-change values) were calculated according to the 2−ΔΔCt method (Livak and Schmittgen, 2001).
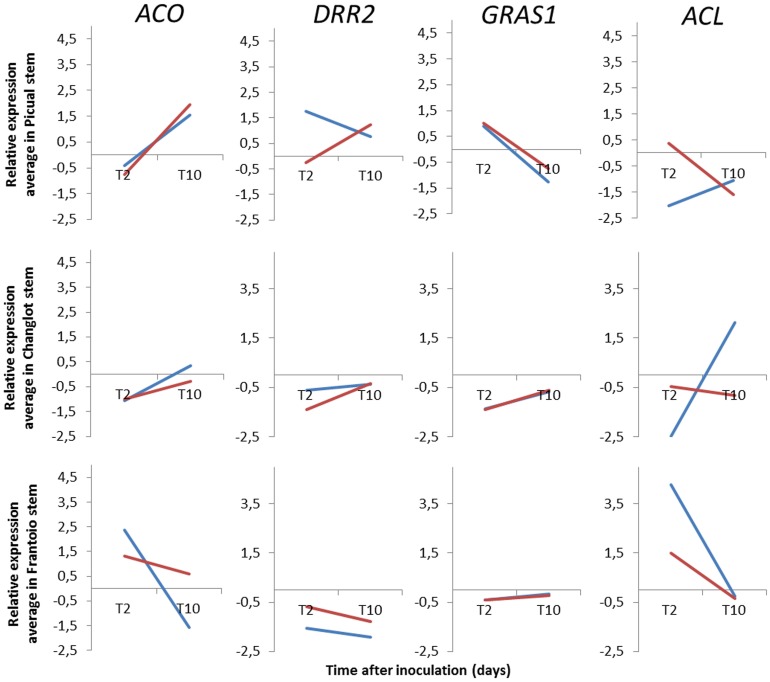
Gene expression pattern of GRAS1, ACO, DRR2 and ACL genes in susceptible and tolerant olive cultivars
In order to compare the gene expression pattern of specific genes in different olive cultivars (“Picual,” susceptible to VWO, and “Frantoio” and “Changlot Real,” tolerant to VWO), four genes (GRAS1, ACO, DRR2, and ACL) were selected (Table 2). Their expression patterns were assessed at 2 and 10DAI. Relative expression for each gene/cultivar combination is shown in Figure 3 (see Table S5 for additional data). Time-course expression of each gene differed among cultivars but the two biological replicas (plants) tested by cultivar did not always show the same expression pattern. ACO expression displayed the same trend in “Picual” and “Changlot Real” in contrast to “Frantoio.” Indeed, this gene showed an induction trend from 2 to 10DAI, particularly in “Picual” plants, whereas it showed a trend to be repressed in “Frantoio” plants along this interval. DRR2 expression is down-regulated in tolerant cultivars but up-regulated in the susceptible “Picual,” the two plants of this cultivar showing discrepant DRR2 expression patterns though (Figure 3). Expression of GRAS1 yielded the most consistent results, this gene being down regulated in tolerant cultivars at both sampling times. On the contrary, “Picual” plants showed a sharp fall in GRAS1 expression levels along time. Finally, results for ACL gene expression were the most contentious since plants of the same cultivar (i.e., “Picual” and “Changlot Real”) showed opposite expression patterns. In contrast, the two biological replicas of “Frantoio” displayed a consistent down regulation trend from 2 to 10DAI. For this gene, however, no clear association between VWO susceptibility and expression of defense genes was found.
Figure 3.
Relative expression (RE) average of four genes from aerial tissues of inoculated tolerant (“Frantoio” and “Changlot Real”) and susceptible (“Picual”) olive cultivars at two different time points after Verticillium dahliae inoculation in roots. ACO, 1-aminocyclopropane-1-carboxylate oxidase; DRR2, disease resistant response protein; ACL, acetone cyanohydrin lyase; GRAS1, transcription factor GRAS1; Relative expression values (log2-fold-change values) were calculated according to the 2−ΔΔCt method (Livak and Schmittgen, 2001). Red and blue lines show RE scored for two different plants sampled at the indicated times after inoculation with the pathogen.
Detection of V. dahliae DNA in roots of inoculated olive plants
Presence of V. dahliae was confirmed in inoculated plants of both tolerant (“Frantoio” and “Changlot Real”) and susceptible (“Picual”) plants. PCR assays allowed the detection of pathogen's DNA in roots of all inoculated plants. Control (non-inoculated) plants yielded negative results for V. dahliae DNA detection (data not shown).
Discussion
Understanding the mechanisms conferring olive tolerance to V. dahliae could provide valuable information for (i) breeding new VWO-tolerant cultivars, (ii) developing novel pathogen diagnostic tools, and (iii) improving integrated management strategies for VWO control. By generating two cDNA libraries, enriched in up- and down-regulated genes respectively, we have explored for the first time the transcriptomic changes taking place in a commercially-relevant woody plant such as olive upon infection by the D pathotype of V. dahliae, the most lethal for olive cultivation. The tolerant olive cultivar Frantoio was selected for such primary aim. We focused on the identification of systemic responses taking place in aerial tissues, particularly those related to defense to different (a)biotic stresses. A secondary objective was to assess the expression pattern of selected genes involved in such responses in olive cultivars showing differential susceptibility to VWO. This could provide valuable information on genetic markers associated to V. dahliae tolerance/susceptibility posing relevant practical implications in VWO management.
Our study has shown that, upon V. dahliae root inoculation, a range of early plant defense responses to (a)biotic stresses are induced (i.e., CO-MT., Hva22) and repressed (i.e., defensin, raffinose synthase) at distant tissues in “Frantoio” plants. Besides, different classes of TF-coding genes involved in (a)biotic stresses (Archana et al., 2009) such as GRAS1 and WRKYs (i.e., WRKY44, WRKY33, and WRKY20) were shown to be systemically up-regulated. For instance, Arabidopsis WRKY33 plays an important role in resistance to necrotrophic pathogens (Zheng et al., 2006), while A. thaliana WRKY20 has been involved in increased resistance to Bacillus amyloliquefaciens (Kjellin, 2012). Homologous genes to both WRKYs have been identified as systemically up-regulated in “Frantoio” upon root colonization by V. dahliae, suggesting the involvement of these TF in defense against a vascular pathogen. On the other hand, different elongation factors such as EF-1α were found in both cDNA libraries. The involvement of EF-1α in translation regulation during abiotic stresses in soybean (Chung et al., 2009), or during cold stress-related expression in barley and maize (Dunn et al., 1993; Berberich et al., 1995) has been reported. Finally, genes coding for β-amylases were also found in both libraries. These enzymes have been recently proposed as negative regulators in Arabidopsis partial resistance against V. dahliae (Gkizi et al., 2015). The role of detected olive elongation factors and β-amylases in the interaction olive-V. dahliae remains to be elucidated.
Seven genes related to defense responses (namely ACO, CO-MT, ACL, GRAS1, DRR2, PR10, and DEF) were selected to analyze their time-course expression patterns in “Frantoio.” ACO (ACC oxidase), involved in ethylene biosynthesis is regulated by a number of environmental factors and (a)biotic stresses (Díaz et al., 2002; Wang et al., 2002; Iwai et al., 2006). Furthermore, silencing of ACO gene was proven to affect the susceptible plant response to pathogen attack (Shan and Goodwin, 2006). CO-MT is involved in the phenylpropanoids pathway which plays important roles in plant development and (a)biotic stress responses (Dixon et al., 2002; Korkina, 2007). ACL is a salicylic acid binding protein involved in cyanogenic glycosides catabolism (Trummler and Wajant, 1997), related to chemical defense systems in plants under different biotic stresses (Ganjewala et al., 2010). GRAS1 is a TF belonging to a large protein family, many of them involved in plant response to (a)biotic stress (Mayrose et al., 2006; Sun et al., 2012). DRR2 is a disease resistance response protein of the dirigent family protein (DIR). Many DIR gene homologs have been detected in various plant species (Davin and Lewis, 2000). PR10 is a pathogenesis-related (PR) protein, a well-known group that constitute a defense response system under (a)biotic stresses (van Loon et al., 2006). Finally, DEF is a protein belonging to a plant defensin family proteins implicated in the first-line host defense against fungal pathogens (Thomma et al., 2002), although certain defensin genes are down-regulated (Moreno et al., 1994; Segura et al., 1998). Results from qRT-PCR revealed an overall decrease of the relative expression in all tested genes at 4DAI. This transient down-regulation was also observed in above-ground tissues during the interaction Pseudomonas fluorescens PICF7-olive (cv. Arbequina) roots (Gómez-Lama Cabanás et al., 2014). One possible explanation could be that plants undergo an overall gene expression repression at this time point, regardless whether they interact with a beneficial endophyte (Gómez-Lama Cabanás et al., 2014) or with a vascular pathogen (this study). Another explanation could be that plants were not exposed to a full day-light period during the first days of the experiment to avoid additional stress after manipulation. It is known that light exposure and intensity can influence the ability of plants to defend themselves from biotic stresses (Graham and Graham, 1996; Asai et al., 2000; Brodersen et al., 2002). qRT-PCR assays also showed that putative olive CO-MT and GRAS1 genes were validated at all tested times indicating that, at least, the phenylpropanoid pathway and one TF related to defense response are systemically induced during a prolonged period of time upon V. dahliae-olive roots. On the contrary, validation of putative olive genes ACL, ACO, DRR2, PR10, and DEF was not achieved at all tested time points. This suggests that these genes are expressed in a transient way and/or at time points other than those chosen in this study.
The expression patterns of ACO, ACL, GRAS1, and DRR2 in “Frantoio” plants in the second experiment showed some differences compared to that observed for this cultivar in the first experiment, particularly for DRR2. Plants used in these experiments showed the same appearance and were at the same phenological stage, although they slightly differed in age (3-month-old vs. 8-month-old). This difference should be irrelevant for a woody, long-living plant such as olive. So far, we do not have any clear answer to explain this outcome. However, commonalities were found depending on the VWO susceptibility/tolerance level of olive cultivars tested. For instance, we found differences between GRAS1 and DRR2 relative expression patterns along time in tolerant cultivars (“Frantoio” and “Changlot Real”) compared to that in susceptible (“Picual”) plants. Indeed, GRAS1 showed a constant down-regulation, although displaying a slight trend to increase its expression from 2 to 10DAI in tolerant cultivars (a situation observed in the first experiment for “Frantoio” plants as well). In contrast, “Picual” plants showed a sharp shift form up-regulation (2DAI) to down-regulation (10DAI). The induction of a GRAS1 homologous gene (SLGRAS1) was shown during the incompatible interaction of tomato (Solanum lycopersicum Mill) plants with Pseudomonas syringe pv. tomato (Mysore et al., 2002; Mayrose et al., 2006). Accumulation of SLGRAS1 transcripts was also found during two incompatible interactions of resistant tomato plants with Xanthomonas campestris pv. vesicatoria (Mayrose et al., 2006). Accordingly, the expression pattern of this TF was different depending on the level of VWO susceptibility. We suggest that expression of olive GRAS1 can be repressed along time after V. dahliae infection in susceptible plants, while in tolerant cultivars systemic expression of this gene will show a trend to be induced after pathogen infection. Likewise, the olive DIR gene DRR2 was mostly observed to be up-regulated in “Picual” plants, in contrast to the overall down-regulation found in tolerant cultivars. This DIR gene was reported to be induced during the interaction of the resistant olive cultivar Moraiolo with Bactrocera oleae (olive fruit fly), although up-regulation of this gene was not validated (Corrado et al., 2012). Furthermore, some similarities can be established to the response of rice (Oryza sativa L.) to heat stress. Thus, Jagadish et al. (2010) reported that the amount of a DIR protein did not change significantly in an O. sativa heat-susceptible genotype, while in a moderately heat-tolerant genotype a significant decrease of this protein was observed. These findings seem to point to a down regulation of these genes to cope with different (a)biotic stresses.
No clear correlation was found between expression pattern of the rest of the evaluated genes and VWO susceptibility degree. Moreover, the two biological replicas (plants) tested per cultivar did not always show the same expression pattern. This could be due to differences in pathogen colonization (spatial and/or temporal) for each plant. However, absence of V. dahliae infection can be ruled out since pathogen DNA was detected in the roots of all sampled plants. Even though the complexity of the pathosystem under study (i.e., large and complex root systems, uneven vascular localization by the pathogen, non-gnotobiotic study system, etc.), consistent results were found in some cases. Thus, ACO expression pattern in “Frantoio” plants was similar in both experiments, with a trend to be down regulated from 2 to 10DAI. However, no clear correlation between its pattern and VWO tolerance was observed since “Picual” and “Changlot Real” plants showed the same expression trend for this gene along time. Accordingly, ACO is not a useful marker to differentiate between VWO susceptible and tolerant cultivars. Difference in ACO expression (and in ACL as well, see below) found between “Changlot Real” and “Frantoio” plants (Figure 3) indicate that level of tolerance to V. dahliae may not be the same for both cultivars, as otherwise observed for the progenies obtained from these cultivars when they are used as genitors (Trapero et al., 2015). Thus, while “Frantoio” produced a large number of VWO tolerant seedlings, even when crossed with the susceptible “Picual,” “Changlot Real” produced many V. dahliae-susceptible descendants.
A putative olive ACL gene was found in both “Frantoio” cDNA libraries. However, different accession numbers were found when searching databases (EYU44351 in FU library and XP_004234906 in FD library). Primer pair used in qRT-PCR experiments did not discriminate between them. A third ACL was found in FD library (FD05-F10T7) but its sequence was different. A possible explanation for the presence of putative olive ACL in both libraries is the existence of two different alleles/genes (74 SNPs in 597 nucleotides overlapped; data not shown). Each library was constructed from a pool of RNAs sampled along a 21-days interval after V. dahliae inoculation, and expression of this gene could undergo fluctuation (up and down regulation) along this time period. While ACL expression pattern showed a consistent trend in “Frantoio” plants in both experiments (a gene repression trend from 2 to 10DAI), results from the second experiment did not allow correlating olive cultivar susceptibility to V. dahliae with expression of this gene. Therefore, as for ACO expression, ACL does not help to differentiate between VWO susceptible and tolerant olive cultivars. Actually, expression of this gene showed, overall, erratic among cultivars tested and even between plants of the same cultivar. Induction of ACL gene has been previously reported at both local (Schilirò et al., 2012) and systemic (Gómez-Lama Cabanás et al., 2014) level in olive (cv. Arbequina) aerial tissues upon root colonization by the beneficial root endophytic bacterium P. fluorescens PICF7. Nevertheless, validation of ACL up-regulation was not possible in above-ground tissues (Gómez-Lama Cabanás et al., 2014), confirming the difficulty to unravel the role of this gene and/or supporting the actual presence of different ACL genes/alleles in olive. These findings encourage more in-depth studies on the actual role of this gene(s). Differential expression of ACL gene(s) in both beneficial (olive-endophytic bacterium) and pathogenic (olive-vascular fungal pathogen) interactions points out to the possibility that this gene may be involved in a general plant defense responses against colonization of different microorganisms.
In conclusion, we report for the first time the broad, systemic transcriptomic changes taking place during the interaction between a devastating vascular fungal pathogen and a VWO tolerant cultivar of a woody host of high commercial relevance (i.e., not a model plant). We have demonstrated that many of these changes are related to defense responses. From the genetic responses detected, the differential expression of GRAS1 and DRR2 genes observed in the olive cultivars here assessed deserves further attention to be explored as markers of the susceptibility/tolerance level of olive genotypes to VWO. Information here reported not only shed light on defense mechanisms operating in olive against V. dahliae attack, but also could pave the way to develop novel tools in breeding for VWO resistance and management of this vascular pathogen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Research supported by grant AGL2009-07275 from Spanish MICINN/MINECO and P12-AGR667 from Junta de Andalucía (Spain), both co-financed by the ERDF of EU.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00928
References
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Archana K., Rama N., Mamrutha H. M., Nataraja K. N. (2009). Down-regulation of an abiotic stress regulated Nicotiana benthamiana WRKY transcription factor induces physiological abnormalities. Indian J. Biotechnol. 8, 53–60. [Google Scholar]
- Arias-Calderón R., León L., Bejarano-Alcázar J., Belaj A., de la Rosa R., Rodríguez-Jurado D. (2015). Resistance to Verticillium wilt in olive progenies from open-pollination. Sci. Hortic. 185, 34–42. 10.1016/j.scienta.2015.01.015 [DOI] [Google Scholar]
- Asai T., Stone J. M., Heard J. E., Kovtun Y., Yorgey P., Sheen J., et al. (2000). Fumonisin B1-induced cell death in Arabidopsis protoplasts requires jasmonate-, ethylene-, and salicylate-dependent signaling pathways. Plant Cell 12, 1823–1836. 10.1105/tpc.12.10.1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asif M. H., Dhawan P., Nath P. (2000). A simple procedure for the isolation of high quality RNA from ripening banana fruit. Plant Mol. Biol. Rep. 18, 109–115. 10.1007/BF02824018 [DOI] [Google Scholar]
- Baídez A. G., Gomez P., Del Rio J. A., Ortuno A. (2007). Dysfunctionality of the xylem in Olea europaea L. Plants associated with the infection process by Verticillium dahliae Kleb. Role of phenolic compounds in plant defense mechanism. J. Agr. Food Chem. 55, 3373–3377. 10.1021/jf063166d [DOI] [PubMed] [Google Scholar]
- Berberich T., Sugawara K., Harada M., Kusano T. (1995). Molecular cloning, characterization and expression of an elongation factor 1 alpha gene in maize. Plant Mol. Biol. 29, 611–615. 10.1007/BF00020988 [DOI] [PubMed] [Google Scholar]
- Brodersen P., Petersen M., Pike H. N., Olszak B., Skov-Petersen S., Oedum N., et al. (2002). Knockout of Arabidopsis Accelerated-cell-death 11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Gene. Dev. 16, 490–502. 10.1101/gad.218202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubici G., Cirulli M. (2012). Control of Verticillium wilt of olive by resistant rootstocks. Plant Soil 352, 363–376. 10.1007/s11104-011-1002-9 [DOI] [Google Scholar]
- Carder J. H., Morton A., Tabrett A. M., Barbara D. J. (1994). Detection and differentiation by PCR of sub-specific groups within two Verticillium species causing vascular wilts in herbaceous hosts, in Modern Assays for Plant Pathogenic Fungi: Identification, Detection and Quantification. Proceedings of COST-88 Meeting, Oxford, 1993, eds Schots A., Dewey F. M., Oliver R. (Wallingford: CAB International; ), 91–97. [Google Scholar]
- Chung E., Cho C. W., So H. A., Yun B. H., Lee J. H. (2009). Differential expression of soybean SLTI100 gene encoding translation elongation factor 1A by abiotic stresses. J. Plant Biotechnol. 36, 255–260. 10.5010/JPB.2009.36.3.255 [DOI] [Google Scholar]
- Conesa A., Götz S., García-Gómez J. M., Terol J., Talón M., Robles M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- Corrado G., Alagna F., Rocco M., Renzone G., Varricchio P., Coppola V., et al. (2012). Molecular interaction between the olive and the fruit fly Bactrocera oleae. BMC Plant Biol. 12:86. 10.1186/1471-2229-12-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daayf F., Nicole M., Boher B., Pando-Bahuon A., Geiger J. P. (1997). Early vascular defense reactions of cotton roots infected with a defoliating mutant strain of Verticillium dahliae. Eur. J. Plant Pathol. 103, 125–136. 10.1023/A:1008620410471 [DOI] [Google Scholar]
- Davin L. B., Lewis N. G. (2000). Dirigent proteins and dirigent sites explain the mystery of specificity of radical precursor coupling in lignan and lignin biosynthesis. Plant Physiol. 123, 453–461. 10.1104/pp.123.2.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derksen H., Rampitsch C., Daayf F. (2013). Signaling cross-talk in plant disease resistance. Plant Sci. 207, 79–87. 10.1016/j.plantsci.2013.03.004 [DOI] [PubMed] [Google Scholar]
- Diatchenko L., Lau Y. F., Campbell A. P., Chenchik A., Moqadam F., Huang B., et al. (1996). Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. U.S.A. 9, 6025–6030. 10.1073/pnas.93.12.6025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz J., ten Have A., van Kan J. A. L. (2002). The role of ethylene and wound signaling in resistance of tomato to Botrytis cinerea. Plant Physiol. 129, 1341–1351. 10.1104/pp.001453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon G. R., Pegg G. F. (1969). Hyphal lysis and tylose formation in tomato cultivars infected by V. albo-atrum. Trans. Br. Mycol. Soc. 53, 109–118. 10.1016/S0007-1536(69)80012-4 [DOI] [Google Scholar]
- Dixon R. A., Achnine L., Kota P., Liu C. J., Reddy M. S. S., Wang L. J. (2002). The phenylpropanoid pathway and plant defense a genomics perspective. Mol. Plant Pathol. 3, 371–390. 10.1046/j.1364-3703.2002.00131.x [DOI] [PubMed] [Google Scholar]
- Dunn M. A., Morris A., Jack P. L., Hughes M. A. (1993). A low-temperature-responsive translation elongation factor 1alpha from barley (Hordeum vulgare L.). Plant Mol. Biol. 23, 221–225. 10.1007/BF00021434 [DOI] [PubMed] [Google Scholar]
- Ganjewala D., Kumar S., Devi S. A., Ambika K. (2010). Advances in cyanogenic glycosides biosynthesis and analyses in plants: a review. Acta Biol. Szegediensis 54, 1–14. 4950985 [Google Scholar]
- Gkizi D., Santos-Rufo A., Rodríguez-Jurado D., Paplomatas E. J., Tjamos S. E. (2015). The β-amylase genes: negative regulators of disease resistance for Verticillium dahliae. Plant Pathol. [Epub ahead of print]. 10.1111/ppa.12360 [DOI] [Google Scholar]
- Gómez-Lama Cabanás C., Schiliró E., Valverde-Corredor A., Mercado-Blanco J. (2014). The biocontrol endophytic bacterium Pseudomonas fluorescens PICF7 induces systemic defense responses in aerial tissues upon colonization of olive roots. Front. Microbiol. 5:427. 10.3389/fmicb.2014.00427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham T. L., Graham M. Y. (1996). Signaling in soybean phenylpropanoid responses (dissection of primary, secondary, and conditioning effects of light, wounding, and elicitor treatments). Plant Physiol. 110, 1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt R. (1999). Phytoalexins: what have we learned after 60 years? Annu. Rev. Phytopathol. 37, 285–306. 10.1146/annurev.phyto.37.1.285 [DOI] [PubMed] [Google Scholar]
- Iwai T., Miyasaka A., Seo S., Ohashi Y. (2006). Contribution of ethylene biosynthesis for resistance to blast fungus infection in young rice plants. Plant Physiol. 142, 1202–1215. 10.1104/pp.106.085258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadish S. V. K., Muthurajan R., Oane R., Wheeler T. R., Heuer S., Bennett J., et al. (2010). Physiological and proteomic approaches to address heat tolerance during anthesis in rice (Oryza sativa L.). J. Exp. Bot. 61, 143–156. 10.1093/jxb/erp289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Yu J., Mahoney N., Chan K. L., Molyneux R. J., Varga J., et al. (2008). Elucidation of the functional genomics of antioxidant based inhibition of aflatoxin biosynthesis. Int. J. Food Microbiol. 122, 49–60. 10.1016/j.ijfoodmicro.2007.11.058 [DOI] [PubMed] [Google Scholar]
- Kjellin J. (2012). Role of WRKY20 Transcription Factor and Raffinose in Plant Defense Responses Upon Bacillus Amyloliquefaciens Strain 5113-Mediated Priming in Arabidopsis thaliana. Bachelor's Thesis, Dept. of Plant Biology and Forest Genetics University of Uppsala.
- Korkina L. G. (2007). Phenylpropanoids as naturally occurring antioxidants: from plant defense to human health. Cell. Mol. Biol. 53, 15–25. 10.1170/T772 [DOI] [PubMed] [Google Scholar]
- Larroque M., Belmas E., Martinez T., Vergnes S., Ladouce N., Lafitte C., et al. (2013). Pathogen-associated molecular pattern-triggered immunity and resistance to the root pathogen Phytophthora parasitica in Arabidopsis. J. Exp. Bot. 64, 3615–3625. 10.1093/jxb/ert195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- López-Escudero F. J., Del Río C., Caballero J. M., Blanco-López M. A. (2004). Evaluation of olive cultivars for resistance to Verticillium dahliae. Eur. J. Plant Pathol. 110, 79–85. 10.1023/B:EJPP.0000010150.08098.2d [DOI] [Google Scholar]
- López-Escudero F. J., Mercado-Blanco J. (2011). Verticillium wilt of olive: a case study to implement an integrated strategy to control a soil-borne pathogen. Plant Soil 344, 1–50. 10.1007/s11104-010-0629-2 [DOI] [Google Scholar]
- Maldonado-González M. M., Schiliró E., Prieto P., Mercado-Blanco J. (2015). Endophytic colonization and biocontrol performance of Pseudomonas fluorescens PICF7 in olive (Olea europaea L.) are determined neither by pyoverdine production nor swimming motility. Environ. Microbiol. 17, 3139–3153. 10.1111/1462-2920.12725 [DOI] [PubMed] [Google Scholar]
- Markakis E. A., Tjamos S. E., Antoniou P. P., Roussos P. A., Paplomatas E. J., Tjamos E. C. (2010). Phenolic responses of resistant and susceptible olive cultivars induced by defoliating and non-defoliating Verticillium dahliae pathotypes. Plant Dis. 94, 1156–1162. 10.1094/PDIS-94-9-1156 [DOI] [PubMed] [Google Scholar]
- Martos-Moreno C., López-Escudero F. J., Blanco-López M. Á. (2006). Resistance of olive cultivars to the defoliating pathotype of Verticillium dahliae. Hort Sci. 41, 1313–1316. [Google Scholar]
- Mayrose M., Ekengren S. K., Melech-Bonfil S., Martin G. B., Sessa G. (2006). A novel link between tomato GRAS genes, plant disease resistance and mechanical stress response. Mol. Plant Pathol. 7, 593–604. 10.1111/j.1364-3703.2006.00364.x [DOI] [PubMed] [Google Scholar]
- Mercado-Blanco J., Rodríguez-Jurado D., Pérez-Artés E., Jiménez-Díaz R. M. (2002). Detection of the defoliating pathotype of Verticillium dahliae pathotype in infected olive plants by nested PCR. Eur. J. Plant Pathol. 108, 1–13. 10.1023/A:1013994827836 [DOI] [PubMed] [Google Scholar]
- Moreno M., Segura A., García-Olmedo F. (1994). Pseudothionin, a potato peptide active against potato pathogens. Eur. J. Biochem. 223, 135–139. 10.1111/j.1432-1033.1994.tb18974.x [DOI] [PubMed] [Google Scholar]
- Mueller W. C., Morgham A. T. (1993). Ultrastructure of the vascular responses of cotton to Verticillium dahliae. Can. J. Bot. 71, 32–36. 10.1139/b93-004 [DOI] [Google Scholar]
- Mysore K. S., Crasta O. R., Tuori R. P., Folkerts O., Swirsky P. B., Martin G. B. (2002). Comprehensive transcript profiling of Pto- and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J. 32, 299–315. 10.1046/j.1365-313X.2002.01424.x [DOI] [PubMed] [Google Scholar]
- Okubara P. A., Paulitz T. C. (2005). Root defense responses to fungal pathogens: a molecular perspective. Plant Soil 274, 215–226. 10.1007/s11104-004-7328-9 [DOI] [Google Scholar]
- Prieto P., Mercado-Blanco J. (2008). Endophytic colonization of olive roots by the biocontrol strain Pseudomonas fluorescens PICF7. FEMS Microbiol. Ecol. 64, 297–306. 10.1111/j.1574-6941.2008.00450.x [DOI] [PubMed] [Google Scholar]
- Robb J. (2007). Verticillium tolerance: resistance, susceptibility, or mutualism? Can. J. Bot. 85, 903–910. 10.1139/B07-09326306814 [DOI] [Google Scholar]
- Schilirò E., Ferrara M., Nigro F., Mercado-Blanco J. (2012). Genetic responses induced in olive roots upon colonization by the biocontrol endophytic bacterium Pseudomonas fluorescens PICF7. PLoS ONE 7:e48646. 10.1371/journal.pone.0048646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura A., Monero M., Molina A., Garcia-Olmedo F. (1998). Novel defensin subfamily from spinach (Spinacia oleracea). FEBS Lett. 435, 159–162. 10.1016/S0014-5793(98)01060-6 [DOI] [PubMed] [Google Scholar]
- Shan X. C., Goodwin P. H. (2006). Silencing an ACC oxidase gene affects the susceptible host response of Nicotiana benthamiana to infection by Colletotrichum orbiculare. Plant Cell Rep. 25, 241–247. 10.1007/s00299-005-0063-8 [DOI] [PubMed] [Google Scholar]
- Sun X., Jones W. T., Rikkerink E. H. A. (2012). GRAS proteins: the versatile roles of intrinsically disordered proteins in plant signaling. Biochem. J. 442, 1–1210. 10.1042/BJ20111766 [DOI] [PubMed] [Google Scholar]
- Thomma B. P., Cammue B. P., Thevissen K. (2002). Plant defensins. Planta 216, 193–202. 10.1007/s00425-002-0902-6 [DOI] [PubMed] [Google Scholar]
- Trapero C., Rallo L., López-Escudero F. J., Barranco D., Díez C. M. (2015). Variability and selection of Verticillium wilt resistant genotypes in cultivated olive and in the Olea genus. Plant Pathol. 64, 890–900. 10.1111/ppa.12330 [DOI] [Google Scholar]
- Trapero C., Serrano N., Arquero O., Trapero A., López-Escudero F. J. (2013). Field resistance to Verticillium wilt in selected olive cultivars grown in two naturally infested soils. Plant Dis. 97, 668–674. 10.1094/PDIS-07-12-0654-RE [DOI] [PubMed] [Google Scholar]
- Trummler K., Wajant H. (1997). Molecular cloning of acetone cyanohydrin lyase fromflax (Linum usitatissimum). Definition of a novel class of hydroxynitrile lyases. J. Biol. Chem. 272, 4770–4774. 10.1074/jbc.272.8.4770 [DOI] [PubMed] [Google Scholar]
- Tsror L. (2011). Epidemiology and control of Verticillium wilt on olive. Isr. J. Plant Sci. 59, 59–69. 10.1560/IJPS.59.1.59 [DOI] [Google Scholar]
- van Loon L. C., Rep M., Pieterse C. M. J. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. 10.1146/annurev.phyto.44.070505.143425 [DOI] [PubMed] [Google Scholar]
- Wang K. L., Li H., Ecker J. R. (2002). Ethylene biosynthesis and signaling networks. Plant Cell 14, S131–S151. 10.1105/tpc.001768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadeta K. A., J Thomma B. P. (2013). The xylem as battleground for plant hosts and vascular wilt pathogens. Front. Plant Sci. 4:97. 10.3389/fpls.2013.00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov E. M., Apweiler R. (2001). InterProScan-an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17, 847–848. 10.1093/bioinformatics/17.9.847 [DOI] [PubMed] [Google Scholar]
- Zheng Z., Qamar S. A., Chen Z., Mengiste T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48, 592–605. 10.1111/j.1365-313X.2006.02901.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.