Abstract
Acinetobacter baumannii is an important opportunistic pathogen associated with a variety of nosocomial infections. A rapid and sensitive molecular detection in clinical isolates is quite needed for the appropriate therapy and outbreak control of A. baumannii. Group 2 carbapenems have been considered the agents of choice for the treatment of multiple drug-resistant A. baumannii. But the prevalence of carbapenem-resistant A. baumannii (CRAB) has been steadily increasing in recent years. Here, we developed a loop-mediated isothermal amplification (LAMP) assay for the rapid detection of A. baumannii in clinical samples by using high-specificity primers of the blaOXA-51 gene. Then we investigated the OXA-carbapenemases molecular epidemiology of A. baumannii isolates in two comprehensive hospitals in Beijing. The results showed that the LAMP assay could detect target DNA within 60 min at 65°C. The detection limit was 50 pg/μl, which was about 10-fold greater than that of PCR. Furthermore, this method could distinguish A. baumannii from the homologous A. nosocomialis and A. pittii. A total of 228 positive isolates were identified by this LAMP-based method for A. baumannii from 335 intensive care unit patients with clinically suspected multi-resistant infections in two hospitals in Beijing. The rates of CRAB are on the rise and are slowly becoming a routine phenotype for A. baumannii. Among the CRABs, 92.3% harbored both the blaOXA-23 and blaOXA-51 genes. Thirty-three pulsotypes were identified by pulsed-field gel electrophoresis, and the majority belonged to clone C. In conclusion, the LAMP method developed for detecting A. baumannii was faster and simpler than conventional PCR and has great potential for both point-of-care testing and basic research. We further demonstrated a high distribution of class D carbapenemase-encoding genes, mainly OXA-23, which presents an emerging threat in hospitals in China.
Keywords: Acinetobacter baumannii, LAMP, rapid diagnosis, carbapenem resistance, molecular epidemiology
Introduction
Acinetobacter baumannii is an important opportunistic pathogen associated with a variety of nosocomial infections, such as ventilator-associated pneumonia, central line-associated bloodstream infections, urinary tract infections, surgical-site infections, and other types of wound infections, especially in intensive care units (ICUs; Peleg et al., 2008). This organism is well adapted to hospital environments, being capable of continually spreading to new patients and making itself a nosocomial pathogen of particular clinical concern and a public health threat (Dijkshoorn et al., 2007).
In recent years, multidrug-resistant A. baumannii (MDR-AB) isolates have been increasingly reported worldwide. MDR-AB strains are associated with an enhanced risk of mortality and prolonged durations of hospitalization (Kempf and Rolain, 2012; Antunes et al., 2014; Lemos et al., 2014). Carbapenems, mainly imipenem (IPM) and meropenem (MPM), have been used to treat MDR-AB infections (Perez et al., 2007). However, the incidence of carbapenem resistance in A. baumannii is growing steadily in China and many other countries (Poirel and Nordmann, 2006; Tiwari et al., 2012a,b). According to the CHINET 2010 year reports, the resistant to IPM and MPM in A. baumannii isolates was 62.3% and 63.8%, respectively (Xi et al., 2012). However, few data are available regarding the molecular epidemiology and antibiotic resistance of A. baumannii infections in hospitals in Beijing, China.
The accurate and rapid identification of A. baumannii is critical for appropriate infection control in hospital settings. To date, the most common and widespread detection methods include characterization via a phenotypic system and commercial phenotypic methods (e.g., the VITEK 2 system [Biomerieux] and the API 20 NE system) or DNA-based testing such as PCR (e.g., 16S rRNA gene amplification), which have been used to successfully identify most Acinetobacter species. However, there are some limitations in these methods (Janda and Abbott, 2007; Karah et al., 2011; Álvarez-Buylla et al., 2012). For example, several days are needed for incubation, and the laboratory diagnosis of A. baumannii is actually for that of the A. calcoaceticus–A. baumannii complex (ABC), which includes A. calcoaceticus, A. baumannii, A. pittii (three species), and A. nosocomialis (species 13TU; Peleg et al., 2008). These bacterial species are different in terms of symptomatology, dissemination patterns, mechanisms of antibiotic resistance, and epidemiology (Golanbar et al., 2011; Chiang et al., 2012).
The loop-mediated isothermal amplification (LAMP) method, which is based on autocycling strand displacement DNA synthesis in the presence of Bst DNA polymerase, can be used to amplify target DNA with high specificity (as four or six specific primers that recognize six or eight different sequences on the DNA target) under isothermal conditions in less than 60 min (Notomi et al., 2000). LAMP is highly sensitive and able to detect DNA at as few as six copies in the reaction mixture, and less prone to the presence of irrelevant DNA than PCR (Notomi et al., 2000). This novel method has been developed widely for the detection of numerous pathogens, including influenza A subtypes H1N1 (Nakauchi et al., 2011), H5N1 (Dinh et al., 2011), H7N9 (Nakauchi et al., 2014), as well as Mycoplasma pneumoniae (Gotoh et al., 2013), Mycobacterium tuberculosis (Kumar et al., 2014), severe acute respiratory syndrome virus coronavirus (Poon et al., 2004), and human immunodeficiency virus (Zeng et al., 2014).
In present study, we describe a LAMP method for the rapid detection of A. baumannii in clinical samples targeting the blaOXA-51 gene, which was based on visual testing. We also investigated molecular-epidemiology and antibiotic-resistance profiles, and detected several common oxacillinase genes from A. baumannii isolates obtained at two comprehensive hospitals in Beijing, China.
Materials and Methods
Bacterial Strains, Antimicrobial Susceptibility Testing, and Pulsed-Field Gel Electrophoresis (PFGE)
A total of 34 strains representing 9 Acinetobacter species (2 A. baumannii strains and 10 other Acinetobacter strains) and 22 non-Acinetobacter species used in this study to develop the LAMP assays. Three hundred and fifty-five clinical sputum samples and nasopharyngeal swabs were obtained from the ICU hospitalized patients with clinically suspected multi-resistant infections in 307th (Affiliated Hospital of Academy of Military Medical Sciences), and the 309th Hospital of PLA in China. Pertinent information and the source of all strains are listed in Table 1. The species identification were identified by the Vitek 2 system (Biomerieux Vitek, Inc., Hazelwood, MO, USA). A. baumannii bacteria were grown overnight at 37°C in Luria-Bertani (LB) broth, while non-Acinetobacter species were cultured at 37°C in brain heart infusion (BHI) broth overnight.
Table 1.
Bacteria strains used in this study.
| Bacteria strains | Source | Strain type |
|---|---|---|
| Acinetobacter species | ||
| A. baumannii | CGMCC | Reference strain |
| A. baumannii ATCC 22933 | CICC | Reference strain |
| A. calcoaceticus | CGMCC | Reference strain |
| A. nosocomialis | CGMCC | Reference strain |
| A. haemolyticus | CGMCC | Reference strain |
| A. Pittii | CICC | Reference strain |
| A. johnsonii | CGMCC | Reference strain |
| A. junii | CGMCC | Reference strain |
| A. calcoaceticus | The Microorganism Centera | Reference strain |
| A. lwoffii | The Microorganism Center (three strains) | Reference strain |
| Non-Acinetobacter species | ||
| Bacillus megatherium 4623 | The Microorganism Center | Reference strain |
| Beta-haemolytic streptococcus group A CMCC32213 | The Microorganism Center | Reference strain |
| Bordetella pertussis ATCC 18530 | The Microorganism Center | Reference strain |
| Brucella suis 3572 | The Microorganism Center | Reference strain |
| Corynebacterium diphtheria CMCC38001 | The Microorganism Center | Reference strain |
| Enteropathogenic Escherichia coli 2348 | The Microorganism Center | Reference strain |
| Enterotoxigenic E. coli 44824 | The Microorganism Center | Reference strain |
| E. coli ATCC25922 | The Microorganism Center | Reference strain |
| Mycobacterium tuberculosis 8362 | The Microorganism Center | Reference strain |
| Neisseria meningitides group B CMCC29022 | The Microorganism Center | Reference strain |
| Pseudomonas aeruginosa ATCC27853 | The Microorganism Center | Reference strain |
| Salmonella aberdeen 9264 | The Microorganism Center | Reference strain |
| Salmonella enteritidis 50326-1 | The Microorganism Center | Reference strain |
| Stenotrophomonas maltophilia 3859 | The Microorganism Center | Reference strain |
| Shigella flexneri 4536 | The Microorganism Center | Reference strain |
| Shigella sonnei 2531 | The Microorganism Center | Reference strain |
| Staphylococcus aureus 2740 | The Microorganism Center | Reference strain |
| Vibrio carchariae 5732 | The Microorganism Center | Reference strain |
| Vibrio cholera 3802 | The Microorganism Center | Reference strain |
| Vibrio parahaemolyticus 5474 | The Microorganism Center | Reference strain |
| Yersinia enterocxolitica 1836 | The Microorganism Center | Reference strain |
| Yersinia pestis 2638 | The Microorganism Center | Reference strain |
CGMCC, China General Microbiological Culture Collection Center; CICC, China Center of Industrial Culture Collection. aThe Microorganism Center of the Institute of Disease Control and Prevention of the Academy of Military Medical Sciences.
Antibiotic susceptibility testing was conducted by disk diffusion in accordance with the guidelines of the Clinical and Laboratory Standards Institute and was confirmed by the Vitek 2 System with the following antibiotics: ampicillin, ampicillin/sulbactam, aztreonam, ciprofloxacin, ceftazidime, ceftriaxone, cefepime, gentamycin, levofloxacin, IPM, MPM, piperacillin/tazobactam, piperacillin, tobramycin, and sulfamethoxazole. Pseudomonas aeruginosa ATCC 27853 was used as a control.
Genomic DNA was prepared in agarose plugs, and digested with the restriction enzyme ApaI (New England BioLabs, Beverly, MA, USA). The DNA restriction fragments were separated using a CHEF-DRIII apparatus (Bio-Rad Laboratories, Richmond, CA, USA). Gel images were analyzed using BioNumerics software, version 6.01 (Applied-Maths, Belgium). The interpretation of gels was performed by visual inspection using the criteria of Tenover et al. (1995).
Preparation of Isolated DNA
Genomic DNA from Acinetobacter species was prepared using the boiling method described by Olive and Bean (1999), with some modifications. Briefly, colonies of each isolate were picked from LB plates and suspended in 100 μl of ddH2O. The bacterial suspensions were boiled at 95°C for 15 min, centrifuged at 12,000 rpm for 5 min, and the supernatants were collected and transferred to fresh Eppendorf tubes as templates for use in LAMP and PCR assays. To determine the sensitivity and specificity of the LAMP assays, bacterial genomic DNA was extracted from A. baumannii using the Wizard® Genomic DNA Purification Kit A1125 (Promega, Madison, WI, USA). The DNA concentration was measured by using OD260 measurements (ND-1000 spectrophotometer, NanoDrop Technologies, Inc, Wilmington, DE, USA) and prepared by serial 10-fold dilutions to yield concentrations ranging from 500 ng/μl to 0.05 pg/μl.
Primer Design for LAMP Assay
The sequence of blaOXA-51 (GenBank No. DQ385606.1) was downloaded from the NCBI GenBank database and used to design blaOXA-51-specific LAMP primers. The sequence was further analyzed by Primer Explorer software (version 4; http://www.primerexplorer.jp/lamp/), and five sets of primers were designed, including the outer forward primer (F3), the outer backward primer (B3), the forward inner primer (FIP), the backward inner primer (BIP), and additional loop primers (loops F and B), as shown in Table 2.
Table 2.
Sequence of primers used for specific amplification of blaOXA-51-like and the OXA carbapenemases PCR detection.
| Primer | Primer type | Sequence (5′–3′) |
|---|---|---|
| OXA-4F3 | Forward outer | CTTATATAGTGACTGCTAATCCAA |
| OXA-4B3 | Backward outer | ATTAAGCATTTTGAAGGTCGA |
| OXA-4FIP | Forward inner | ACCCGTAGTGTGTACTTCGTTAAATTTTTACAGCGCTTCAAAATCTGA |
| OXA-4BIP | Backward inner | TTAGTTATCCAACAAGGCCAAACTTTTTAGCAGGTACATACTCGGTC |
| OXA-4LB | Loop forward | AAAGCTATGGTAATGATCTTGCTCG |
| blaOXA-23-F | Forward | ATGAATAAATATTTTACTTG |
| blaOXA-23-R | Reverse | TTAAATAATATTCAGCTGTT |
| blaOXA-24-F | Forward | ATGAAAAAATTTATACTTCCTATATTCAGC |
| blaOXA-24-R | Reverse | TTAAATGATTCCAAGATTTTCTAGC |
| blaOXA-51-like-F | Forward | TAATGCTTTGATCGGCCTTG |
| blaOXA-51-like-R | Reverse | CTATAAAATACCTAATTKTTCTAA |
| blaOXA-58-F | Forward | ATGAAATTATTAAAAATATTGAGT |
| blaOXA-58-R | Reverse | ATAAATAATGAAAAACACCCAA |
LAMP Reaction and Product Detection
LAMP reactions were performed in a total volume of 25 μl and contained 12.5 μl reaction mixtures (DNA Amplification Kit; Eiken Chemical Co., Ltd., Tochigi, Japan), 2.6 μl primer mixture (40 pmol for FIP and BIP, 20 pmol for LF and LB, and 5 pmol for F3 and B3), and 1 μl of Bst polymerase (eight units, New England BioLabs, Ipswich, MA, USA). Finally, 1-μl DNA of genomic template DNA (the concentration is above 50 ng/μl) was added to the reaction tubes. Reaction was performed in a LA-320CE instrument (Eiken Chemical Co., Ltd., Tochigi, Japan) at 65°C for 60 min and stopped by heating to 80°C for 5 min, according to the manufacturer’s instructions. The LA-320CE instrument can monitor the turbidity of real-time LAMP products through spectrophotometric analysis by recording the optical density at 650 nm every 6 s with the help of a Loopamp real-time turbidimeter. In addition, a visual color-change detection method was employed. Briefly, 1 μl of fluorescence-detection reagent (Eiken Chemical Co., Ltd., Tochigi, Japan) was added to each 25 μl LAMP-reaction mixture prior to initiating the reactions. Positive reactions were identified by a green color change, while negative reactions remained orange in color. The color change could be observed by the naked eye under natural light, or with the aid of ultraviolet light excitation at 365 nm.
To compare the sensitivity and specificity of LAMP with PCR, normal PCR reactions were performed using the blaOXA-51-like-F and blaOXA-51-like-R primers (Table 2). The primers were synthesized commercially by Sangon Biotech Co., Ltd. (Beijing, China). PCR was performed as described previously (Kuo et al., 2012).
PCR Detection of Carbapenemase Genes
The class D OXA carbapenemases of Acinetobacter sp. are represented by 4 main phylogenetic subgroups: OXA-23-like, OXA-24-like, OXA-51-like, and OXA-58. The primers used for detecting these genes by PCR are listed in Table 2. PCR was performed in a 25-μl reaction mixtures containing 12.5 μl PCR Master Mix Reagent (Tiangen Biotech Co., Ltd., Beijing, China); 1 μl of forward primer (10 pmol), 1 μl of reverse primer (10 pmol), and 1 μl of DNA template. Reaction mixtures were initially heated to 94°C for 5 min, followed by 30 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 90 s. The final extension step was performed at 72°C for 7 min. The PCR-amplified products were analyzed by 1% agarose gel electrophoresis and stained with ethidium bromide. Images were acquired using a Bio-Rad Gel Doc EQ imaging system.
Results
Optimization of the LAMP Assay Targeting blaOXA-51-like
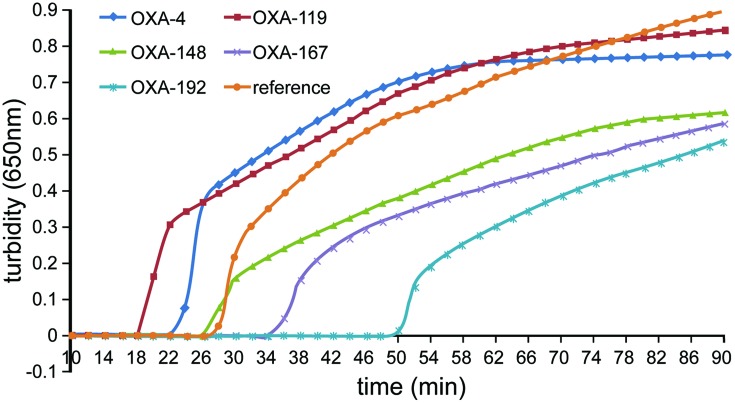
Five sets of different primers were initially tested for detection of the blaOXA-51-like gene, and the primers for pgaD gene of A. baumannii were added as a control (Wang et al., 2013). Four of the five primer sets enabled successful amplification (Figure 1). The OXA-4 primer set amplified the target sequence within the shortest time and was, therefore, chosen as the optimal primer set (Table 2).
FIGURE 1.
Five sets of primers were used to amplify the indicated target genes of Acinetobacter baumannii under the same conditions. Turbidity was monitored using a Loopamp real-time turbidimeter by measuring the absorbance at 650 nm every 6 s. The OXA-4 primer set was chosen as the most appropriate primers for the rapid detection of A. baumannii.
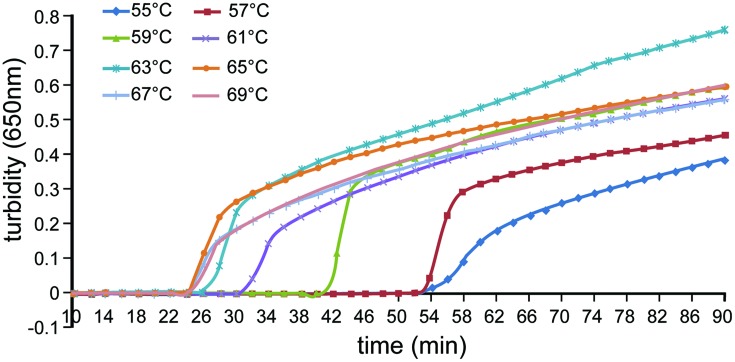
Reaction temperatures ranging from 55 to 69°C at a 2°C intervals were compared to determine optimal amplification conditions. The amplification efficiency was highest at 65°C (Figure 2) and was, therefore, used in subsequent experiments.
FIGURE 2.
Effect of differing temperatures on the efficiency of detection of A. baumannii by loop-mediated isothermal amplification (LAMP). Turbidity was monitored using a Loopamp real-time turbidimeter by measuring the absorbance at 650 nm every 6 s.
Specificity and Sensitivity of the LAMP Assay
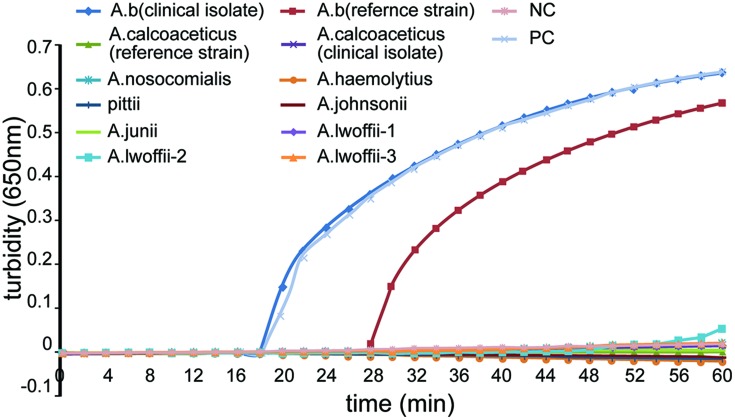
To evaluate the specificity of LAMP detection for A. baumannii, genomic DNA was extracted from 2 A. baumannii strains, as well as other 7 Acinetobacter species (10 strains) and 22 non-A. baumannii reference strains, and tested using real-time turbidity or visual detection of color changes as readouts. Genomic A. baumannii DNA (ATCC 22933) and distilled water were used as positive and negative controls, respectively. As shown in Figure 3, both methods of analysis positively identified the A. baumannii isolates. All other strains (as well as the blank control) tested negatively, indicating that the LAMP assay was specific for A. baumannii. Interestingly, the LAMP assay could differentiate A. baumannii from other Acinetobacter species.
FIGURE 3.
Specificity of the LAMP reactions in detecting the blaOXA-51-like gene. Turbidity was monitored using a Loopamp real-time turbidimeter by measuring the absorbance at 650 nm every 6 s. Amplification was performed at 65°C for 60 min.
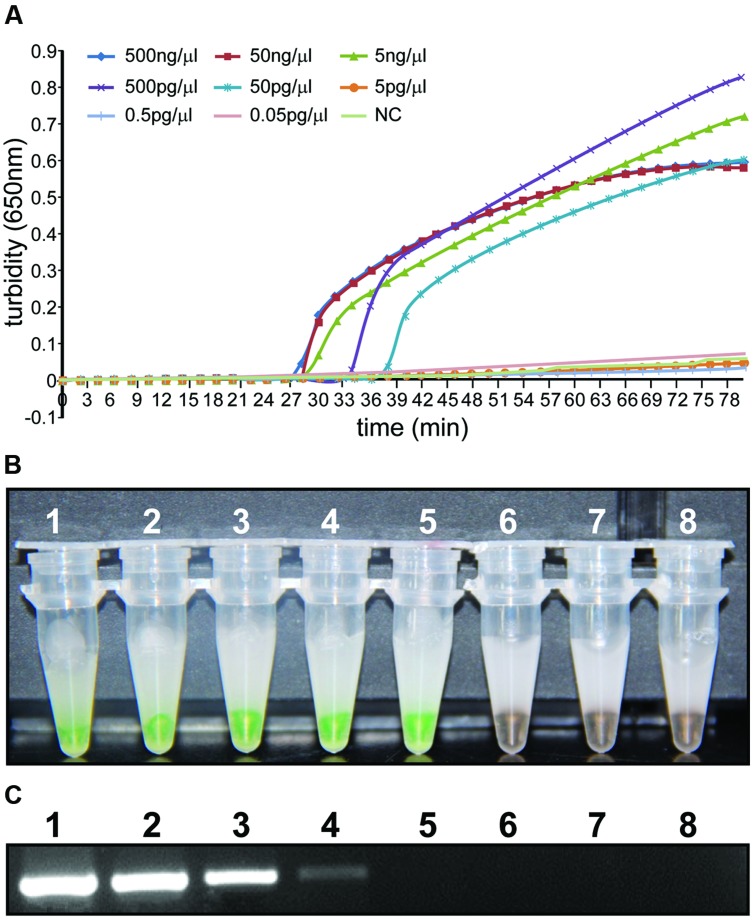
To compare the detection limit of traditional PCR with that of LAMP using either real-time turbidity or color-change measurements, 10-fold serial dilutions (50 ng/μl–5 pg/μl) were tested using genomic DNA extracted from A. baumannii ATCC 22933. As shown in Figure 4, the detection limits of real-time turbidity and visual detection were both 50 pg/μl, which was 10-fold more sensitive than traditional PCR assay.
FIGURE 4.
Sensitivity of the LAMP reaction and PCR for detection of the blaOXA-51-like gene. Genomic DNA was diluted in a set of serial 10-fold dilutions. Both LAMP reactions (A,B) and PCR (C) were performed in duplicate for each dilution. Tubes and lanes: 1: 500 ng/μl, 2: 50 ng/μl, 3: 5 ng/μl, 4: 500 pg/μl, 5: 50 pg/μl, 6: 5 pg/μl, 7: 0.5 pg/μl, 8: 0.05 pg/μl. (A) Turbidity was monitored using a Loopamp real-time turbidimeter by measuring the absorbance at 650 nm every 6 s. (B) The direct visual method for the detection of LAMP. One microliter of fluorescent detection reagent was added to 25 μl of LAMP reaction mixture before the LAMP reactions were initiated. (C) PCR products were separated by 1% agarose gel electrophoresis and stained with ethidium bromide.
Detection of A. baumannii in Clinical Samples
A total of 355 clinical sputum samples and nasopharyngeal swabs were collected for LAMP-based surveillance of blaOXA-51-like from ICU patients suspected of having multidrug-resistant infections in two hospitals in Beijing, China. Ten pairs of sputum samples and nasopharyngeal swabs from healthy people were collected as controls. All clinical samples were simultaneously analyzed by LAMP and PCR. Of the 355 clinical samples, the LAMP assay detected 228 positive samples and 127 negative samples, while the PCR assay detected 221 positive samples, and 134 negative samples. Then, A. baumannii was successfully cultured from all 228 samples that were positively identified by LAMP. Samples from the healthy control subjects all tested negatively by LAMP and PCR. The LAMP assay showed 100% specificity compared to 91.89% by PCR assay. Thus, the results showed the LAMP assays were more sensitive and specific than PCR for the diagnosis of A. baumannii in clinical samples.
Antibiotic Susceptibility and Oxacillinase Distribution
A total of 228 clinical A. baumannii isolates were characterized by antibiotic susceptibility testing using both the VITEK®2 system and disk diffusion. Resistance rates were very high, with nearly 90% isolates being resistant to ampicillin, 68.2% being MDR-AB, 66.5% being resistant to IPM, and 63.6% being resistant to MPM.
Among all 145 isolates resistant to both IPM and MPM, 100% harbored the blaOXA-51-like gene, 92.3% (134 strains) co-occurred with the blaOXA-23 gene, 1 isolate carrying both the blaOXA-51-like gene and blaOXA-58 gene, and no isolate carried the blaOXA-24 gene. Ten isolates were negative for the tested plasmid-mediated OXA-carbapenemase genes.
We also investigated the presence of the blaOXA-51-like gene in A. baumannii isolates susceptible to carbapenem treatment. All of these isolates harbored the naturally occurring blaOXA-51-like gene, making it an excellent candidate for the identification of Acinetobacter species.
Molecular Epidemiology of Carbapenem-Resistant A. baumannii
Forty-one out of 145 carbapenem-resistant A. baumannii isolates were analyzed by PFGE. Genomic DNA digested with ApaI showing similar patterns were designated as the same type, and those with 1–3 different bands compared with this type were designated as subtypes. Digested DNA with >3 different bands compared with one type were designated as other types. Bionumerisc software V6.01 was used to perform cluster analysis of these data.
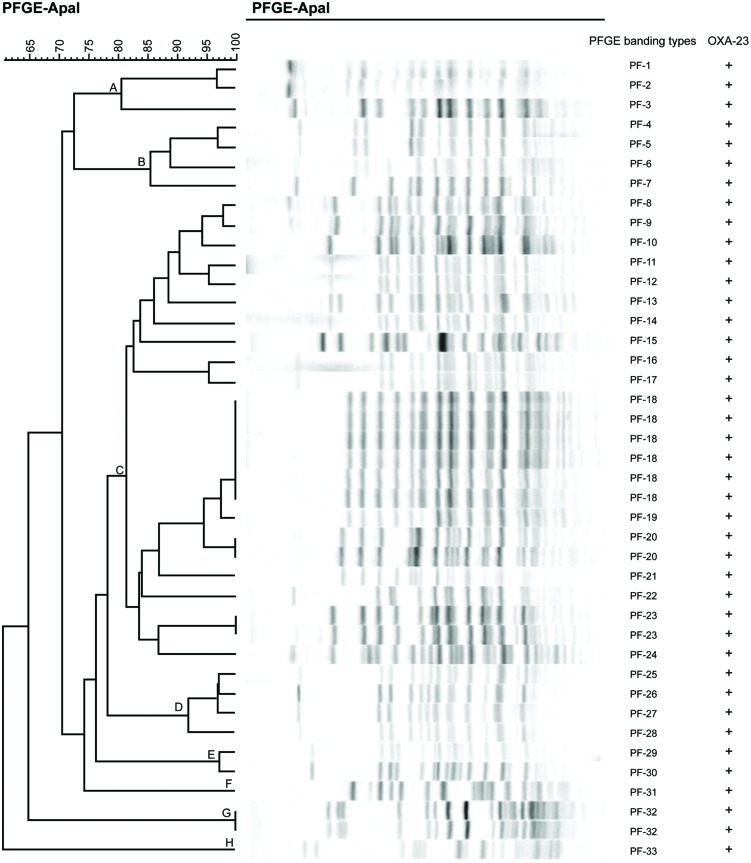
A total of 33 PFGE-banding types were identified, suggesting a diverse population of A. baumannii rather than the spread of a specific clone. According to the dice similarity index (80%), these types could be clustered into eight distinct PFGE patterns (clones A–H), and clone C was the dominant clone with 24 isolates (Figure 5). Clinical isolates with PFGE-banding type 18 (PF-18) represented the dominant epidemic strain in the ICU during the testing period.
FIGURE 5.
Pulsed-field gel electrophoresis (PFGE) of Apa I-digested DNA from 41 A. baumannii isolates. The dendrogram shown was generated via the unweighted pair-group method using arithmetic averages approach, based on Dice similarity coefficients that were determined using BioNumerics software, V6.01.
Discussion
Carbapenems including IPM and MPM are used as a last resort for treating MDR-AB. The incidence of carbapenem resistance in A. baumannii has increased steadily over the past decade and now represents a global problem. The Meropenem Yearly Susceptibility Test Information Collection program revealed a considerable worldwide increase in IPM and MPM resistance rates, which increased from 10 and 35% in 1999 to 47.9 and 57.4% in 2008, respectively (Rhomberg and Jones, 2009). Similarly, the SENTRY program documented an overall increase in IPM resistance from 34.5% in 2006 to 59.8% in 2009 (Gales et al., 2011). In the USA and Europe, carbapenem resistance accounted for 65% of A. baumannii-related pneumonia in 2012 (Farrell et al., 2014). In Asia, more than 60% of A. baumannii isolates causing hospital-acquired pneumonia were pan-resistant bacteria and resistant to carbapenem (Zarrilli et al., 2013; Tiwari and Moganty, 2014).
In this study, we observed that 66.5% (152 strains) or 63.6% (145 strains) of test isolates were resistant to IPM or MPM, respectively. The 228 clinical A. baumannii isolates were mainly collected from the ICUs of two hospitals; thus, the incidence of carbapenem resistance in A. baumannii was relatively high. Prolonged hospitalization, invasive medical procedures, and the prior use of broad-spectrum antibiotics (third-generation cephalosporins and fluoroquinolones) are risk factors for the acquisition of MDR-AB (Karageorgopoulos and Falagas, 2008). These findings indicated that treatment with carbapenem may potentially induce carbapenem resistance.
During PFGE analysis, 33 PFGE-banding patterns were identified and classified into 8 distinct clones, based on Tenover’s criteria (Tenover et al., 1995). Clone C was the dominant clone, making it possibly responsible for the A. baumannii epidemic in the ICUs of the two comprehensive hospitals from which we obtained the isolates. Clinical isolates of subtype PF-18 represented the main epidemic strain during the testing period. Previous results have demonstrated that carbapenem-resistant A. baumannii isolates showing the same PFGE patterns possess the same carbapenemase-associated genes (Yan et al., 2010; Park et al., 2012); however, our results were not consistent with these observations. Three isolates carrying only the blaOXA-51-like gene, but not the blaOXA-23 gene, were grouped into different clones. However, isolates carrying both the blaOXA-51-like gene and the blaOXA-23 gene were clustered into distinct clones, either. This may be due to the complicated resistance mechanism that underlies carbapenem resistance in A. baumannii.
Carbapenem susceptibility in A. baumannii is compromised by a variety of mechanisms. By far the most common mechanism are carbapenemases (Pogue et al., 2013). Carbapenem-hydrolyzing class D β-lactamases (CHDLs, Ambler class D), also known as oxacillinases, are the most common mediator of carbapenem resistance in A. baumannii (Poirel and Nordmann, 2006), although they are relatively inefficient compared with other types of carbapenemases, such as class B metallo-β-lactamases (MBLs; Ambler class B) represented by the VIM, IMP, SIM families, and the recently discovered New Delhi metallo-β-lactamase (NDM).
Among CHDLs in A. baumannii, there are four major distinct groups, including OXA-23, OXA-24, OXA-51-like, and OXA-58, which are encoded by the blaOXA-23, blaOXA-24, blaOXA-51-like, and blaOXA-58 genes, respectively (Poirel and Nordmann, 2006; Zavascki et al., 2010). The blaOXA-23 gene has been reported in A. baumannii isolates worldwide and is mediated both by chromosomal integration and plasmids. The blaOXA-24 gene can also be localized to chromosomes or plasmids, appears to be less widespread than the blaOXA-23 gene, and is relatively rare in China (Poirel et al., 2010). The chromosomally located blaOXA-51-like gene is unique and naturally occurring in A. baumannii; thus, this gene is becoming an important genetic marker for the identification of organisms at the species level (Turton et al., 2006,). The plasmid-mediated blaOXA-58 gene has been reported in Acinetobacter sp. from many countries around the world, but is rare in China.
In this research, we found that in all the 145 CRAB isolates, the co-occurrence of blaOXA-51-like gene and blaOXA-23 gene could be detected in 134 isolates (92.3%). This result indicated that the plasmid-mediated transfer of blaOXA-23 gene might be the most common mechanism by which A. baumannii becomes carbapenem resistance in these two comprehensive military hospitals in Beijing. Although the blaOXA-58 gene and blaOXA-24 gene have been reported in many countries, but the prevalence of these two genes is relatively rare in China. That could be explained that only one isolate carried the blaOXA-58 gene and no blaOXA-24 gene was detected in the CRAB isolates. However, 10 isolates could detect none of the plasmid-mediated OXA-carbapenemase genes including blaOXA-23, blaOXA-24 and blaOXA-58, which indicated that these isolates might harbored other resistant genes such as MBLs against carbapenem. These data are consistent with other reports from China and other countries (Turton et al., 2006; Woodford et al., 2006; Zavascki et al., 2010; Tiwari et al., 2012a).
In the conventional PCR assay, the blaOXA-51-like gene could not be amplified from seven clinical isolates. We thought that the low bacterial contents and incorrect pathogen identification may account for the failure to amplify these samples. Thus, the seven strains were analyzed for the presence of the blaOXA-51-like gene by the LAMP method described here, yielding positive results in each case, which was confirmed using the Vitek 2 system. Our results showed that these clinical isolates were actually A. baumannii. These results suggest that occasionally the sensitivity of conventional PCR is not satisfactory and may introduce errors into diagnostic test results.
Compared with traditional methods for pathogen diagnosis, LAMP assays can generate results more easily and rapidly, with high sensitivity and specificity (Notomi et al., 2000). The LAMP reaction does not require temperature cycling, with just a temperature-controlled water bath or a constant-temperature environment being sufficient. LAMP primers are designed to match four or six of the six or eight independent target-sequence regions, so the specificity and sensitivity are enhanced greatly. Under optimum conditions, the LAMP reaction can be completed in 1 h. In this study, we described a LAMP assay based on the blaOXA-51-like gene for detecting A. baumannii. This LAMP method could detect the target DNA within 60 min at an isothermal temperature of 65°C. The detection limit of the LAMP assay was 50 pg/μl, which was about 10-fold greater than that of PCR. Furthermore, the LAMP method described here could distinguish A. baumannii from A. nosocomialis (genospecies 13TU), A. pittii (genospecies 3), and the A. Baumannii complex (A. baumannii, A. nosocomialis, A. pittii, and A. calcoaceticus). Previously, there were two reports on the use of an A. baumannii LAMP assay, based on targeting the 16S–23S rRNA intergenic spacer sequence (Soo et al., 2013) and the conserved regions of the pgaD gene of A. baumannii (Wang et al., 2013). However, the former could not effectively distinguish A. baumannii from A. nosocomialis, and A. pittii, and the specificity of A. baumannii LAMP assay of the latter was only 75%.
Conclusion
A rapid, sensitive, specific, and effective LAMP assay was established for the detection of A. baumannii. The LAMP assay will be very useful for the rapid detection of pathogens in clinical samples. Meanwhile, this report provides some therapeutic recommendations for the treatment of A. baumannii infection and a warning that the growing emergence of carbapenem-resistant A. baumannii has led to an inadequacy of therapeutic choices in treating MDR-AB infections among patients in China. Our findings further emphasize that when investigating outbreaks caused by carbapenem-resistant A. baumannii, both the detection of carbapenemase-associated genes and PFGE are needed.
Author Contributions
Puyuan Li and Wenkai Niu wrote the main manuscript text. Puyuan Li, Huan Li, and Wei Liu prepared Figures 1–4. Leijing Guo prepared Figure 5. Puyuan Li, Wenkai Niu, Hong Lei, Xiangna Zhao, Dayang Zou, Xin Yuan, and Huiying Liu executed the experiments. Changqing Bai and Jing Yuan helped conceive the project and designed the experiments. All authors reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the 309th Hospital of PLA and Institute of Disease Control and Prevention, Academy of Military Medical Sciences for kindly providing clinical isolates and reference strains. We are grateful to Yin Xiuyun from the Department of Clinical Laboratory of the 307th Hospital of PLA for performing the collection of isolates. The analysis of PFGE by Wang Jian from the Institute of Disease Control and Prevention, Academy of Military Medical Sciences (Beijing, China) is gratefully appreciated. We also thank Wei Xiao, Cui Qian, Yang Zhan, Yan Xiabei, and Dong Derong for assistance during the course of this research.
Footnotes
Funding. This project was supported by the Beijing Natural Science Foundation (Grant No.7142118), a Translational Medicine Project from the Academy of Military Medical Science in 2013, Mega-projects of Science and Technology Research of China Grant 2013ZX10004-203, and Capital Characteristic Clinic project of Beijing (Z121107001012127).
References
- Álvarez-Buylla A., Culebras E., Picazo J. J. (2012). Identification of Acinetobacter species: is Bruker biotyper MALDI-TOF mass spectrometry a good alternative to molecular techniques? Infect. Genet. Evol. 12 345–349. 10.1016/j.meegid.2012.01.002 [DOI] [PubMed] [Google Scholar]
- Antunes L., Visca P., Towner K. J. (2014). Acinetobacter baumannii: evolution of a global pathogen. Pathog. Dis. 71 292–301. 10.1111/2049-632X.12125 [DOI] [PubMed] [Google Scholar]
- Chiang M.-C., Kuo S.-C., Chen S.-J., Yang S.-P., Lee Y.-T., Chen T.-L., et al. (2012). Clinical characteristics and outcomes of bacteremia due to different genomic species of Acinetobacter baumannii complex in patients with solid tumors. Infection 40 19–26. 10.1007/s15010-011-0187-4 [DOI] [PubMed] [Google Scholar]
- Dijkshoorn L., Nemec A., Seifert H. (2007). An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5 939–951. 10.1038/nrmicro1789 [DOI] [PubMed] [Google Scholar]
- Dinh D. T., Le M. T. Q., Vuong C. D., Hasebe F., Morita K. (2011). An updated loop-mediated isothermal amplification method for rapid diagnosis of H5N1 Avian Influenza Viruses. Trop. Med. Health 39 3–7. 10.2149/tmh.2010-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell D. J., Sader H. S., Flamm R. K., Jones R. N. (2014). Ceftolozane/tazobactam activity tested against Gram-negative bacterial isolates from hospitalised patients with pneumonia in US and European medical centres (2012). Int. J. Antimicrob. Agents 43 533–539. 10.1016/j.ijantimicag.2014.01.032 [DOI] [PubMed] [Google Scholar]
- Gales A. C., Jones R. N., Sader H. S. (2011). Contemporary activity of colistin and polymyxin B against a worldwide collection of Gram-negative pathogens: results from the SENTRY Antimicrobial Surveillance Program (2006–09). J. Antimicrob. Chemother. 66 2070–2074. 10.1093/jac/dkr239 [DOI] [PubMed] [Google Scholar]
- Golanbar G. D., Lam C. K., Chu Y.-M., Cueva C., Tan S. W., Silva I., et al. (2011). Phenotypic and molecular characterization of Acinetobacter clinical isolates obtained from inmates of California correctional facilities. J. Clin. Microbiol. 49 2121–2131. 10.1093/jac/dkr239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh K., Nishimura N., Takeuchi S., Hattori F., Horiba K., Isaji M., et al. (2013). Assessment of the loop-mediated isothermal amplification assay for rapid diagnosis of Mycoplasma pneumoniae in pediatric community-acquired pneumonia. Jpn. J. Infect. Dis. 66 539–542. 10.1007/s10156-012-0388-5 [DOI] [PubMed] [Google Scholar]
- Janda J. M., Abbott S. L. (2007). 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45 2761–2764. 10.1128/jcm.01228-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karageorgopoulos D. E., Falagas M. E. (2008). Current control and treatment of multidrug-resistant Acinetobacter baumannii infections. Lancet Infect. Dis. 8 751–762. 10.1016/S1473-3099(08)70279-2 [DOI] [PubMed] [Google Scholar]
- Karah N., Haldorsen B., Hegstad K., Simonsen G. S., Sundsfjord A. Samuelsen Ø et al. (2011). Species identification and molecular characterization of Acinetobacter spp. blood culture isolates from Norway. J. Antimicrob. Chemother. 66 738–744. 10.1093/jac/dkq521 [DOI] [PubMed] [Google Scholar]
- Kempf M., Rolain J.-M. (2012). Emergence of resistance to carbapenems in Acinetobacter baumannii in Europe: clinical impact and therapeutic options. Int. J. Antimicrob. Agents 39 105–114. 10.1016/j.ijantimicag.2011.10.004 [DOI] [PubMed] [Google Scholar]
- Kumar P., Pandya D., Singh N., Behera D., Aggarwal P., Singh S. (2014). Loop-mediated isothermal amplification assay for rapid and sensitive diagnosis of tuberculosis. J. Infect. 69 607–615. 10.1016/j.jinf.2014.08.017 [DOI] [PubMed] [Google Scholar]
- Kuo H. Y., Chang K. C., Kuo J. W., Yueh H. W., Liou M. L. (2012). Imipenem: a potent inducer of multidrug resistance in Acinetobacter baumannii. Int. J. Antimicrob. Agents 39 33–38. 10.1016/j.ijantimicag.2011.08.016 [DOI] [PubMed] [Google Scholar]
- Lemos E., de la Hoz F., Einarson T., McGhan W., Quevedo E., Castañeda C., et al. (2014). Carbapenem resistance and mortality in patients with Acinetobacter baumannii infection: systematic review and meta-analysis. Clin. Microbiol. Infect. 20 416–423. 10.1111/1469-0691.12363 [DOI] [PubMed] [Google Scholar]
- Nakauchi M., Takayama I., Takahashi H., Tashiro M., Kageyama T. (2014). Development of a reverse transcription loop-mediated isothermal amplification assay for the rapid diagnosis of avian influenza A (H7N9) virus infection. J. Virol. Methods 204 101–104. 10.1016/j.jviromet.2014.03.028 [DOI] [PubMed] [Google Scholar]
- Nakauchi M., Yoshikawa T., Nakai H., Sugata K., Yoshikawa A., Asano Y., et al. (2011). Evaluation of reverse transcription loop-mediated isothermal amplification assays for rapid diagnosis of pandemic influenza A/H1N1 2009 virus. J. Med. Virol. 83 10–15. 10.1002/jmv.21934 [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28 E63 10.1093/nar/28.12.e63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive D. M., Bean P. (1999). Principles and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 37 1661–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y. K., Jung S.-I., Park K.-H., Kim S. H., Ko K. S. (2012). Characteristics of carbapenem-resistant Acinetobacter spp. other than Acinetobacter baumannii in South Korea. Int. J. Antimicrob. Agents 39 81–85. 10.1016/j.ijantimicag.2011.08.006 [DOI] [PubMed] [Google Scholar]
- Peleg A. Y., Seifert H., Paterson D. L. (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21 538–582. 10.1128/CMR.00058-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez F., Hujer A. M., Hujer K. M., Decker B. K., Rather P. N., Bonomo R. A. (2007). Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51 3471–3484. 10.1128/aac.01464-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue J. M., Mann T., Barber K. E., Kaye K. S. (2013). Carbapenem-resistant Acinetobacter baumannii: epidemiology, surveillance and management. Expert Rev. Anti Infect. Ther. 11 383–393. 10.1586/eri.13.14 [DOI] [PubMed] [Google Scholar]
- Poirel L., Naas T., Nordmann P. (2010). Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrob. Agents Chemother. 54 24–38. 10.1128/AAC.01512-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Nordmann P. (2006). Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12 826–836. 10.1111/j.1469-0691.2006.01456.x [DOI] [PubMed] [Google Scholar]
- Poon L. L., Leung C. S., Tashiro M., Chan K. H., Wong B. W., Yuen K. Y., et al. (2004). Rapid detection of the severe acute respiratory syndrome (SARS) coronavirus by a loop-mediated isothermal amplification assay. Clin. Chem. 50 1050–1052. 10.1373/clinchem.2004.032011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhomberg P. R., Jones R. N. (2009). Summary trends for the meropenem yearly susceptibility test information collection program: a 10-year experience in the United States (1999–2008). Diagn. Microbiol. Infect. Dis. 65 414–426. 10.1016/j.diagmicrobio.2009.08.020 [DOI] [PubMed] [Google Scholar]
- Soo P.-C., Tseng C.-C., Ling S.-R., Liou M.-L., Liu C.-C., Chao H.-J., et al. (2013). Rapid and sensitive detection of Acinetobacter baumannii using loop-mediated isothermal amplification. J. Microbiol. Methods 92 197–200. 10.1016/j.mimet.2012.11.020 [DOI] [PubMed] [Google Scholar]
- Tenover F. C., Arbeit R. D., Goering R. V., Mickelsen P. A., Murray B. E., Persing D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari V., Kapil A., Moganty R. R. (2012a). Carbapenem-hydrolyzing oxacillinase in high resistant strains of Acinetobacter baumannii isolated from India. Microb. Pathog. 53 81–86. 10.1016/j.micpath.2012.05.004 [DOI] [PubMed] [Google Scholar]
- Tiwari V., Vashistt J., Kapil A., Moganty R. R. (2012b). Comparative proteomics of inner membrane fraction from carbapenem-resistant Acinetobacter baumannii with a reference strain. PLoS ONE 7:e39451 10.1371/journal.pone.0039451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari V., Moganty R. R. (2014). Conformational stability of OXA-51 β-lactamase explains its role in carbapenem resistance of Acinetobacter baumannii. J. Biomol. Struct. Dyn. 32 1406–1420. 10.1080/07391102.2013.819789 [DOI] [PubMed] [Google Scholar]
- Turton J. F., Woodford N., Glover J., Yarde S., Kaufmann M. E., Pitt T. L. (2006). Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 44 2974–2976. 10.1128/jcm.01021-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhou Y., Li S., Zhuo C., Xu S., Huang L., et al. (2013). Real-time fluorescence loop mediated isothermal amplification for the detection of Acinetobacter baumannii. PLoS ONE 8:e66406 10.1371/journal.pone.0066406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford N., Ellington M. J., Coelho J. M., Turton J. F., Ward M. E., Brown S., et al. (2006). Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27 351–353. 10.1016/j.ijantimicag.2006.01.004 [DOI] [PubMed] [Google Scholar]
- Xi H., Xu Y., Zhu D., Wang F., Ni Y., Sun J., et al. (2012). CHINET 2010 surveillance of antibiotic resistance in Acinetobacter baumannii in China. Chin. J. Infect. Chemother. 2 006. [Google Scholar]
- Yan Z.-Q., Shen D.-X., Cao J.-R., Chen R., Wei X., Liu L.-P., et al. (2010). Susceptibility patterns and molecular epidemiology of multidrug-resistant Acinetobacter baumannii strains from three military hospitals in China. I. J. Antimicrob. Agents 35 269–273. 10.1016/j.ijantimicag.2009.10.016 [DOI] [PubMed] [Google Scholar]
- Zarrilli R., Pournaras S., Giannouli M., Tsakris A. (2013). Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int. J. Antimicrob. Agents 41 11–19. 10.1016/j.ijantimicag.2012.09.008 [DOI] [PubMed] [Google Scholar]
- Zavascki A. P., Carvalhaes C. G., Picão R. C., Gales A. C. (2010). Multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii: resistance mechanisms and implications for therapy. Expert Rev. Anti Infect. Ther. 8 71–93. 10.1586/eri.09.108 [DOI] [PubMed] [Google Scholar]
- Zeng Y., Zhang X., Nie K., Ding X., Ring B. Z., Xu L., et al. (2014). Rapid quantitative detection of Human immunodeficiency virus type 1 by a reverse transcription-loop-mediated isothermal amplification assay. Gene 541 123–128. 10.1016/j.gene.2014.03.015 [DOI] [PubMed] [Google Scholar]