Abstract
In previous work, we showed that coinoculating Rhizobium leguminosarum bv. viciae 128C53 and Bacillus simplex 30N-5 onto Pisum sativum L. roots resulted in better nodulation and increased plant growth. We now expand this research to include another alpha-rhizobial species as well as a beta-rhizobium, Burkholderia tuberum STM678. We first determined whether the rhizobia were compatible with B. simplex 30N-5 by cross-streaking experiments, and then Medicago truncatula and Melilotus alba were coinoculated with B. simplex 30N-5 and Sinorhizobium (Ensifer) meliloti to determine the effects on plant growth. Similarly, B. simplex 30N-5 and Bu. tuberum STM678 were coinoculated onto Macroptilium atropurpureum. The exact mechanisms whereby coinoculation results in increased plant growth are incompletely understood, but the synthesis of phytohormones and siderophores, the improved solubilization of inorganic nutrients, and the production of antimicrobial compounds are likely possibilities. Because B. simplex 30N-5 is not widely recognized as a Plant Growth Promoting Bacterial (PGPB) species, after sequencing its genome, we searched for genes proposed to promote plant growth, and then compared these sequences with those from several well studied PGPB species. In addition to genes involved in phytohormone synthesis, we detected genes important for the production of volatiles, polyamines, and antimicrobial peptides as well as genes for such plant growth-promoting traits as phosphate solubilization and siderophore production. Experimental evidence is presented to show that some of these traits, such as polyamine synthesis, are functional in B. simplex 30N-5, whereas others, e.g., auxin production, are not.
Keywords: coinoculations, Bacillus simplex, genome studies, rhizosphere, legumes
Introduction
Rhizosphere bacteria function as a consortium, synergistically protecting plants from disease (Kloepper et al., 2004), providing plants with essential nutrients (Pradhan and Sukla, 2005; Martínez-Hidalgo et al., 2014), and stimulating plant growth by producing growth-promoting factors (El-Tarabily et al., 2008; Merzaeva and Shirokikh, 2010). Rhizosphere bacteria are analogous to gut bacteria in mammals, which perform similar functions, and like gut bacteria, the microbes that live on and within plant tissues are indispensable for plant survival. Although the microbial composition of the root microbiomes for many plants is known (Schlaeppi et al., 2014), defining the mechanisms driving the microbe/plant synergism in the soil is challenging. This is because soil is complex and the experiments are difficult to perform. Thus, simpler models have been employed, such as using microcosms or rhizotrons and also limiting the number of plant and microbial species to be studied. This is especially true for specific interactions such as those involved in nitrogen fixation, where investigations of the interactions between nitrogen-fixing bacteria and other soil bacteria or fungi consist of coinoculating a legume plant with a rhizobium and a single plant growth promoting bacterial (PGPB) species. Such interactions usually result in an enhancement of plant growth over inoculation solely with rhizobia (see references in Schwartz et al., 2013).
The most frequent bacterial partners in coinoculation studies involving rhizobia are Bacillus species, including among others, B. subtilis, B. amyloliquefaciens, B. licheniformis, and B. pumilus. Earlier, we showed that coinoculating Pisum sativum L. with Rhizobium leguminosarum bv. viciae 128C53 and B. simplex 30N-5 resulted in better nodulation and an overall increase in plant dry weight (Schwartz et al., 2013). B. simplex 30N-5 is a relatively new player in the panoply of bacteria that positively influence plant growth. This species is mainly known for its phenotypic adaptations with respect to growing on the sun compared to shade walls of “Evolution Canyon” in Israel (Koeppel et al., 2008). However, a number of publications, including our own, have reported that B. simplex also functions as a PGPB species (Ertruk et al., 2010; Hassen and Labuschagne, 2010). Recently, the sequenced genomes of several B. simplex strains became available and allowed prediction of possible molecular mechanisms for the observed interactions. The essential extension of such genome comparisons include the identification of the expressed proteins, and perhaps most importantly, the identification of the small molecule products of their activity.
In this study, we coinoculated B. simplex 30N-5 with either Sinorhizobium (Ensifer) meliloti 1021 (alpha-rhizobium) or Burkholderia tuberum STM678 (beta-rhizobium), on their respective hosts. To our knowledge, Bu. tuberum STM678 (Moulin et al., 2001; Vandamme et al., 2002) has not been previously employed in coinoculation studies. To obtain a better understanding of the traits that are important for the plant responses in the coinoculation experiments, we analyzed the B. simplex 30N-5 genome for genes known to encode PGPB traits. To do this, we compared B. simplex 30N-5 with the well-established PGP Bacillus strains, namely B. subtilis GB03, B. amyloliquefaciens subsp. plantarum FZB42, and others. In this report, we also demonstrate that several of these PGPB traits are functional in B. simplex 30N-5.
Materials and methods
Phylogenetic analysis
Nucleotide sequences were obtained from the Joint Genome Institute (IMG/ER) database for microbial genomes (Markowitz et al., 2012). Five housekeeping genes atpD, urvA, rpoB, lepA, and recA were used to construct concatenated sequences (Table S1). The concatenated gene sequences were aligned with Clustal X (Thompson, 1997), and phylogenetic distances were calculated according to the Kimura two-parameter model (Kimura, 1980). The phylogenetic tree topology was inferred from the maximum-likelihood method employing MEGA5 (Tamura et al., 2011). Confidence levels on each node are the product of 1000 bootstrap replicates.
Growth of bacteria
Bacillus strains were grown on LB (Luria-Bertani; Miller, 1972), Tryptic Soy Agar (TSA; Difco®, Becton Dickenson) or Tryptone Yeast Extract (TY; Beringer, 1974) medium at 30°C or 37°C. Rhizobial strains were cultured at 30°C on either Yeast Mannitol Agar (YMA; Somasegaran and Hoben, 1994) or on TY medium with or without 10 μg/mL tetracycline. Bu. tuberum STM678 was grown on LB minus salt or on BSE medium (Caballero-Mellado et al., 2007) with or without antibiotics. Cell density was determined from the OD600 nm of the cultures. The bacterial strains studied in this report are listed in Table 1.
Table 1.
Strains and plasmids used in this study.
| Strain number | Species name and relevant characteristics | Source or reference |
|---|---|---|
| 30N-5 | Bacillus simplex | Schwartz et al., 2013 |
| 237 | Bacillus simplex | Kaplan et al., 2013 |
| 11 | Bacillus simplex | Kaplan et al., 2013 |
| FZB42 | Bacillus amyloliquefaciens subsp. plantarum | Bacillus Stock Center |
| DSM13 Goettingen/ATCC 14580 | Bacillus licheniformis | Bacillus Stock Center |
| GB03 | Bacillus subtilis | Bacillus Stock Center |
| NRRL B-4317 | Paenibacillus polymyxa | Bacillus Stock Center |
| 60b4 | Bacillus subtilis | Flora Pule-Meulenberg |
| 26a1 | Bacillus cereus | Flora Pule-Meulenberg |
| HB101 | E. coli | Cathy C. Webb |
| Rm1021 | Wild-type Sinorhizobium meliloti | Lab strain |
| Rm1021/pHC60 | GFP+, Tetr derivative of wild-type S. meliloti | This study |
| STM678 | Wild-type Burkholderia tuberum | Moulin et al., 2001; Vandamme et al., 2002 |
| STM678/TnGFP | Tetr derivative of wild-type Bu. tuberum | Elliott et al., 2007 |
| Plasmids | Relevant characteristics | Source or Reference |
| pHC60 | GFP plasmid, Tetr | Cheng and Walker, 1998 |
To introduce fluorescent markers into Sinorhizobium (Ensifer) meliloti 1021, the plasmid pHC60 (Cheng and Walker, 1998) carrying a green fluorescent protein (GFP) construct was mobilized into S. meliloti using a triparental mating procedure (Figurski and Helinski, 1979) as adapted by Schwartz et al. (2013). The Bu. tuberum STM678 GFP+ strain was a gift from Dr. J. Peter Young (University of York).
The Voges-Proskauer test (Voges and Proskauer, 1898) was performed as modified by Werkman (1930) and Barritt (1936). Each strain was tested three times.
Chemical analysis
Cell pellets (1.8 × 109 cells/sample) from B. simplex 30N-5 were lysed in 5 ml of either methanol or aqueous trifluoroacetic acid (TFA, 10%) or aqueous trichloroacetic acid (TCA, 8.3%). The homogenates were centrifuged (16,000 × g, 5 min, room temperature) and the supernatants were taken to dryness in a vacuum centrifuge. The dried residue was resuspended in water (500 μL), centrifuged (16,000 × g, 5 min, room temperature) and the supernatant transferred to LC injector vials. For the polyamines, aliquots of the supernatant were injected (8 μl) onto a reverse phase HPLC column (Phenomenex Kinetex C18 100 × 2.1 mm, 1.7 μ particle size and 100 Å) equilibrated in 80% solvent A (0.1 mM perfluoro-octanoic acid in water) and 20% solvent B (0.1 mM perfluoro-octanoic acid in methanol), and eluted (100 μL/min) with an increasing concentration of solvent B (min/%B; 0/20, 5/20, 15/75, 20/75, 22/20, 30/20). The effluent from the column was directed to an electrospray ion source connected to a triple quadrupole mass spectrometer (Agilent 6460) operating in the positive ion tandem mass spectrometric (MS/MS) mode, and the time-dependent intensity of multiple reaction monitoring (MRM) transitions were recorded at previously optimized settings [spermine, m/z (MH+) 203 129, 112, 84, fragmentor 55, collision energy 16; spermidine, m/z (MH+) 146
129, 112, 84, fragmentor 55, collision energy 16; spermidine, m/z (MH+) 146 129, 112, and 72, fragmentor 55, collision energy 12; putrescine, m/z (MH+) 89
129, 112, and 72, fragmentor 55, collision energy 12; putrescine, m/z (MH+) 89 72, fragmentor 40, collision energy 4]. Peak areas for each compound at the corresponding retention times (spermine, spermidine, and putrescine at 16.4, 16.0, and 15.6 min, respectively) were computed with instrument manufacturer-supplied software (Agilent MassHunter). A standard curve was prepared with each experiment from samples containing known concentrations of all three compounds using the signals for the most intense MRM transitions (203
72, fragmentor 40, collision energy 4]. Peak areas for each compound at the corresponding retention times (spermine, spermidine, and putrescine at 16.4, 16.0, and 15.6 min, respectively) were computed with instrument manufacturer-supplied software (Agilent MassHunter). A standard curve was prepared with each experiment from samples containing known concentrations of all three compounds using the signals for the most intense MRM transitions (203 112, 146
112, 146 112, and 89
112, and 89 72 for spermine, spermidine, and putrescine, respectively), and the amount of each amine in each biological sample was calculated by interpolation from the standard curves. Under the prescribed conditions, the limit of detection for the amines was about 1 pmol injected for spermine and spermidine and 10 pmol injected for putrescine.
72 for spermine, spermidine, and putrescine, respectively), and the amount of each amine in each biological sample was calculated by interpolation from the standard curves. Under the prescribed conditions, the limit of detection for the amines was about 1 pmol injected for spermine and spermidine and 10 pmol injected for putrescine.
For indole acetic acid (IAA, auxin), aliquots of the supernatants were injected (8 μl) onto a mixed cationic/anionic/reverse phase HPLC column (Imtakt Scherzo SS-C18, 100 × 2 mm, 3 μ particle size and 130 Å pore size) equilibrated in 40% solvent C (water/acetonitrile/formic acid, 97/3/0.1, all by vol) and 60% solvent D (45 mM aqueous ammonium formate/acetonitrile, 65/35, v/v), and eluted (200 μL/min) with an increasing concentration of solvent D (min/%D; 0/60, 5/60, 20/100, 22/60, 30/60). The effluent from the column was directed to the same ESI mass spectrometer as described above, and the time-dependent intensity of the IAA MRM transition was recorded at previously optimized settings [m/z (MH+) 176 130, fragmentor 45, collision energy 12]. Peak areas for the transition response at the corresponding retention time (10.1 min) were computed as described above. A standard curve was prepared with each experiment from samples containing known concentrations of IAA. Under the prescribed conditions, the limit of detection (LOD) for IAA was about 5 pmol injected, which was about four-fold lower than what could be achieved in the negative ion mode also under previously optimized conditions by monitoring the transition of the (M-H)− ion at m/z 174
130, fragmentor 45, collision energy 12]. Peak areas for the transition response at the corresponding retention time (10.1 min) were computed as described above. A standard curve was prepared with each experiment from samples containing known concentrations of IAA. Under the prescribed conditions, the limit of detection (LOD) for IAA was about 5 pmol injected, which was about four-fold lower than what could be achieved in the negative ion mode also under previously optimized conditions by monitoring the transition of the (M-H)− ion at m/z 174 130. Also, the LOD using combined liquid chromatography-MS/MS-MRM (LC/MS/MS-MRM) in either the positive or negative ion mode was significantly lower than what could be achieved by combined gas chromatography/mass spectrometry (GC/MS) in the selected ion-monitoring (SIM) mode (Waters GCT) of the trimethylsilyl derivative.
130. Also, the LOD using combined liquid chromatography-MS/MS-MRM (LC/MS/MS-MRM) in either the positive or negative ion mode was significantly lower than what could be achieved by combined gas chromatography/mass spectrometry (GC/MS) in the selected ion-monitoring (SIM) mode (Waters GCT) of the trimethylsilyl derivative.
Cross-streaking experiments
Fresh samples of each bacterial strain were taken from frozen cultures and grown on either LB minus NaCl, LB, or TY agar for cross streaking (Lertcanawanichakul and Sawangnop, 2008). A single colony from one strain was first streaked vertically down the middle of the plate and 24 h later the second strain was streaked perpendicularly to the first. The order of microbes was changed in each experiment, which was repeated 4 times with 3 or 4 biological replicates. Qualitative data were obtained by photographing the plates daily for 7 days. For the pairs of S. meliloti and B. simplex, we also performed parallel streaking and overlapping streaking experiments. As before, the order of microbes was changed in each experiment, and the plates were followed for 10 days.
Plant coinoculation experiments
Macroptilium atropurpureum (siratro), Medicago truncatula A17, and Melilotus alba L. U389 (white sweetclover) seeds were planted in black polyethylene boxes (Really Useful Boxes®) or Magenta® jars (Magenta Corp.). The substrate used for the black boxes was Seramis® (Mars GmbH) and perlite, and for the Magenta jars, a 2:1 mixture of vermiculite and perlite. The substrates were autoclaved and then watered with ¼ strength Hoagland's medium minus nitrogen (Machlis and Torrey, 1956). Prior to planting seeds were scarified for 1 min, soaked in 95% ethanol for 5 min, and then in full-strength commercial bleach for 30–45 min. The conditions for siratro seed sterilization and inoculation with Bu. tuberum are detailed in Angus et al. (2013). The imbibed seeds were transferred to the boxes or Magenta jars using sterile tools, and inoculated singly with Bu. tuberum (siratro) or S. meliloti (Medicago and Melilotus) and B. simplex 30N-5, together and alone. The bacteria were diluted with sterile water to a final OD600 nm of 0.1–0.2. The siratro seeds were coinoculated with a 1:1 mixture of Bu. tuberum and B. simplex 30N-5 whereas the Melilotus and Medicago seeds were coinoculated with a 1:1 mixture of S. meliloti 1021 and B. simplex 30N-5. Each Magenta jar or black box was inoculated with 4 mL of the inoculum. Controls were included for all experiments and were used as a phenotypic reference for −N, +N, and no nutrient conditions (water). The control sample size was smaller than the experimental due to space limitation, but the controls consistently gave the same phenotype (see Angus et al., 2013, 2014). The siratro plants were grown in a temperature controlled Conviron growth chamber at 24°C and the S. meliloti hosts in a Percival growth cabinet at 21°C. The Medicago and Melilotus species were harvested 5 weeks after inoculation and the siratro plants 5–6 weeks after inoculation. Each experiment was repeated three times. The plants were photographed, their shoot height and nodule numbers were recorded, and they were then dried (48 h, 65°C) before dry weight measurements were made. Statistical significance of the data was validated using One-way ANOVA with Tukey's post hoc test (Figure 3A) and multiple comparison procedure. Jittered boxplot and family-wise error rates (Figure 3B and Supplementary Figure 2) were used for assessment (Herberich et al., 2010).
Genome analysis
Selection of strains
Draft and finished genome sequences of several PGPB from the Joint Genome Institute IMG/ER (Markowitz et al., 2012) or from NCBI (http://www.ncbi.nlm.nih.gov) were queried by BLAST (Altschul et al., 1990) using sequences of genes encoding known PGPB traits. Thirteen bona fide PGP Bacillus and two Paenibacillus strains were chosen for comparison against B. simplex 30N-5 (Figure 1). The JGI genomes queried included B. simplex 30N-5 (permanent draft), B. simplex II3b11 (permanent draft), B. firmus DS1 (permanent draft), B. licheniformis DSM 13T/ATCC 14580 (finished), B. kribbensis DSM 17871 (permanent draft), B. megaterium DSM 319 (finished), B. amyloliquefaciens subsp. plantarum FZB42T (finished), B. subtilis GB03 (permanent draft), B. subtilis subtilis 168 (finished), B. cereus JM-Mgvxx-63 (permanent draft), B. thuringiensis sv. israelensis (permanent draft), Paenibacillus polymyxa ATCC 12321 (permanent draft), Paenibacillus pini JCM16418 (permanent draft), Pseudomonas fluorescens strains A506 and CHAO (finished), and Azospirillum brasilense FP2 (permanent draft) and Azospirillum sp. B510 (permanent draft). B. simplex 30N-5 was isolated from the Mildred E. Mathias Botanical Garden at UCLA and strain II3b11 belongs to the Putative Ecotype 9 (Koeppel et al., 2008). It originates from the south facing, hot or “African savannah-like” slope of “Evolution Canyon II” in Nahal Keziv, Israel (Sikorski and Nevo, 2005). In addition, the genome of B. simplex strains P558 (Croce et al., 2014) and BA2H3 (Khayi et al., 2015), both from NCBI, were queried when a gene from one or both IMG/ER B. simplex strains was missing. Details of the Bacillus strains studied for the genomic analysis are found in Table 2.
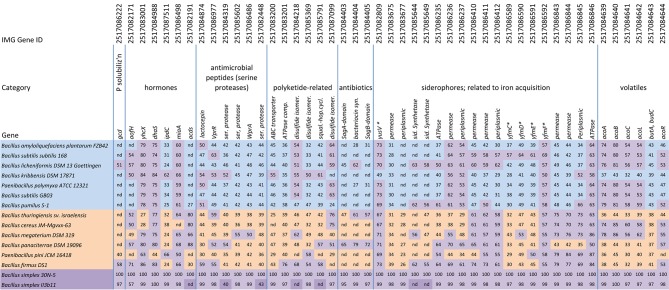
Figure 1.
Homologs of reference set of Plant Growth Promoting (PGP) genes identified in Bacillus and Paenibacillus. Top row, IMG gene ID number; second row, general categories of PGP genes; third row, PGP genes identified in each Bacillus (or Paenibacillus) strain (left column) following comparison with the B. simplex 30N-5 gene (always 100%) and clustered using the K-means algorithm. The highest sequence identity alignment from blastp searches (% values in cells), sequence identities passing the 50% cutoff (cells highlighted in purple) show 3 clusters: cluster 1 (blue), cluster 2 (orange), and cluster 3 (purple). Genes not detected (nd). *Gene names from B. megaterium, B. licheniformis, and B. amyloliquefaciens (see text).
Table 2.
Genomic features of the B. simplex 30N-5 genome and comparison with the genomes of other Bacillus spp.
| B. simplex 30N-5 | B. simplex II3b11 | B. thuringiensis sv. israelensis | B. pumilus S-1 | B. kribbensis DSM 17871 | B. firmus DS1 | B. amyloliquifaciens pl. FZB42 | B. licheniformis DSM 13/ATCC 14580 | B. subtilis GB03 | B. megaterium DSM 319 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Genome size (bp) | 5459036 | 5582948 | 5643051 | 3692073 | 5054217 | 4971242 | 3918589 | 4222645 | 3849547 | 5097447 |
| G+C content (mol%) | 40.43 | 40.27 | 35.18 | 41.26 | 42.93 | 41.46 | 46.4 | 46.19 | 46.55 | 38.13 |
| Protein-coding sequences | 5288 | 4841 | 5349 | 3786 | 4914 | 4922 | 3693 | 4196 | 3705 | 5100 |
| % of coding region | 81.54 | 70.30 | 81.27 | 88.92 | 81.66 | 85.02 | 88.0 | 88.13 | 89.61 | 83.04 |
Homologous gene identification
A curated set of more than 50 PGP genes (47 are displayed in Figure 1), all manually annotated in the B. simplex 30N-5 genome, were selected for use as the reference query genes for blastp homolog searches. The genes represented a wide variety of PGP functions. Because several of the genomes investigated have permanent draft status, our analyses of gene homologies occasionally found no matching gene; these “missing” genes are indicated by “n.d.”. We used conservative criteria to compare the protein sequences. The blastp searches were filtered to include alignments with an e < 10−5, and with a sequence identity of ≥50%, although homologs having smaller percentages and greater e-values were present in the other Bacillus genomes. B. simplex 30N-5 was the reference genome for the comparisons and the value of its gene identities for the comparison was set at 100%. Although both Gram-negative and Gram-positive bacteria were initially screened for PGP traits, the values for the Gram-negative bacteria as well as for the more distantly related Gram-positive species were generally low and hence deleted from the final data set.
We also used BAGEL3 (http://bagel2.molgenrug.nl/) to query the B. simplex 30N-5 genome for bacteriocins, and a stand-alone version of ANTIsmash (http://antismash.secondarymetabolites.org/) to search for non-ribosomal peptide synthetases (NRPSs).
K-means clustering
The sequence identity matrix (Figure 1) contains 13 Bacillus strains and 2 Paenibacillus strains (rows) with the highest detected sequence identity for each reference PGP gene displayed in columns. Despite the 50% cutoff sequence identity limit, some sequence identity scores under 50% were also recorded because blastp sequence alignments with low sequence identity may still exhibit homology or contain conserved functional domains. This scheme enabled clustering, using the K-means clustering algorithm, to place Bacillus strains into groups that had overall similar profiles of PGP genes either as present or absent. The algorithm was implemented with an objective function to minimize the within-cluster Euclidean distance of the sequence identity vectors (rows) from their assigned clusters.
Our implementation of the K-means algorithm used a two-step iterative algorithm (Lloyd, 1982; Slonim et al., 2013). In the assignment step, the sequence identity vectors (rows) were assigned to the nearest cluster (measured by Euclidean distance between each sequence identity vector and centroid corresponding to each cluster). In the update step, the coordinates of each centroid were updated to the mean of the respectively assigned sequence identity vectors. The maximum number of iterations permitted was 10,000. Initial centroids were randomly assigned to the sequence identity vectors, and 100 random centroid initializations were run. Of the 100 K-means runs, the cluster arrangement minimizing the total Euclidean distance of the sequence identity vectors (rows) from their assigned clusters was retained for visualization (Figure 1).
Results
Phylogenies
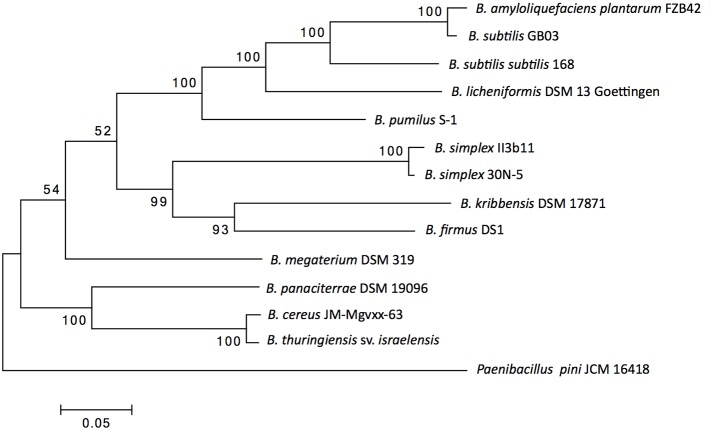
A concatenated gene Maximum Likelihood phylogeny of the selected strains is shown in Figure 2. The housekeeping genes used for the tree are representative of the differences between the genomes of the species tested, as the most closely related species to B. simplex are also those with most similarities found in the PGPB genes studied (see colors in Figure 1). Although the topology within the clade was supported by high bootstrap values, the two subclades in the top part of Figure 2 were supported by a low bootstrap value (52%). One subclade contained B. amyloliquefaciens subsp. plantarum FZB42, B. subtilis GB03, B. subtilis subtilis 168, B. licheniformis DSM13 Goettingen (ATCC 14580), and B. pumilus S-1, whereas the second subclade included the two B. simplex strains, and B. firmus DS1 and B. kribbensis DSM 17871. In this tree, B. subtilis GB03 and B. amyloliquefaciens subsp. plantarum FZB42 clustered together. A not-as-strongly supported branch of the top clade (54% bootstrap support) included B. megaterium DSM 319. The clade (bottom part of Figure 2) brought together with strong support, B. panaciterrae DSM 19096, B. cereus JM-Mgvxx-63, and B. thuringiensis sv. israelensis. P. pini JCM 16418 was the outgroup.
Figure 2.
Phylogenetic tree. Maximum-likelihood phylogenetic tree based on concatenated gene sequences of five housekeeping genes (atpD, urvA, rpoB, lepA, and recA). Paenibacillus pini JCM 16418 was used as the outgroup. Numbers at branch points indicate bootstrap values (based on 1000 replicates); only those above 50% are indicated. Bar, 0.05 substitutions per nucleotide position.
Pre-coinoculation (cross-streaking) assays
Earlier we reported positive effects on pea growth when B. simplex 30N-5 was coinoculated with R. leguminosarum bv. viciae 128C53 (Schwartz et al., 2013). Before setting up coinoculation experiments with a different set of bacteria, cross-streaking assays were used to detect incompatibility or interference between the nodulating strains, S. meliloti 1021 (data not shown) and Bu. tuberum STM678 (Supplementary Figure 1A), to be used in the coinoculation study with B. simplex 30N-5. No inhibition of growth was found. An additional Bacillus strain previously isolated and studied (Schwartz et al., 2013), B. subtilis 30VD-1, was also tested in these experiments. B. subtilis 30VD-1 inhibited B. simplex 30N-5 growth and vice versa, suggesting that one or both synthesized bacteriocins or other antimicrobial agents (Supplementary Figure 1A and see later section).
For S. meliloti, the results from the initial cross-streak experiments were less clear because although the S. meliloti streak was not touching the B. simplex one, it was closer to it than the distance observed for the B. subtilis and B. simplex cross-streaks (Supplementary Figure 1A). When we repeated the experiments by either doing a side-by-side streak or inoculating one strain over the other in a cross pattern, we observed no incompatibility between the two strains (data not shown).
Coinoculation studies
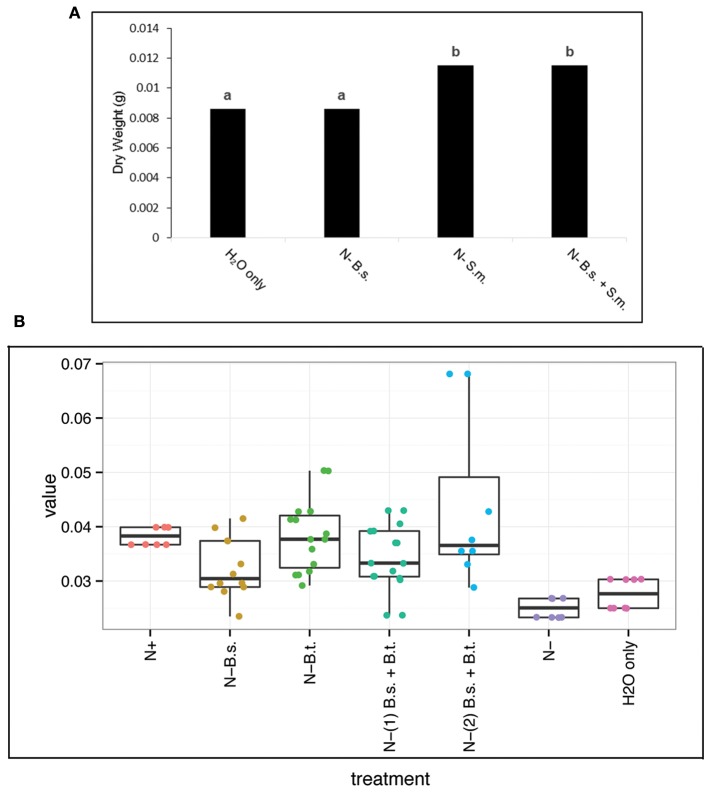
Because B. simplex 30N-5 demonstrated a positive effect on both plant growth and rhizobial nodulation on pea (Schwartz et al., 2013), we tested whether or not this was a general phenomenon by coinoculating B. simplex 30N-5 and S. meliloti Rm1021 onto roots of M. truncatula and M. alba. In contrast to our previous results with pea, M. alba exhibited no significant growth enhancement when inoculated with B. simplex alone over the uninoculated control (Figure 3A). Although shoot height and nodule number were measured for all the conditions examined, no statistical significance was observed when the experimental treatments were compared with their respected controls (data not shown). Moreover, when single inoculations with S. meliloti and coinoculations with both strains were compared, the treatments (measured as dry weight increase) did not differ from each other although both were statistically different from the uninoculated and B. simplex alone-inoculated plants (Figure 3A). M. truncatula exhibited a similar response (data not shown). Overall, we found that dry weight increases were a more reliable measurement of plant biomass accumulation than any other parameters (see next section).
Figure 3.
Biomass measurements of Melilotus alba and Macroptilium atropurpureum 35 days post inoculation. (A) Melilotus alba plants were singly or coinoculated with Bacilus simplex (B.s.) and Sinorhizobium meliloti (S.m.); Different letters represent values that differ significantly, p < 0.01. (B) Jittered boxplot. Macroptilium atropurpureum plants were singly or coinoculated with Bacillus simplex (B.s.) and Burkholderia tuberum (B.t). The first coinoculation (1) introduced both bacteria species at the same time, whereas the second (2) was inoculated with B.s. first followed by B.t. inoculation 5 days later. Harvesting was performed as described in Methods. Boxes indicate minimum, maximum, 1st and 3rd quartiles and the median value.
Siratro plants were coinoculated with B. simplex and Bu. tuberum; the latter nodulates siratro effectively (Angus et al., 2013). In contrast to the S. meliloti host plants, simultaneous coinoculation with B. simplex and Bu. tuberum, or coinoculation with B. simplex first and then Bu. tuberum 5 days later resulted in significant changes over the controls and were comparable to or better than the +N control. The siratro plants inoculated with B. simplex alone also exhibited an increase in dry weight over the −N control and were comparable to the +N control (Figure 3B, Supplementary Figure 2).
Nutrient acquisition
Although B. simplex was isolated on a solidified N-free medium, it is not a diazotroph because it lacks nifH, a structural gene essential for nitrogenase function (Schwartz et al., 2013). In an N-free liquid medium, B. simplex 30N-5 ceased growing unless the medium was supplemented with 1-aminocyclopropane-1-carboxylate (ACC), which is broken down into 2-oxobutanoate and ammonia; the latter sustained bacterial growth for a short time. This finding suggested that B. simplex had acdS activity (see later section).
Phosphate solubilization
B. simplex 30N-5 effectively solubilized mineralized phosphate as measured by activity on PVK plates (Schwartz et al., 2013). Although we detected a gene encoding a soluble quinoprotein glucose/sorbosone dehydrogenase, which is important for gluconic acid production (de Werra et al., 2009) (Figure 1, column 1), no additional genes involved in the breakdown of either inorganic or organic phosphates were found. However, the putative B. simplex (gcd) gene was not overly similar to the comparable genes in either B. licheniformis (51%) or B. firmus (58%) (Figure 1). We were unable to detect an equivalent gene in the other PGPB strains. Species of B. amyloliquefaciens (Kim et al., 1998), B. licheniformis (Tye et al., 2002), and B. subtilis (Kerovuo et al., 1998) have been reported to produce phytase (myo-inositol-hexaphosphate 3-phosphohydrolase), which degrades organic phosphates. A phytase gene was not detected in the B. simplex strains.
Siderophores
Siderophores secreted by bacteria also support the development and growth of plants by helping them sequester iron from the environment. Previously, we showed that B. simplex 30N-5 exhibited a positive reaction in a CAS assay, which detects siderophore activity (Schwartz et al., 2013). We identified several siderophore operons in B. simplex 30N-5 (Figure 1). The iron-dicitrate transporter genes were similar to yfmCDEF of B. megaterium and several other bacilli, whereas the iron-compound transport system had genes conserved with yfiZ and yfiY of B. licheniformis as well as with B. amyloliquefaciens genes.
Production of volatiles
Many PGPB emit volatiles that positively enhance growth, e.g., B. amyloliquefaciens subsp. plantarum FZB42 and B. subtilis GB03. The latter was reported to acidify the rhizosphere of Arabidopsis in response to volatiles (Zhang et al., 2007), which may help in phosphate solubilization. The most commonly studied volatiles are acetoin (Xiao and Xu, 2007), and 2,3-butanediol, which is known to be involved in Arabidopsis defense induction (Ryu et al., 2003). The Voges-Proskauer test was used to demonstrate acetoin synthesis as well as the potential for production of 2,3-butanediol (Xiao and Xu, 2007) by means of a colorimetric reaction. We used this test on a number of Bacillus strains known to produce volatiles and included B. simplex 30N-5 along with two additional strains of B. simplex (Kaplan et al., 2013) for the analysis. Escherichia coli was the negative control. Although the known PGPB strains tested positive for acetoin, including P. polymyxa NRRL B-4317, the three B. simplex strains were negative even after a long incubation period (Table 3).
Table 3.
Voges-Proskauer test results for selected strains.
| Strain | 30′ | 1 h |
|---|---|---|
| E. coli HB101* | − | − |
| B. amyloliquefaciens plantarum FZB42 | + | + |
| B. cereus 26a1 | − | − |
| B. licheniformis DSM 13/ATCC 14580 | + | + |
| B. simplex 237 | − | − |
| B. simplex 11 | − | − |
| B. simplex 30N-5 | − | − |
| B. subtilis GB03 | + | + |
| B. subtilis 60b4** | + | + |
| P. polymyxa NRRL B-4317 | + | + |
Negative control
Positive control.
Measurements were taken 30 min and 1 h after the addition of the colorimetric reagents.
Genes from B. amyloliquefaciens subsp. plantarum FZB42 were used to search for sequences in the B. simplex genome that could encode proteins for acetoin synthesis. We detected five genes, several aco genes, as well as one encoding alsS, which is important for acetolactate synthesis (Xiao and Xu, 2007). However, no gene for alsD, which encodes an alpha-acetolactate decarboxylase, was detected although it was present in the reference PGPB genomes. Also, a butA/budC gene was detected in the aco operon and showed greater than 95% identity to the other B. simplex strain (II3b11) (Figure 1). The butA/budC gene encodes meso-butanediol dehydrogenase.
Root colonization and growth promotion factors
Motility
In response to root exudates, many bacteria migrate toward root surfaces and colonize roots. In B. amyloliquefaciens subsp. plantarum FZB42 and other Bacillus species, a number of genes are expressed (Chen et al., 2007), including flagellar genes (De Weger et al., 1987; Croes et al., 1993). In the B. simplex genome, flagellar and chemotaxis genes are located in two apparently unlinked areas (Figure 4). A similar arrangement exists for other PGPB Bacillus strains, such as B. firmus DS1, B. kribbensis DSM 17871, B. megaterium DSM 319 (Supplementary Figure 2), and the two Paenibacillus species included in our analysis (data not shown). However, a major difference in arrangement was observed in B. cereus, which contains strains that can be either beneficial or pathogenic (Bottone, 2010). For example, five chemotaxis-associated genes (cheABCD and W) within the flagellar gene operon are conserved among B. simplex and related PGP strains (Figure 4, Supplementary Figure 3). However, the chemotaxis genes, cheB and cheD, were not detected within the flagellar gene region of B. cereus JM-Mgvxx-63, which differs in organization (Figure 4). In addition, the flagellar genes are not highly related. For example, the flhF genes of B. simplex 30N-5 and B. cereus JM-Mgvxx-63 are 36% identical, but only 24% DNA identity is observed when the two fliS genes were compared.
Figure 4.
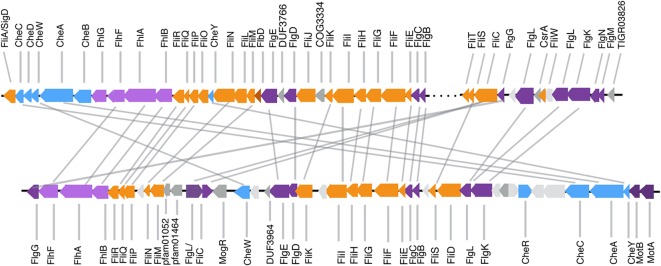
Comparison of flagellar open reading frame (ORF) clusters. The top cluster is from B. simplex 30N-5, which consists of two unlinked regions. The bottom cluster is from B. cereus JM-Mgvxx-63 where most of the genes related to B. simplex are within a single region on the chromosome.
Many PGPB, e.g., alpha-rhizobia (Amaya-Gómez et al., 2015) and Bacillus spp. (Dietel et al., 2013), swarm as well as swim prior to colonization. B. simplex 30N-5 cells swarm on 0.8% agar suggesting that genes for this behavior are present and expressed. The swrC gene shares 100% identity among the B. simplex strains. Although originally annotated as a cation/multidrug efflux pump, it is orthologous and 61 and 57% identical to genes annotated as swrC in B. thuringiensis and B. amyloliquefaciens, respectively.
Plant growth-promoting traits: hormones
One of the most prominent features of PGPB is their ability to produce compounds that directly influence plant growth, e.g., the phytohormones. PGPB also synthesize gene products that affect plant growth in a more indirect way.
Auxin
Many bacteria are known to synthesize auxin (involved in lateral root proliferation), and at least five tryptophan-dependent or tryptophan-independent biosynthetic pathways are employed by bacteria to synthesize auxin (Patten and Glick, 1996; Spaepen and Vanderleyden, 2011). Because we obtained positive results with the Salkowski test following the addition of tryptophan (Schwartz et al., 2013), we hypothesized that the genes for auxin synthesis might be present in the genome of this species.
One of the most studied of the auxin biosynthetic pathways includes the genes encoding IAA monooxygenase (iaaM) and indole acetamide hydrolyase (iaaH), found in the gall-forming Agrobacterium tumefaciens and Pseudomonas savastonoi, but these genes were not detected in the B. simplex 30N-5 genome. However, putative ipdC genes were identified in the B. simplex strains with greater than 97% DNA identity among them, following a query with an indole pyruvate carboxylase gene from Enterobacter cloacae (Koga, 1995). Although < 50% DNA identity to ipdC from several other PGPB was observed (Figure 1), ipdC genes in B. kribbensis and P. pini were 62 and 66% identical in DNA sequence, respectively, and orthologous to the B. simplex gene. A gene encoding a putative indole-3-acetaldehyde dehydrogenase, which is involved in tryptophan-dependent IAA synthesis and is the last step in the pathway, was also detected in B. simplex 30N-5. The gene is ca. 75% identical to dhaS of several bacilli, and has >84% identity to genes in B. firmus DS1 and B. kribbensis DSM 17,871.
Other auxin-related genes were also found in B. simplex 30N-5. A gene orthologous to aofH, which codes for an indole-3-acetic oxidase [suggesting that tryptamine, rather than tryptophan, is converted to indole-3-acetaldehyde (IAAld)], was uncovered using AZL.b03560 from Azospirillum sp. B510 (Wisniewski-Dyé et al., 2011) to query the B. simplex genome. Although the different B. simplex strains have almost identical aofH gene sequences, this gene is not well conserved with genes from other bacilli with the exception of B. firmus DS1 (71% identity) (Figure 1). Lastly, a B. simplex gene with 79% DNA identity and orthologous to a predicted nitrilase, the yhcX gene in B. amyloliquefaciens, was detected in the B. simplex 30N-5 genome. A similar gene identity was observed for many of the other PGP bacilli (Figure 1).
The lack of a complete pathway and the low identity for ipdC made us question whether IAA is actually synthesized by B. simplex. To address this question, we performed a chemical analysis. No signals for IAA were found in the LC/MS/MS-MRM assay for this compound. With a limit of detection of about 5 pmol injected, it is concluded the concentration of this hormone, if present, is less than 174 pmol/109 cells.
Phenylacetic acid
In Azospirillum brasilense, IpdC is also involved in the production of phenylacetic acid (PAA), which has weak auxin activity and is also antimicrobial against both bacteria and fungi (Somers et al., 2005). As in Azospirillum, the B. simplex genome has the paa operon (data not shown), which is important for the degradation of PAA.
Cytokinin
Many PGP bacilli have been reported to produce cytokinins, but few cytokinin biosynthetic genes have been detected (see Vacheron et al., 2013). Querying various Bacillus genomes with tzs (trans-zeatin synthase) from A. tumefaciens, where it is required for tumor formation, yielded no hits. On the other hand, miaA, which encodes tRNA dimethylallyltransferase that removes a zeatin precursor from tRNA, is common among the PGP bacilli, including B. simplex.
Polyamines
Many bacteria produce polyamines such as spermine, spermidine, and putrescine, which in B. subtilis OKB105 have PGP properties (Xie et al., 2014). A number of genes involved in polyamine synthesis (Sekowska et al., 1998) were detected in the B. simplex 30N-5 genome including speA, which results in agmatine synthesis; speB, putrescine synthesis; and speE and speD, which encode the stages for spermidine synthesis. Also, metK, responsible for the conversion of methionine to S-adenosyl-methionine, was detected (data not shown). Many of these were ≤ 80% identical to genes in the PGP bacilli (speD), whereas others, e.g., speE, are less similar ( < 50%) albeit orthologous. Genes for various binding proteins, permeases, and transporters for polyamines are also present in the genome of B. simplex 30N-5.
Clear strong signals were obtained for cell lysates prepared in either TCA or TFA showing the presence of significant quantities of spermine, spermidine, and putrescine in all samples examined (Figure 5). Verification of the assignments was made with co-chromatography experiments in which known amounts of authentic standards were added to cell lysate samples prior to LC/MS/MS-MRM. In these experiments, single peaks for each amine were obtained for the spiked samples, with appropriate area and intensity enhancement of the signals. The signals were slightly more intense for spermine and spermidine when TFA was used compared to TCA during cell homogenization (1.6- and 1.2-fold, respectively), and slightly more intense for putrescine when TCA compared to TFA was used (2.8-fold) (Table 4). The MRM chromatograms from methanol extracts were less clear with peaks at other retention times and significantly less intense peaks for the polyamines. Quantitation based on external standards shows spermine, spermidine and putrescine concentrations in the range of 333, 222, and 2.2 nmol/109 cells, respectively, although for more precise measurements, the work requires repeating using an internal standard to correct for losses during extraction. To this end, it was noted that the cell extracts did not contain any detectable amount of hexamethylediamine (MRM transition (MH+) m/z 117 100) that could be used for this purpose.
100) that could be used for this purpose.
Figure 5.
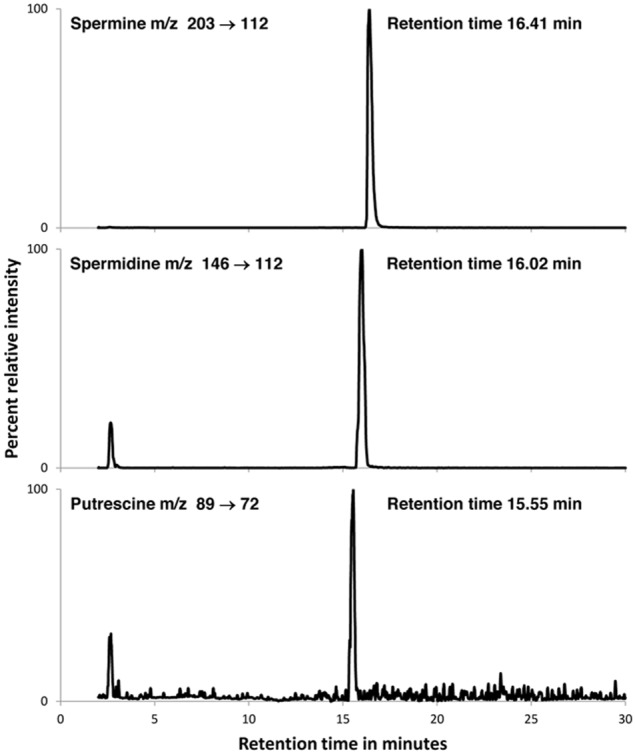
LC/MS/MS-MRM traces for the TCA extract of B. simplex 30N-5. Peaks for spermine (top), spermidine (middle) and putrescine (bottom) are shown. Samples were prepared and analyzed as described in Methods. Co-chromatography experiments in which the authentic compounds were added to the bacterial extract showed single peaks for each trace with appropriate augmentation of the peak areas. A quantitative summary of the results is presented in Table 4.
Table 4.
The concentrations of spermine, spermidine, and putrescine in methanol, TFA, and TCA extracts of B. simplex 30N-5 measured by LC/MS/MS-MRM using external standards.
| Sample | nmol/sample | nmol/sample | nmol/sample |
|---|---|---|---|
| 30N-5 methanol extract 1 | 0.92 | 1.46 | 1.09 |
| 30N-5 methanol extract 2 | 1.03 | 5.38 | 1.18 |
| 30N-5 TFA extract 1 | 613.75 | 355.15 | 2.02 |
| 30N-5 TFA extract 2 | 622.45 | 462.71 | 1.18 |
| 30N-5 TCA extract 1 | 400.68 | 388.64 | 5.28 |
| 30N-5 TCA extract 2 | 380.60 | 312.82 | 3.53 |
AcdS
Ethylene is inhibitory to root development and also induces plant defense response pathways. We detected a sequence in B. simplex 30N-5 that is similar to genes annotated as acdS in B. thuringiensis and in B. cereus JM-Mgvxx-63 (80%) and as D-cysteine desulfhydrase in B. panaciterrae DSM 19096 (88%) (Figure 1). The comparable genes for the two additional B. simplex strains available at NCBI were annotated as a cytochrome C biogenesis protein/D-cysteine desulfhydrase. These proteins are part of the PLP-dependent ACC family. However, genes homologous to this sequence were not detected in the typical PGPB group (blue group; Figure 1).
Antibiotics and related compounds
Bacteria synthesize a number of compounds that contribute to their survival in the rhizosphere. B. subtilis has been a paradigm for studying these antimicrobial compounds, which fall into two major classes: those synthesized on ribosomes (e.g., bacteriocins and lantibiotics) and post-translationally modified and those produced on large multienzymes known as Nonribosomal Peptide Synthetases (NRPSs), e.g., iturin and fengycin (Stein, 2005).
Bacteriocins
Many nonpathogenic bacteria produce bacteriocins, molecules used to compete with closely related bacteria, and many classes of these antimicrobial peptides are known. We scanned the B. simplex genome for genes potentially encoding bacteriocins and found three candidates, but genes comparable to those in B. simplex were not detected (n.d.) in any of the other PGP bacilli (Figure 1). The highest DNA sequence identity of genes encoding proteins for bacteriocin synthesis (Figure 1) was to the sequences found in B. panaciterrae. The highest DNA sequence identity of the bacteriocin biosynthesis gene, based on amino acid sequence, was 79% (Figure 1). These same gene sequences were picked up using the BAGEL3 website and a gene map is depicted in Supplementary Figure 1B.
Another protein with 99 and 100% identity to the two B. simplex strains in NCBI (B. simplex P558, CEG34010.l; B. simplex BA243, WP 034090.1, respectively) was also found. It matched to a protein described as a colicin V production protein. Although the gene neighborhoods were well conserved among the Bacillus species in Figure 2, the percentage DNA identity was 70% or lower (data not shown).
Additional secondary metabolites
Many PGPB synthesize diverse secondary metabolites, which have antibiotic activity, including lantibiotics, nonribosomally synthesized peptides, and polyketides. For example, subtilin is a 32-amino acid pentacyclic lantibiotic produced by B. subtilis (Stein, 2005). Although several subtilisin-like serine protease (AprE-like) genes are present in the B. simplex genome as well as proteins involved in subtilin processing (WprA and Vpr-like), no evidence was found in the B. simplex genome for the presence of genes similar to spaS and spaBTC, the subtilin structural genes and the genes promoting subtilin expression, respectively, nor to the genes spaIFEG, which confer immunity. Similarly, we saw no matches to genes encoding lantibiotic-like peptides such as sublanchin or subtilisin A produced by B. subtilis.
We looked for, but did not find, genes for the synthesis of nonribosomally synthesized peptide antibiotics in B. simplex, such as surfactin (see next section), iturin, or bacillomycin or for antimicrobial polyketides such as macrolactin, bacillaene, or difficidin, which are found in many PGPB Bacillus strains.
Other nonribosomal peptide synthetase (NRPS) products
Genes were found for the synthesis of koranimine, a cyclic imine. The genes involved are: korA, korB, korC, korD, as well as genes encoding a phosphopantetheinyl transferase (kfp) and a type II thioesterase (korTE) (Supplementary Figure 4). These genes had been detected earlier in an environmental Bacillus strain (NK2003) using a proteomics-based approach (Evans et al., 2011). We found them using a B. amyloliquefaciens subsp. plantarum FZB42 gene sequence (srfAA) in a blastp search, and although only 34% sequence identity was found between korA and srfAA, their amino acid adenylation domains were highly conserved (89.7%). Genome analysis led to the discovery of an orthologous gene in Bacillus sp. NK2003, which was annotated as a nonribosomal peptide synthetase. Koranimine synthetic genes were also found in the B. simplex II3b11 genome, each gene having greater than 95% DNA sequence identity to the kor genes of B. simplex 30N-5. An amino acid adenylation domain sequence that lines up with the middle part of korC was found in another part of the B. simplex genome (data not shown).
Although we utilized a gene encoding a surfactin to uncover the kor genes, we could not find any genes for surfactin production itself or any other NRPS-produced metabolites in B. simplex. Moreover, using a modification of a published procedure of the drop-collapsing assay (Kuiper et al., 2004) to determine surfactant activity, we found none of the three B. simplex strains tested exhibited a change in the diameter of the drops due to decrease in surface tension of the droplet (data not shown), suggesting that B. simplex 30N-5 lacks surfactant activity.
We also detected the polyketides described above using the ANTIsmash webserver. In addition, evidence for a gene encoding squalene synthetase and highly conserved with the genes of other B. simplex strains, but not found in the PGPB bacilli. An orthologous gene with 60 and 57.5% DNA identity was detected in B. panaciterrae and B. megaterium, respectively (data not shown). Similarly, a gene encoding chalcone synthase that is orthologous and 63–61% identical to genes in B. kribbensis and B. firmus, respectively, was found using ANTIsmash. The genes of the PGPB bacilli are orthologous and 51% identical to the B. simplex gene (data not shown).
Other pathways
Vitamins
More than one-third of the bacteria that have been sequenced possess genes for cobalamin (vitamin B12) synthesis (Raux et al., 2000), including B. simplex 30N-5. The B. simplex 30N-5 and the other B. simplex genomes also contain genes for riboflavin (vitamin B2) synthesis as previously described for B. subtilis (Stahmann et al., 2000). Riboflavin subunit alpha (ribF) was also found in all genomes. Similarly, the menaquinone (vitamin K2) pathway genes found in B. subtilis (Sato et al., 2001) were detected in the genomes of the B. simplex strains (chorismate synthase, isochorismate synthase, demethylmenaquinone methyltransferase, and 2-heptaprenyl-1,4-naphthoquinone methyltransferase).
Protein secretion systems
Gram-positive bacteria secrete proteins usually by translocation across the single membrane by the Sec pathway or via the two-arginine (Tat) pathway. B. simplex also possesses the genes, with a 99% identity, tatA, tatC and a third gene from the same family. In addition, a specialized secretion system, which is responsible for protein translocation across both the membrane and the cell wall, called a type VII secretion system (Tseng et al., 2009), was detected in the B. simplex 30N-5 genome.
Discussion
B. simplex has been shown in a number of reports to be an effective PGPB (Ertruk et al., 2010; Hassen and Labuschagne, 2010; Schwartz et al., 2013). In this report, we show that B. simplex strains 30N-5 and II3b11 are phylogenetically and genetically different from the known PGPB bacilli. Based on an analysis of 5 different housekeeping genes, they cluster in a separate subclade from most PGPB bacilli and their PGP-related genes. We thus placed them into a group separate from other PGPB (purple, Figure 1). Because interest in this species as a PGPB species and producer of novel enzymes has been increasing (Velivelli et al., 2015; Venkatachalam et al., 2015), we undertook a detailed investigation of the potential of B. simplex 30N-5 to act as a PGPB.
A survey of the three legume hosts used in the coinoculation studies suggests that B. simplex may behave differently among plant species. In our study on pea, we observed that simultaneous inoculation of B. simplex and R. leguminosarum bv. viciae resulted in an enhancement of nodulation and plant dry weights (Schwartz et al., 2013). In contrast, coinoculation of S. meliloti Rm1021 and B. simplex did not produce a significantly different dry weight measurement for either Melilotus alba or Medicago truncatula compared to the single inoculation. Moreover, B. simplex alone did not enhance the growth of the S. meliloti hosts compared to pea (Figure 3A).
In contrast, siratro responded positively to single inoculation with B. simplex and showed an increase in dry weight, as we had observed for pea (Schwartz et al., 2013). Whether or not this difference between the S. meliloti hosts and siratro and pea in response is a consequence of having smaller seeds vs. larger seeds is not known. The larger-seeded legumes contain more stored N, which results in a protracted growth response under N-deficient conditions. In addition, some PGPB strains appear to exhibit host specificity toward different plants (Kloepper, 1996). We are investigating these possibilities further.
Flagella are important for root colonization via cell motility, swarming, and biofilm formation. We found that B. simplex and the PGPB bacilli of the blue group (Figure 1) have an identical flagellar gene arrangement (Figure 4, Supplementary Figure 3). In contrast, members of the orange group (Figure 1), B. thuringiensis and B. cereus, have a different flagellar arrangement (Figure 4). It is not known if this difference is significant. In our previous studies of plant-associated Burkholderia species, the arrangement of the flagella genes between the plant-associated Burkholderia species and the mammalian and opportunistic pathogens was also dissimilar (Angus et al., 2014). In Burkholderia, the flagellar genes were linked together on a chromosome in the plant-associated species whereas they were located in different parts of the genome in the pathogen-clade. Flagella from pathogenic species are well known for triggering induced systemic resistance (ISR) in numerous plant species, but information about whether flagella from beneficial bacteria, especially from PGPB Bacillus spp., affect host responses is not available.
Volatiles are also important for inducing a systemic response. Although several genes associated with the acetoin pathway are present in B. simplex 30N-5, this strain did not produce detectable quantities of acetoin based on the Voges-Proskauer test. We observed that none of the B. simplex genomes that have been sequenced contain the alsD gene, whereas the genomes of the typical PGPB bacilli do. Because AlsD is missing, alpha-acetolactate, which is unstable following synthesis by AlsS via the condensation of two pyruvate molecules, cannot be converted to 3-hydroxy-2-butanone (acetoin), the precursor of 2,3-butanediol (Xiao and Xu, 2007). B. simplex M3-4, which has a positive effect on potato tuber yields, was also found to be negative for the Voges-Proskauer test (Velivelli et al., 2015). Nonetheless, this strain produces volatiles, such as 2-hexen-1-ol, 2,5-dimethylpyrazine, and several others, which inhibit Rhizoctonia solani growth. Thus, B. simplex strains are effective PGPB even though they do not emit 2,3-butanediol (this work; Velivelli et al., 2015). Future studies will investigate whether B. simplex 30N-5 produces similar volatiles.
Plant growth promotion frequently results from the action of hormones, and auxin synthesis is a common trait that is associated with PGPB bacilli. In it, tryptophan is converted to IPA by L-tryptophan aminotransferase (Patten and Glick, 1996). Although several aminotransferases were detected in the B. simplex genome, none could be specifically designated as this enzyme or as tryptophan transferase. In addition, even though genes were found for ipdC (indole pyruvic acid carboxylase), dhaS (indole-3-acetaldehyde dehydrogenase), aofH (indole-3-acetaldehyde oxidase), and yhcX (a nitrilase) based on similarities to other Bacillus spp., the gene identities were low except for dhaS and yhx (Figure 1). Using a sensitive and specific LC/MS/MS-MRM assay, we could not detect IAA in cell culture homogenates, further supporting the absence of an active biosynthetic pathway for this hormone in B. simplex 30N-5. These results lead us to propose that the commonly used Salkowski test is not definitive for the synthesis of auxin by bacteria.
Polyamines are also PGP compounds. Studies by Xie et al. (2014) showed that mutations in yecA (a permease) and speB (encoding one of the first steps in the conversion of agmatine to putrescine and then spermidine) eliminated the PGP activities of B. subtilis OKB105, e.g., root elongation. Reverse phase HPLC with UV detection of chemically-derivatized samples detected spermidine in the OKB105 culture filtrate (Xie et al., 2014). Our data using a more specific and sensitive assay show that all three polyamines are present in the culture medium of B. simplex 30N-5, and indeed based on the genome information, the entire pathway for polyamine production is present.
The enzyme acdS is thought to improve plant growth by interfering with ethylene formation. It does this by deaminating 1-aminocyclopropane-1-carboxylic acid (ACC), a direct precursor to ethylene production. Previously, we cloned a potential ACC deaminase gene from B. simplex 30N-5 by using acdS primers and found a sequence that was closely related to a gene encoding a pyridoxal phosphate-dependent enzyme. Experimental evidence will be needed to determine if this gene product has AcdS activity, but based on the fact that ACC deaminase is a member of the above protein family, it might be acdS. Nevertheless, because this gene in the B. simplex genomes (P558 and BA2H3) available at NCBI was annotated as a cytochrome C biogenesis protein/cysteine desulfhydrase, we cannot be completely certain that it encodes AcdS. However, ethylene synthesis is inhibited in plant root cells in response to B. subtilis OKB105 in response to polyamine synthesis (Xie et al., 2014), suggesting that polyamines may be an additional or alternative mechanism used by certain bacilli for reducing ethylene content in plants.
Genes encoding surfactin or related polyketides were not detected in B. simplex 30N-5, but the entire pathway for the synthesis of koranimine, a newly identified NRPS-synthesized peptide (Evans et al., 2011) was found. Although the function of this compound is unknown, cyclic imines are well known marine-based bacterial compounds that accumulate in crustaceans and fish possibly for defense purposes because of their toxicity to predators when ingested (Otero et al., 2011). Hence, the possibility exists that koranimine may play an antibiotic role in its interactions with other microbes and in protecting the plant. Again, additional studies are needed.
In summary, B. simplex 30N-5 exhibits potentially novel PGPB properties that are shared, but also dissimilar from some of the more commonly studied PGP bacilli. To be certain that this microbe has no deleterious effects on plants, we are testing it and related strains on both legumes and nonlegumes as well as on Caenorhabditis elegans to determine its lack of virulence. Published data showed that B. simplex 237 did not have a detrimental effect on C. elegans (Angus et al., 2014), and our preliminary results with B. simplex 30N-5 demonstrate no toxic effects on nematodes or onions (M. Arrabit and A.M. Hirsch, unpubl.). Also, based on our investigations of the genome, no obvious virulence genes are observed. Thus, B. simplex 30N-5 may be an excellent candidate to be added to the group of beneficial bacilli that help plants grow and survive under sustainable agriculture conditions.
Author contributions
MM, PM, KF, and AH designed and conducted experiments, and wrote the manuscript. ES and AH conceived the work, and ES made critical revisions to the manuscript. ES, WV, PM, KC, TA, JS, and AH acquired the genomics data, and ST and AH interpreted it. MM, PM, KF, TI, LH, MC, TS, NF, and JV acquired experimental data. MM and JS did the statistical analyses. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Kris Reddi (UCLA) for isolating the B. simplex 30N-5 DNA for genome sequencing and Dr. Flora Pule-Meulenberg (Botswana College of Agriculture) for additional Bacillus strains. The following UCLA undergraduates, Leah Briscoe, Fiorella Candamo, Ariga Gharibian, Ethan Humm, Sofianne Kajouke, Suzie Martikyan, and Darren Shum, are acknowledged for their contributions. This research was supported in part by NSF Award IOS 1201735, and a grant from the Shanbrom Family Foundation to AH. Much of the genomics work was performed through the IMG/ER website of the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, which is supported under Contract No. DE-AC02-05CH11231. Additional funding came from a postdoctoral fellowship awarded to PM from “Fundación Ramón Areces” (Spain) and an award to MC from the State Foundation for Studying Abroad of China. WV was funded in part by NSF DUE grant 1022918. Funding to sequence the B. simplex 30N-5 genome was provided by the Howard Hughes Medical Institute (Award No. 52006944).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00784
References
- Altschul S. F., Gish W., Miller E. W., Myers D. J., Lipman J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Amaya-Gómez C. V., Hirsch A. M., Soto M. J. (2015). Biofilm formation assessment in Sinorhizobium meliloti reveals interlinked control with surface motility. BMC Microbiol. 15:58. 10.1186/s12866-015-0390-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus A. A., Agapakis C. M., Fong S., Yerrapragada S., Estrada-de los Santos P., Yang P., et al. (2014). Plant-associated symbiotic Burkholderia species lack hallmark strategies required in mammalian pathogenesis. PLoS ONE 9:e83779. 10.1371/journal.pone.0083779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus A. A., Lee A. S., Lum M. R., Shehayeb M., Hessabi R., Fujishige N. A., et al. (2013). Nodulation and effective nitrogen fixation of Macroptilium atropurpureum (siratro) by Burkholderia tuberum, a beta-proteobacterium, are influenced by environmental factors. Plant Soil 369, 543–562. 10.1007/s11104-013-1590-7 [DOI] [Google Scholar]
- Barritt M. M. (1936). The intensification of the Voges-Proskauer reaction by the addition of a-naphthol. J. Pathol. Bacteriol. 42, 441–454. 10.1002/path.1700420212 [DOI] [Google Scholar]
- Beringer J. E. (1974). R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84, 188–198. 10.1099/00221287-84-1-188 [DOI] [PubMed] [Google Scholar]
- Bottone E. J. (2010). Bacillus cereus, a volatile human pathogen. Clin. Microbiol. Rev. 23, 382–398. 10.1128/CMR.00073-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero-Mellado J., Onofre-Lemus J., Estrada-de los Santos P., Martínez-Aguilar L. (2007). The tomato rhizosphere, an environment rich in nitrogen-fixing Burkholderia species with capabilities of interest for agriculture and bioremediation. Appl. Environ. Microbiol. 73, 5308–5319 10.1128/aem.00324-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.-H., Koumoutsi A., Scholz R., Eisenreich A., Schneider K., Heinemeyer I., et al. (2007). Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 25, 1007–1014. 10.1038/nbt1325 [DOI] [PubMed] [Google Scholar]
- Cheng H.-P., Walker G. C. (1998). Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 180, 5183–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce O., Hugon P., Lagier J.-C., Bibi F., Robert C., Azhar E. I., et al. (2014). Genome sequence of Bacillus simplex strain P558, isolated from a human fecal sample. Genome Announc. 2:e01241-14. 10.1128/genomeA.01241-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croes C. L., Moens S., van Bastelaere E., Vanderleyden J., Michiels K. W. (1993). The polar flagellum mediates Azospirillum brasilense adsorption to wheat roots. J. Gen. Microbiol. 139, 2261–2269. 10.1099/00221287-139-9-2261 [DOI] [Google Scholar]
- De Weger L. A., Van der Vlugt C. I. M., Wijfjes A. H. M., Bakker P. A. H. M., Schippers B., Lugtenberg B. (1987). Flagella of a plant-growth-stimulating Pseudomonas fluorescens strain are required for colonization of potato roots. J. Bacteriol. 169, 2769–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Werra P., Péchy-Tarr M., Keel C., Maurhofer M. (2009). Role of gluconic acid production in the regulation of biocontrol traits of Pseudomonas fluorescens CHA0. Appl. Environ. Microbiol. 75, 4162–4174. 10.1128/AEM.00295-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietel K., Beator B., Budiharjo A., Fan B., Borriss R. (2013). Bacterial traits involved in colonization of Arabidopsis thaliana roots by Bacillus amyloliquefaciens FZB42. Plant Pathol. J. 29, 59–66. 10.5423/PPJ.OA.10.2012.0155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott G. N., Chen W.-M., Bontemps C., Chou J.-H, Young, J. P. W., Sprent J. I., et al. (2007). Nodulation of Cyclopia spp. (Leguminosae, Papilionoideae) by Burkholderia tuberum. Ann. Bot. 100, 1403–1411. 10.1093/aob/mcm227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Tarabily K. A., Nassar A. H., Sivasithamparam K. (2008). Promotion of growth of bean (Phaseolus vulgaris L.) in a calcareous soil by a phosphate-solubilizing, rhizosphere-competent isolate of Micromonospora endolithica. Appl. Soil Ecol. 39, 161–171. 10.1016/j.apsoil.2007.12.005 [DOI] [Google Scholar]
- Ertruk Y., Ercisli S., Haznedar A., Cakmakci R. (2010). Effects of plant growth rhizobacteria (PGPR) on rooting and root growth of kiwifruit (Actinidia deliciosa) stem cuttings. Biol. Res. 43, 91–98. 10.4067/s0716-97602010000100011 [DOI] [PubMed] [Google Scholar]
- Evans B. S., Ntai I., Chen Y., Robinson S. J., Kelleher N. L. (2011). Proteomics-based discovery of koranimine, a cyclic imine natural product. J. Amer. Chem. Soc. 133, 7316–7319. 10.1021/ja2015795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. (1979). Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U.S.A. 76, 1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassen A. I., Labuschagne N. (2010). Root colonization and growth enhancement in wheat and tomato by rhizobacteria isolated from the rhizoplane of grasses. World J. Microbiol. Biotechnol. 26, 1837–1846. 10.1007/s11274-010-0365-z [DOI] [Google Scholar]
- Herberich E., Sikorski J., Hothorn T. (2010). A Robust procedure for comparing multiple means under heteroscedasticity in unbalanced designs. PLoS ONE 5:e9788. 10.1371/journal.pone.0009788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D., Maymon M., Agapakis C. M., Lee A., Wang A., Prigge B. A., et al. (2013). A survey of the microbial community in the rhizosphere of the dominant plant of the Negev Desert, Zygophyllum dumosum Boiss., using cultivation-dependent and –independent methods. Am. J. Bot. 100, 1713–1725. 10.3732/ajb.1200615 [DOI] [PubMed] [Google Scholar]
- Kerovuo J., Lauraeus M., Nurminen P., Kalkkinen N., Apajalahti J. (1998). Isolation, characterization, molecular gene cloning and sequencing of a novel phytase from Bacillus subtilis. Appl. Environ. Microbiol. 64, 2079–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khayi S., Raoul des Essarts Y., Mondy S., Moumni M., Hélias V., Beury-Cirou A., et al. (2015). Draft genome sequences of the Three Pectobacterium-antagonistic bacteria Pseudomonas brassicacearum PP1-210F and PA1G7 and Bacillus simplex BA2H3. Genome Announ. 3:e01497-14. 10.1128/genomeA.01497-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. O., Lee J. K., Kim H. K., Yu J. H., Oh T. K. (1998). Cloning of the thermostable phytase gene (phy) from Bacillus sp. DS11 and its over expression in Escherichia coli. FEMS Microbiol. Lett. 162, 185–191. 10.1111/j.1574-6968.1998.tb12997.x [DOI] [PubMed] [Google Scholar]
- Kimura M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- Kloepper J. W. (1996). Host specificity in microbe-microbe interactions. Bioscience 46, 406–409. 10.2307/1312874 [DOI] [Google Scholar]
- Kloepper J. W., Ryu C.-M., Zhang S. (2004). Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology. 94, 1259–1266 10.1094/PHYTO.2004.94.11.1259 [DOI] [PubMed] [Google Scholar]
- Koeppel A., Perry E. B., Sikorski J., Krizanc D., Warner A., Ward D. M., et al. (2008). Identifying the fundamental units of bacterial diversity: A paradigm shift to incorporate ecology into bacterial systematics. Proc. Nat. Acad. Sci. U.S.A. 105, 2504–2509. 10.1073/pnas.0712205105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga J. (1995). Structure and function of indolepyruvate decarboxylase, a key enzyme in indole-3-acetic acid biosynthesis. Biochim. Biophys. Acta 1249, 1–13. 10.1016/0167-4838(95)00011-I [DOI] [PubMed] [Google Scholar]
- Kuiper I., Lagendijk E. L., Pickford R., Derrick J. P., Lamers G. E., Thomas-Oates J. E., et al. (2004). Characterization of two Pseudomonas putida lipopeptide biosurfactants, putisolvin I and II, which inhibit biofilm formation and break down existing biofilms. Mol. Microbiol. 51, 97–113. 10.1046/j.1365-2958.2003.03751.x [DOI] [PubMed] [Google Scholar]
- Lertcanawanichakul M., Sawangnop S. (2008). A comparison of two methods used for measuring the antagonistic activity of Bacillus species. Walailak J. Sci. Technol. 5, 161–171. 10.2004/wjst.v5i2.86 [DOI] [Google Scholar]
- Lloyd S. P. (1982). Least squares quantization in pcm. IEEE Transact. Inform. Theory. 28, 129–137. 10.1109/TIT.1982.1056489 [DOI] [Google Scholar]
- Machlis L, Torrey, J. G. (1956). Plants in Action. San Francisco, CA: W.H Freeman. [Google Scholar]
- Markowitz V. M., Chen I. M., Palaniappan K., Chu K., Szeto E., Grechkin Y., et al. (2012). IMG: the integrated microbial genomes database and comparative analysis system. Nucl. Acids Res. 40, D115–D122. 10.1093/nar/gkr1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Hidalgo P., Galindo-Villardón P., Igual J. M., Martínez-Molina E. (2014). Micromonospora from nitrogen fixing nodules of alfalfa (Medicago sativa L.). A new promising plant probiotic bacteria. Sci. Rep. 4:6389. 10.1038/srep06389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzaeva O. V., Shirokikh I. G. (2010). The production of auxins by the endophytic bacteria of winter rye. Appl. Biochem. Microbiol. 46, 44–50. 10.1134/S0003683810010072 [DOI] [PubMed] [Google Scholar]
- Miller J. H. (1972). Experiments in Molecular Genetics, Cold Spring Harbor Laboratory Press. New York, NY: Cold Spring Harbor. [Google Scholar]
- Moulin L., Munive A., Dreyfus B., Boivin-Masson C. (2001). Nodulation of legumes by members of the beta-subclass of Proteobacteria. Nature 411, 948–950. 10.1038/35082070 [DOI] [PubMed] [Google Scholar]
- Otero A., Chapela M.-J., Atanassova M., Vieites J. M., Cabado A. G. (2011). Cyclic imines: chemistry and mechanism of action: a review. Chem. Res. Toxicol. 24, 1817–1829. 10.1021/tx200182m [DOI] [PubMed] [Google Scholar]
- Patten C. L., Glick B. R. (1996). Bacterial biosynthesis of indole-3-acetic acid. Can. J. Microbiol. 42, 207–220. [DOI] [PubMed] [Google Scholar]
- Pradhan N., Sukla L. B. (2005). Solubilization of inorganic phosphates by fungi isolated from agriculture soil. Afr. J. Biotechnol. 5, 850–854. 10.5897/AJB2006.000-5050 [DOI] [Google Scholar]
- Raux E., Schubert H. L., Warren M. J. (2000). Biosynthesis of cobalamin (vitamin B12): a bacterial conundrum. Cell. Mol. Life. Sci. 57, 1880–1893. 10.1007/PL00000670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu C-M., Farag M. A., Hu C.-H., Reddy M. S., Wei H-X., Paré P. W., et al. (2003). Bacterial volatiles promote growth in Arabidopsis. Proc. Nat. Acad. Sci. U.S.A. 100, 4927–4932. 10.1073/pnas.0730845100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Yamada Y., Ohtani Y., Mitsui N., Murasawa H., Araki S. (2001). Production of Menaquinone (vitamin K2)-7 by Bacillus subtilis. J. Biosci. Bioeng. 91, 16–20. 10.1016/S1389-1723(01)80104-3 [DOI] [PubMed] [Google Scholar]
- Schlaeppi K., Dombrowski N., Oter R. G., Ver Loren van Themaat E., Schulze-Lefert P. (2014). Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc. Natl. Acad. Sci. U.S.A. 111, 585–592. 10.1073/pnas.1321597111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz A. R., Ortiz I., Maymon M., Fujishige N. A., Hanamoto K, Diener A., et al. (2013). Bacillus simplex alters legume root architecture and nodule morphology when co-inoculated with Rhizobium. Agronomy. 3, 595–620. 10.3390/agronomy3040595 [DOI] [Google Scholar]
- Sekowska A., Bertin P., Danchin A. (1998). Characterization of polyamine synthesis pathway in Bacillus subtilis 168. Mol. Microbiol. 29, 851–858. 10.1046/j.1365-2958.1998.00979.x [DOI] [PubMed] [Google Scholar]
- Sikorski J., Nevo E. (2005). Adaptation and incipient sympatric speciation of Bacillus simplex under microclimatic contrast at “Evolution Canyons” I and II, Israel. Proc. Natl. Acad. Sci. USA. 102, 15924–15929. 10.1073/pnas.0507944102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slonim N., Aharoni E., Crammer K. (2013). Hartigan's K-means versus Lloyd's K-means: is it time for a change?, in Proceedings of the 23rd International Joint Conference on Artificial Intelligence (Menlo Park, CA: AAAI Press; ), 1677–1684. [Google Scholar]
- Somasegaran P., Hoben H. J. (1994). Handbook of Rhizobia. New York, NY; Berlin; Heildelberg: Springer-Verlag. 337. [Google Scholar]
- Somers E., Ptacek D., Gysegom P., Srinivasan M., Vanderleyden J. (2005). Azospirllum brasilense produces the auxin-like phenylacetic acid by using the key enzyme for indole-3-acid biosynthesis. Appl. Environ. Microbiol. 71, 1803–1810. 10.1128/AEM.71.4.1803-1810.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaepen S., Vanderleyden J. (2011). Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 3:a001438. 10.1101/cshperspect.a001438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahmann K.-P., Revuelta J. L., Seulberger H. (2000). Three biotechnical processes using Ashbya gossypii, Candida famata, or Bacillus subtilis compete with chemical riboflavin production. Appl. Microbiol. Biotechnol. 53, 509–516. 10.1007/s002530051649 [DOI] [PubMed] [Google Scholar]
- Stein T. (2005). Bacillus subtilis antibiotics: structure, syntheses and specific functions. Mol. Microbiol. 56, 845–857. 10.1111/j.1365-2958.2005.04587.x [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl. Acids Res. 25, 4876–4882. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng T.-T., Tyler B. M., Setubal J. C. (2009). Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol. 9:S2. 10.1186/1471-2180-9-S1-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye A. J., Siu F. K., Leung T. Y., Lim B. L. (2002). Molecular cloning and the biochemical characterization of two novel phytase from Bacillus subtilis 168 and Bacillus licheniformis. Appl. Microbiol. Biotechnol. 59, 190–197. 10.1007/s00253-002-1033-5 [DOI] [PubMed] [Google Scholar]
- Vacheron J., Desbrosses G., Bouffaud M.-L., Touraine B., Moënne-Loccoz Y., Muller D., et al. (2013). Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 4:356. 10.3389/fpls.2013.00356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme P., Goris J., Chen W.-M., de Vos P., Willems A. (2002). Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov., nodulate the roots of tropical legumes. Syst. Appl. Microbiol. 25, 507–512. 10.1078/07232020260517634 [DOI] [PubMed] [Google Scholar]
- Velivelli S. L. S., Kromann P., Lojan P., Rojas M., Franco J., Suarez J. P., et al. (2015). Identification of mVOCs from Andean rhizobacteria and field evaluation of bacterial and mycorrhizal inoculants on growth of potato in its center of origin. Microb. Ecol. 69, 652–667. 10.1007/s00248-014-0514-2 [DOI] [PubMed] [Google Scholar]
- Venkatachalam S., Gowdaman V., Prabagaran S. R. (2015). Culturable and culture-independent bacterial diversity and the prevalence of cold-adapted enzymes from the Himalayan Mountain Ranges of India and Nepal. Microb. Ecol. 69, 472–491. 10.1007/s00248-014-0476-4 [DOI] [PubMed] [Google Scholar]
- Voges O., Proskauer B. (1898). Beitraege zur Ernaehrungsphysiologie und zur Differential Diagnose der Bakterien der hemmorrhagischen Septicamie. Z. Hyg. 28, 20–32. 10.1007/BF02285362 [DOI] [Google Scholar]
- Werkman C. H. (1930). An improved technic for the Voges-Proskauer test. J. Bacteriol. 20, 121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski-Dyé F., Borziak K., Khalsa-Moyers G., Alexandre G., Sukharnikov L. O., Wuichet K., et al. (2011). Azospirillum genomes reveal transition of bacteria from aquatic to terrestrial environments. PLoS Genet. 7:e1002430. 10.1371/journal.pgen.1002430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z., Xu P. (2007). Acetoin metabolism in bacteria. Crit. Rev. Microbiol. 33, 127–140. 10.1080/10408410701364604 [DOI] [PubMed] [Google Scholar]
- Xie S-S., Wu H-J., Zang H-Y., Wu L-M., Zhu Q-Q., Gao X-W. (2014). Plant growth promotion by spermidine-producing Bacillus subtilis OKB105. Mol. Plant Microbe Int. 27, 655–663. 10.1094/MPMI-01-14-0010-R [DOI] [PubMed] [Google Scholar]
- Zhang H., Kim M.-S., Krishnamachari V., Payton P., Sun Y., Grimson M., et al. (2007). Rhizobacterial volatile emissions regulate auxin homeostasis and cell expansion in Arabidopsis. Planta 226, 839–851. 10.1007/s00425-007-0530-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.