Abstract
Biotic and abiotic stresses affect plant development and production through alternation of the gene expression pattern. Gene expression itself is under the control of different regulators such as miRNAs and transcription factors (TFs). MiRNAs are known to play important roles in regulation of stress responses via interacting with their target mRNAs. Here, for the first time, seven conserved miRNAs, associated with drought, heat, salt and cadmium stresses were characterized in sunflower. The expression profiles of miRNAs and their targets were comparatively analyzed between leaves and roots of plants grown under the mentioned stress conditions. Gene ontology analysis of target genes revealed that they are involved in several important pathways such as auxin and ethylene signaling, RNA mediated silencing and DNA methylation processes. Gene regulatory network highlighted the existence of cross-talks between these stress-responsive miRNAs and the other stress responsive genes in sunflower. Based on network analysis, we suggest that some of these miRNAs in sunflower such as miR172 and miR403 may play critical roles in epigenetic responses to stress. It seems that depending on the stress type, theses miRNAs target several pathways and cellular processes to help sunflower to cope with drought, heat, salt and cadmium stress conditions in a tissue-associated manner.
Keywords: abiotic stress, miRNA, sunflower (Helianthus annuus), regulatory network
Introduction
Abiotic and biotic stresses impose challenging physiological hurdles to plants. As a response to adverse environmental conditions, plants re-program their cellular activities through multiple gene regulatory mechanisms including post-transcriptional regulation of gene expression. Transcription factors (TFs) and non-coding RNAs are the two major regulatory elements in functional genomics (Deihimi et al., 2012; Mahdi et al., 2013; Panahi et al., 2013; Chiasson et al., 2014).
Small RNAs, particularly microRNAs (miRNAs), have emerged as key post-transcriptional gene regulatory molecules in plants and animals (Trindade et al., 2010; Giacomelli et al., 2012; Sunkar et al., 2012). MiRNAs are 18–22 nucleotides in length and regulate expression of their target genes by degradation or translational repression (Voinnet, 2009; Sunkar et al., 2012). In particular, miRNAs play important roles in plant responses to abiotic and biotic stresses including cold, salt, heat, dehydration, oxidative and mechanical stresses (Jin et al., 2013; Panahi et al., 2013). For example, some of the genes in ARF transcription factor family are targeted by miR160 and miR167 through auxin regulation interference (Gutierrez et al., 2009), and seem to be important for the attenuation of plant growth and development under stress conditions (Sunkar et al., 2012). Another microRNA, miR398, is associated with response to abiotic and biotic stress condition (Zhu et al., 2011) and its expression is up-regulated in response to water deficit in Medicago truncatula (Trindade et al., 2010). MiR403 controls the expression of AGO2 (Allen et al., 2005), which is known to have an antiviral role (Harvey et al., 2011). Srivastava et al. (2013) reported that miR426 is up-regulated in response to arsenic stress in Brassica juncea. In another study, miR842 expression levels were observed to be up-regulated in Arabidopsis (Moldovan et al., 2010). These are handful examples of miRNAs that are differentially expressed in response to various stress conditions.
Sunflower (Helianthus annuus L.) is one of the most important oilseed crops and is resistant to various abiotic stresses, due to its metabolic, physiological and morphological adaptation strategies. This crop is of special interest for its adaptation to high temperatures, limited water availability, high salinity and heavy metal concentrations in the soil (Merah et al., 2012). Using bioinformatics methods, previous studies have identified and reported several miRNAs in sunflower. A number of these were experimentally verified (Barozai et al., 2012; Monavar Feshani et al., 2012). However, their expression patterns under different stress conditions have not been studied.
We have examined the roles of seven of these sunflower miRNAs in response to different abiotic stresses including drought, high temperature, salinity and cadmium. Seven mentioned miRNAs, miR160, miR167, miR172, miR398, miR403, miR426 and miR842, play crucial roles in plant growth and show different expression patterns under both biotic and abiotic stress conditions in several plant species. Simultaneously, we analyzed the expression levels of these miRNAs concurrently with their potential targets to corroborate the putative miRNA-target relationship. Based on our results, several gene regulatory networks were constructed to gain a comprehensive overview of the role of these miRNAs in stress responses.
Materials and methods
Prediction of miRNA targets
Most plant miRNAs bind to their targets with perfect or near-perfect complementarity (Jones-Rhoades and Bartel, 2004). We have used this characteristic for predicting potential miRNA targets. Sequences of pre-miRNAs were obtained from the public database, miRBase (Kozomara and Griffiths-Jones, 2011). The number of mismatches at complementary sites between miRNA sequence and potential mRNA target were kept below 4 and no gap at the complementary sites was tolerated. The miRNA targets were predicted using psRNAtarget online sever (http://plantgrn.noble.org/psRNATarget/).
Plant material, growth conditions and stress treatments
Sunflower (Helianthus annuus, variety Sirna) seeds were surface sterilized and placed in petri dishes containing two layers of damp sterile filter paper. For germination, the seeds were incubated at room temperature for 4 days.
For applying drought stress treatment, five germinated seeds were sown in round pots (10.5 × 13.5 cm, D × H; five plants/pot) filled with soil (¾ river sand and ¼ soil). Plants were grown in the glasshouse under the controlled condition (16 h of light, 24°C; 8 h of dark, 15°C) and well watered.
For heat, salinity and cadmium stress treatments, five seedlings were grown in a hydroponic system. The system contained aerated Hoagland solutions, pH 5.8 (Hoagland and Arnon, 1950) in plastic pots (1.5 liters volume). One-week-old plantlets were planted in ¼ strength Hoagland's nutrient for 1 more week to immerse the growing roots in the nutrient solution. Plantlets were then transferred to half-strength nutrient solution and grown until 6-leave stage (about 4 weeks). Four weeks old plantlets were then submitted to different stresses as specified in the following sections.
Drought stress
Four weeks old plantlets (6 leaf stage) of sunflower were subjected to water stress by withholding water for 12, 24 and 48 h. Relative water content (RWC) was measured in leaves to determine the plant water status following Faatsky method (Čatský, 1960).
Heat stress
Four weeks old plantlets in ½ strength Hoagland solution were transferred to humidity growth chamber (Memmert, Germany) with 70% relative humidity and maintained at 42 ± 1°C for 1.5, 3 and 6 h (Senthil-Kumar et al., 2003; Mashkina et al., 2010; Mangelsen et al., 2011). We selected stress-related HSP70-related protein gene (Gene Bank ID: AAB57695.1) to investigate the effect of heat stress in root tissue of sunflower.
Salt stress
Plantlets were moved into solution contained either 75 or 150 mM of NaCl (Di Caterina et al., 2007). Leaf and root samples from stressed plants were collected 24 h later alongside control samples. Potassium to sodium ratio was measured as criteria for examining different salinity stress levels.
Cadmium stress
Plantlets were transferred into containers with either 5 or 20 mg/L of CdCl2 (Niu et al., 2007). Stress and control samples were collected after 24 h from leaf and root tissue and immediately frozen in liquid nitrogen before they stored at −80°C. Cadmium content was measured as an indicative of stress.
Total RNA isolation from leaves and roots
Total RNAs were extracted from sunflower root and leaf tissues using TRIZOL Reagent (Invitrogen, USA) according to the manufacturer's instructions. The extractions were performed separately for two independent biological replicates. Equal amount of total RNA was subsequently pooled for each sample based on their concentration. The quantity and quality of purified RNAs were assessed with a Biophotometer spectrophotometer (Eppendorf, Germany) and the integrity was evaluated by conducting a denaturing agarose gel electrophoresis. All RNA samples were stored at −80°C until further processing.
Stem-loop reverse transcription
Stem-loop RT primers for H. annuus miR160, miR167, miR172, miR398, miR403, miR426 and miR842 were designed according to Varkonyi-Gasic et al. (2007) (Table S1). The miRNA specific RT reactions were carried out using prime script RT reagent kit (Cat. #RR037A, TAKARA, Japan). The miRNA stem–loop reverse transcription was accomplished according to Varkonyi-Gasic et al. (2007) with minor modifications at cDNA synthesis, using 200 ng of total RNA sample of treated leaf and root tissues (1 μL), 1 μL of 1 μM miRNA primer RT and 5.5 μL nuclease-free water. The mix was incubated at 65°C for 5 min followed by incubation on ice for 2 min. Two microliters of Primescript buffer (5×) and 0.5 μL Primescript RT enzyme mix were added to each tube and the RT reaction was performed for 30 min at 16°C followed by 60 cycles at 30°C for 30 s, 42°C for 30 s, 50°C for 1 s and terminated by incubation at 85°C for 5 min. Control reactions were performed with no RT primer and no RNA samples.
Quantitative real-time PCR
To measure and compare the expression levels of the selected H. annuus miRNAs in root and leaf tissues under different stress treatments, RT-qPCR was conducted using SYBR Premix Ex Taq II (Cat. #RR820A, TAKARA) in a Rotor-GeneQ Real-Time PCR system (Qiagen).
For RT-qPCR analysis, 6 μL SYBR Premix Ex Taq II (2×), 1 μL (10 pmol) each of forward and reverse primers, 3 μL nuclease-free water and 1 μL RT stem-loop cDNA product were mixed. Forward primers were specifically designed for each miRNA, and 5′-GTGCAGGGTCCGAGGT-3′ sequence was used as the universal reverse primer (Varkonyi-Gasic et al., 2007) (Table S1). The RT-qPCR reactions were performed using following conditions; initial denaturation at 95°C for 30 s, followed by 40 cycles at 95°C for 5 s and 60°C for 20 s. The melting curves were generated during denaturation step at 95°C followed by the cooling of PCR products at 50°C and the fluorescence signals were collected continuously from 50 to 95°C as the temperature increased at 0.2°C per second. All reactions were repeated four times. For each condition, the RT-qPCR experiments were run as pooled biological duplicates and expression levels were normalized according to the previous studies (Schmittgen and Livak, 2008). Relative fold changes in miRNA expression were calculated using the comparative Ct (2−ΔΔCt) method with 18S rRNA as the endogenous control (Schmittgen and Livak, 2008). The miRNAs clustering analysis, based on their relative expression, and Pearson correlation between expression miRNAs and target genes in each stress were performed using Minitab 16.0. The heat map of expression was visualized using R 3.0 package.
Analysis of miRNAs target expression with RT-qPCR
To determine the expression of predicted miRNA targets and possible discovery of novel miRNA target genes under the drought, heat, salinity and cadmium stresses in sunflower, expression levels of miRNA-related target genes were measured with quantitative real-time PCR. ESTs for target quantification analysis were selected based on two criteria: (1) protein found in BLAST search should be a previously published target of the related miRNA in other plant species. (2) The EST should have a possible protein-coding ORF that can allow us to design RT-qPCR primers for the conserved 3′ UTR region. RT-qPCR primers for these selected genes (Table S1) were designed using Primer3 according to following criteria: (1) Primers 3′ self-complementary = 0. (2) Primer annealing temperatures = 62 ± 0–3°C. (3) Product size limit for primer pairs = 150 bp.
Total cDNAs were synthesized from 1 μg RNA using RevertAid™ First Strand cDNA Synthesis Kit (Fermentas) according to the manufacturer's instructions. Further expression analysis was performed for all miRNA targets using the same batch of RNA samples for miRNA RT-qPCR assay. One microliter of this cDNA was amplified with 0.6 μM of specific primers in a total of 10 μl volume using SYBER Premix Ex Taq II (Cat. #RR820A, TAKARA) with Rotor-GeneQ Real-Time PCR system (Qiagen). The quantification was performed using 18S rRNA (Gene ID: 18250984, forward: TTCAGACTGTGAAACTGCGAATGG /reverse: TCATCGCAGCAACGGGCAAA) as a normalizer and four independent PCR results with acceptable efficiency (1.8–2.2) were averaged. Specified RT-qPCR thermal setup was adjusted as follows: pre-denaturation step at 95°C for 1 min, followed by 40 cycles of 95°C for 30 s, 60°C for 1 min. The melting curves were generated as mentioned for miRNAs.
Network analysis of miRNAs
To construct miRNAs and target genes interaction network, RESNET Plant database of Pathway Studio software v.9 (Elsevier) was used. This database includes new aliases for genes in the model and non-model plant species including barely, corn, tomato, potato and tobacco and collects data through MedScan (text mining tool) to extract functional relationships between miRNAs, proteins, stresses and cellular processes (Nikitin et al., 2003; Alimohammadi et al., 2013; Ebrahimie et al., 2014). In addition to adding the result of this study to prediction database of RESNET database, this database was also updated by MedScan, especially from literature on miRNAs/target genes and drought, heat, salt and cadmium stress conditions before network construction. To predict the interaction networks, the software makes different groups of miRNAs and finds the relations between a protein and its group using algorithms such as Fisher's Exact Test (Alanazi et al., 2013; Ebrahimie et al., 2014).
Network constructed by union selected and physical interaction algorithms were used to make statistical subnetworks based on miRNAs and their target genes. Also, GO (gene ontology) analysis was performed through DAVID Functional Annotation web-tool (https://david.ncifcrf.gov/) and separate tables were produced for biological process, molecular function and cellular component categories. MiRNAs and putative target genes are marked with yellow circles (Figures 6, 7).
Results
In this study, expression levels of seven miRNAs including miR160, miR167, miR172, miR398, miR403, miR426 and miR842 and their targets were investigated in response to four abiotic stress conditions including drought, heat, salt and cadmium stresses. In addition, the impacts of these stresses on the physiological and molecular characteristics of plants were evaluated by measuring the changes in RWC, expression of HSP70-related protein, sodium, potassium and cadmium concentrations in different tissues of plant, grown under stress (Supplementary 1). The expression level of HSP70-related protein up-regulated over time compared to control in root tissue with the highest level at 6 h (Table S2). Moreover, alteration of sodium, potassium and cadmium concentration were remarkably higher in root tissue compared to leaf, in particular in later stage of stress (Table S3A). The average RWC of plants in response to drought stress showed slight decline at 12 and 24 h, and it started to reduce sharply subsequently (Table S3B). The results indicated that plants were significantly affected by stress exposure and were able to initiate stress related responses.
Prediction and determination of H. annuus miRNA targets
The mRNA targets of these miRNAs (Table 1) were predicted via psRNAtarget database using selected miRNAs as queries. Targets of miR167 and miR403 were previously reported in the other plant species (Table 1). We predicted and analyzed new targets for miR160, miR398, miR426 and miR842 for the first time in sunflower. Protein sequence analysis of the new putative Helianthus miR160 target, QHG18J04.yg.ab1 showed the presence of ARF, phosphorylase and kinase domains (data not shown) in this protein. Interestingly, COX5b, which was previously introduced as miR398 target in Arabidopsis, was identified as a predicted target for miR172 in sunflower.
Table 1.
Putative Helianthus annuus stress-inducible miRNA target genes.
| miRNA | miRNA sequence | Predicted targets | Accession number |
|---|---|---|---|
| miR160g | UGCCUGGCUCCCUGUAUGCCA | QHG18J04.yg.ab1 | BU026935 |
| miR167e | UGAAGCUGCCAGCAUGAUCU | Auxin Response Factors6 (ARF6) | 839913 |
| miR172d | AGAAUCCUGAUGAUGCUGC | Cytochrome-c oxidase activity (COX5B-2) | 844363 |
| miR398 | GUGUUCUCAGGUCGCCCC | Glycosyltransferase (NtGT5b) | AB176524.1 |
| miR403 | UUAGAUUCACGCACAAACUCG | Argonaute 2 (AGO2) | 840016 |
| miR426 | CUUUGGAAGUUUGUCCUUAGU | Probable trehalose-phosphate phosphatase 2 (OsTPP2) | AB277360.1 |
| miR842 | UCAUGGUCAGAUUCAUCAUCC | (R)-mandelonitrile lyase | AT1G73050 |
Expression of miRNAs and putative targets under drought stress
MiRNAs expression levels were significantly down-regulated (P < 0.05) in leaves of plants grown under drought stress with the lowest expression levels were at 48 h (except miR403). MiR172 expression modulation was not significant (P < 0.05) in all period of stress in root tissue. The expression patterns of miR160, miR426 and miR842 were similar in both tissues, except miR160 which showed opposite pattern at 48 h after drought stress in root tissue. The miRNAs, miR167 and miR398 showed similar trend under drought stress in both tissues. They were down-regulated at all-time points within range of 2- to 19-fold change, whereas expression of miR167 was slightly up-regulated at 48 h compared to moderate stress in root tissue. MiR403 showed the opposite pattern in leaf and root tissues at 24 h. Its expression abruptly decreased (24-fold compared to control) in leaf tissue. It, however, exhibited an increasing trend in root tissue while its expression was still lower than control condition (Table 2; Figure 1).
Table 2.
Expression of miRNAs and their target genes response to abiotic stress in Helianthus annuus.
| miRNA/Target name | Drought stress | Heat stress | Salt stress | Cadmium stress | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tissue | Leaf | Root | Leaf | Root | Leaf | Root | Leaf | Root | ||||||||||||
| 12 h | 24 h | 48 h | 12 h | 24 h | 48 h | 1.5 h | 3 h | 6 h | 1.5 h | 3 h | 6 h | 75 mM | 150 mM | 75 mM | 150 mM | 5 mg | 20 mg | 5 mg | 20 mg | |
| miR160 | ↓1.80** | ↓2.87* | ↓8.04* | ↓1.41ns | – | ↑1.80** | ↑2.22** | ↓1.75** | ↓1.54* | ↑2.54* | ↓10.02** | ↓16.27** | ↑2.25** | ↓2.7** | ↓1.81* | ↑1.16ns | ↑2.21** | ↓2.3538** | ↑14.35** | ↑14.85** |
| T160 | ↓8.57* | ↓4.75* | ↑2.54* | ↓2.40ns | ↓2.92ns | ↑4.41** | ↓6.49** | ↑7.81** | ↓1.09ns | ↓1.36ns | ↓1.23ns | ↑2.15** | ↑4.26** | ↑1.58* | ↑1.14n.s | ↑1.67** | ↑2.62** | ↑5.15** | ↑32.62** | ↑47.42** |
| miR167 | ↓2.41* | ↓10.19* | ↓13.22* | ↓4.28* | ↓19.02* | ↓9.67* | ↑1.56* | ↓1.39ns | ↓2.10** | ↑4.04** | ↓9.96** | ↓3.97** | ↓1.56* | ↓2.0** | ↑1.68* | ↑2.1** | ↑3.69** | ↑6.96** | ↑322.35** | ↑25.24** |
| T167 | ↓13.45* | ↓5.12* | ↓8.13* | ↓1.55* | ↓2.10* | ↑1.86** | ↑1.34ns | ↓1.94** | ↓2.33** | ↓1.39* | ↓39.39** | ↓57.68** | ↓1.67** | ↓1.03** | ↓1.03ns | ↑1.23ns | ↑1.46ns | ↓1.57ns | ↑22.51** | ↑30.75** |
| miR172 | ↓6.38* | ↓5.46* | ↓6.64* | ↑1.20ns | ↑1.56ns | ↑1.62ns | – | ↓2.43** | ↓1.20ns | ↑2.37** | ↓12.62** | ↓2.16** | ↓1.42ns | ↑1.22ns | ↓2.0ns | ↓1.07ns | ↑1.78** | ↑1.005ns | ↑14.27** | ↑57.38** |
| T172 | ↓5.85* | ↓10.37* | ↓18.06* | ↓3.86* | ↓2.0* | ↑3.03* | ↑1.36ns | ↓1.32ns | ↓1.47ns | ↑2.92** | ↓9.18** | ↓6.84** | ↓2.44** | ↓4.76** | ↓1.70** | ↓1.17ns | ↓3.58** | ↓2.03** | ↑5.34** | ↑32.96** |
| miR398 | ↓1.65** | ↓3.60* | ↓16.85* | ↓3.32* | ↓2.08** | ↓7.63* | ↑3.42** | ↑1.82ns | ↑2.68** | ↑1.65* | ↓8** | ↓2.92** | ↑11.31** | ↑3.92* | ↑20.40** | ↑1.23ns | ↓1.62** | ↑1.62** | ↑385.34** | ↑123.43** |
| T398 | ↓5.55* | ↓5.65* | ↓10.02* | ↓2.25** | ↓4.51* | ↓2.63* | ↓10.07** | ↑3.23** | ↑1.01ns | ↓2.28** | ↑3.17** | ↓1.28ns | ↑1.65* | ↓2.08** | ↑1.44ns | ↓2.27** | ↓16.28** | ↑1.37ns | ↓3.83** | ↑10.93** |
| miR403 | ↓2.87* | ↓24.67* | ↓5.46* | ↓8.77* | ↓1.21ns | ↓10.99* | ↑3.42** | ↑1.72** | ↑1.16ns | ↑1.20ns | ↓8.72** | ↓2.96** | ↑2.91** | ↑1.18ns | ↓1.19ns | ↓1.41* | ↑6.28** | ↑1.02ns | ↑52.62** | ↑35.94** |
| T403 | ↓3.60* | ↓6.72* | ↓2.29ns | ↓1.93** | ↓2.33* | ↑16.85* | ↑2.63** | ↑1.18ns | ↓1.12ns | ↑3.19** | ↓1.31ns | ↓1.360ns | ↑2.83** | ↓2.51** | ↑2.46** | ↑3.25** | ↑1.07ns | ↓1.03ns | ↑22.12** | ↑65.91** |
| miR426 | ↓3.19* | ↓2.87* | ↓5.06* | ↓6.61* | ↓1.41ns | ↓4.0* | ↑1.36ns | ↓1.66ns | ↑1.28ns | ↑1.68** | ↓38.71** | ↓7.90** | ↓1.46** | ↓2.25** | ↓1.11ns | ↑9.19** | ↑1.80** | ↑1.17ns | ↑11.65** | ↑16.77** |
| T426 | ↓3.30* | ↓3.08* | ↓2.46* | ↓1.83ns | ↓3.03* | ↑4.83* | ↓1.59** | ↓1.52* | ↓1.96** | ↑1.46** | ↓14.84** | ↓13.29** | ↓1.94** | ↓3.38** | ↑1.04ns | ↑2.11** | ↓2.13** | ↓2.03** | ↑10.27** | ↑14.75** |
| miR842 | ↓6.06* | ↓10.02* | ↓15.81* | ↓5.37* | ↑1.05ns | ↓3.86* | ↑1.93ns | ↓1.47** | ↓1.65ns | ↓1.49* | ↓24.11** | ↓4.72** | ↑1.19ns | ↓10.7** | ↓1.01ns | ↑3.34** | ↑1.34** | ↑2.26** | ↑10.87** | ↑4.65** |
| T842 | ↓1.71** | ↓1.168ns | ↓3.30* | ↓3.73* | ↑1.53* | ↑3.40* | ↑1.09ns | ↓1.16ns | ↓5.99** | ↓1.43ns | ↓2.37** | ↓1.67ns | ↑1.65* | ↑2.60** | ↑4.50** | ↑29.86** | ↓2.34** | ↓1.60ns | ↑7.96** | ↑32.96** |
Sunflower seedling at the six leaf stages grew under normal conditions and leaves and roots of them were harvested as control then they treated by withholding the water for 12, 24 and 48 h. Plants treated at 42 ± 1°C for 1.5, 3, and 6 h. Plant subjected under NaCl treatment in two concentration, 75 and 150 mM. Plants treated with cadmium in 5 and 20 mg/L concentration. Number shows the fold change in relative to control. ↑ The expression is up-regulated. ↓ The expression is down-regulated.
Show that the t-test is significant as p < 0.05.
Show that the t-test is highly significant as p < 0.01.
show that the t-test is not significant.
Figure 1.
Expression patterns of miRNAs and their target genes under drought stress. Sunflower seedling at the six leaf stages grew on soil (¾ river sand and ¼ soil) under normal conditions and leaves and roots of them were harvested as control then they treated by withholding the water for 12, 24 and 48 h. (A) Expression patterns of miRNAs in the leaf. (B) Expression patterns of miRNAs in the root. (C) Expression patterns of target gene in the leaf. (D) Expression patterns of target genes in the root.
The expression of QHG18J04.yg.ab1 gene as target for the miR160, was down- regulated at 12 h and 24 h (2- to 8-fold), and up-regulated at 48 h in both tissues. Except at 48 h in leaf tissue, correlation of miR160 and its target showed similar expression pattern over time and tissue. The expression of ARF6 (target of miR167), COX5b (target of miR172), AGO2 (target of miR403) and TPP2 (target of miR426) displayed similar patterns in both tissues; their expression was induced only in the root tissue at 48 h of drought stress. Interestingly, the coherent trend was observed only at 48 h in the root tissue for miR167, miR403 and miR426 and their respective targets. The transcript of NtGT5b (target of miR398) was constantly decreased in both tissues with 2- to 10-fold under drought conditions where the minimum peak was at 48 h and 24 h after treatment in root and leaf tissue, respectively. The expression of (R)-mandelonitrile lyase (target of miR842) increased at 12 h and decreased at subsequent time points in leaf tissue whereas it showed opposite pattern in root. In general, most targets exhibited their highest expression level at 48 h after stress depending on the tissue (Table 2; Figure 1).
Expression of miRNAs and putative targets under heat stress
The expression levels of all seven miRNAs in leaf tissue were slightly up-regulated after 1.5 h exposure to heat stress, except for miR172 which had constant expression level. However, expression of miRNAs in leaves indicated a mixed pattern at 3 and 6 h after treatment. The expression levels of miR167 and miR172 were down-regulated within a range of 2- to 3-fold at 3 and 6 h after stress. But in root tissues, their reduction was between 2 and 12 fold at these two time points. MiR398 and miR403 showed similar trend in leaf and root tissues. Their expression was immediately up-regulated in leaf at the initial time point, although, their expressions were decreased at 3 and 6 h after stress. The levels of their expression were higher than control condition. However, in root tissue, they were up-regulated at 1.5 h but down-regulated subsequently compared to control with a 2- to 8-fold change at 3 and 6 h. The miR160, miR426 and miR842 exhibited similar trend under heat stress in leaf tissue. Their expressions were induced at 1.5 h and down-regulated at 3 h and again induced at severe stress. Nevertheless, their expressions were lower compared to the control. In root tissue, except 1.5 h after stress, miR842 and miR426 showed similar pattern within a range of 4- to 38-fold change after stress. The expression of miR160 instantly decreased at 3 h (10-fold change compared to control) and its decrease continued constantly at 6 h (Table 2; Figure 2).
Figure 2.
Expression patterns of miRNAs and their target genes under heat stress. Plants treated at 42 ± 1°C for 1.5, 3, and 6 h. (A) Expression patterns of miRNAs in the leaf. (B) Expression patterns of miRNAs in the root. (C) Expression patterns of target gene in the leaf. (D) Expression patterns of target genes in the root.
The expression of QHG18J04.yg.ab1 was significantly down-regulated at 1.5 h and was reversed at 3 h, while its expression was only up-regulated at 6 h in root. The pattern of its transcript had negative correlation with miR160. Transcript of ARF6 was down-regulated at all-time points with a sharp decrease in the root tissue at 3 h. The temporal variation of COX5b was significant only in the root tissue while it decreased sharply at 3 h after heat stress. AGO2 expressed at a significant level at 1.5 h in both tissues where its expression was up-regulated with more than two-fold change, compared to the control. NtGT5b displayed similar pattern in both tissues. The expression level declined at an initial time point and was induced at 3 h after treatment, whereas it showed coherent type with miR398. In leaves, the expression of (R)-mandelonitrile lyase was abruptly down-regulated at 6 h, whereas its expression was declined slightly over time in the root. Expression of TPP2 dropped in leaf tissue, but it decreased in root only at 3 h with 14-fold changes after stress. However, there was a slight increase in its expression at 6 h in the root. The expression was still lower compared with control (Table 2; Figure 2).
Expression of miRNAs and putative targets under salt stress
The expression of miR167 showed opposite pattern in both tissues. In leaves, salt stress reduced the expression of miR167 by two-fold while the expression level was up-regulated in the root tissues with a two-fold change. Interestingly, miR403 exhibited opposite pattern of expression in leaf and root, whereas its expression had increasing and declining trend in leaf and root, respectively. The expression trends of miR160, miR426 and miR842 were similar in both tissues at severe stress. Their expression showed decreasing pattern in leaf and increasing pattern in root tissue. Interestingly, an abrupt gradient was observed for miR842 in leaf tissue at 150 mM NaCl. None of the tissues disclosed a significant alteration for miR172 expression in response to salt stress. Salt stress induced the miR398 expression in both tissues with the sharp increase observed after 75 mM NaCl treatment in both tissues. In spite of this, there was a considerable reduction in its expression at 150 mM concentration in leaf and root, and the expression level was higher than control condition (Table 2; Figure 3).
Figure 3.
Expression patterns of miRNAs and their target genes under salt stress. Plant subjected under NaCl treatment in two concentration, 75 and 150 mM. (A) Expression patterns of miRNAs in the leaf (B) Expression patterns of miRNAs in the root. (C) Expression patterns of target gene in the leaf. (D) Expression patterns of target genes in the root.
Under salt stress, the transcripts of QHG18J04.yg.ab1 slightly increased in the root. Although, there was a decline pattern at 150 mM of NaCl in comparison with an early stage of stress in the leaf tissue, its expression was higher compared to control. The expression of ARF6 was not significant in the root. However, its expression showed down-regulation at both stages of treatment in leaf tissue. Its expression level revealed a slightly increasing trend at 150 mM compared with 75 mM of NaCl. COX5b expression was lower in both tissues at all stages of treatment with sharp reduction at 150 mM of NaCl in the leaf tissue. NtGT5b exhibited similar expression pattern in both tissues where its mRNA level was up-regulated at 75 mM concentration and was abruptly decreased at 150 mM. The AGO2 transcript was highly accumulated at all stages in the root tissue, but in the leaf tissue, was induced at 75 mM of NaCl, and down-regulated subsequently compared with control. The expression pattern of TPP2 was opposite in leaf and root tissue, as decreased in leaf and increased in root tissue with its peak of expression at severe stress. The expression of (R)-mandelonitrile lyase was induced in both tissues with its peak at 150 mM concentration (Table 2; Figure 3).
Expression of miRNAs and putative targets under cadmium stress
Cadmium treatment resulted in up-regulation of all miRNAs in roots, with fold-changes between 4 and 385. The lowest and highest peaks were for miR842 in 20 mg/L and miR398 in 5 mg/L concentration of cadmium, respectively. The expression of miR426 was induced only at 5 mg/L in leaf, while in root induced at all stages of stress. The opposite pattern for miR160 and miR842 observed only at 20 mg/L in leaf tissue. The level of miR160 was drastically decreased in leaf tissue, but the expression of miR842 was slightly raised. In root tissue, their expression was increased although; the levels of miR842 at 20 mg/L were declined in comparison to 5 mg/L, where its expression was still higher compared to control. MiR403 expression was increased in both tissues whereas expression levels in root tissue were higher than leaf tissue. The highest expression of miR403 was in 5 mg/L concentration with 52-fold change compared to control. The expression pattern of miR172 was induced at all stages of treatment in both tissues with its peak at 20 mg/L concentration in root. Interestingly, the level of miR398 was up-regulated at 5 mg/L, but down-regulated at 20 mg/L with 1.6-fold change in leaf tissue. Similar trend was observed for miR167 and miR398 in root tissue, with the highest peak at 5 mg/L concentration (Table 2; Figure 4).
Figure 4.
Expression patterns of miRNAs and their target genes under cadmium stress. Plants treated with cadmium in 5 and 20 mg/L concentration. (A) Expression patterns of miRNAs in the leaf. (B) Expression patterns of miRNAs in the root. (C) Expression patterns of target gene in the leaf. (D) Expression patterns of target genes in the root.
Transcript of QHG18J04.yg.ab1 showed up-regulation trend in both tissues where in root, it was over 10-fold higher than leaf. The target of miR167 and miR403 exhibited similar trend in both tissues as their expressions were significantly induced in the root with their highest peak at 20 mg/L of CdCl2. The expression of COX5b and TPP2 was declined in leaf and was increased in root tissue in both concentrations of cadmium with its peak in 20 mg/L. (R)-mandelonitrile lyase highly accumulated in the root with abrupt increase at 20 mg/L. In contrast, its expression was decreased in leaf tissue after treatment. Expression of NtGT5b revealed temporal variation where it was down-regulated at 5 mg/L and up-regulated at 20 mg/L in both tissues. Interestingly, miR172, miR398, miR426 and miR842 showed inverse correlation with their targets in the leaf tissue at 5 mg/L (Table 2; Figure 4).
Cluster analysis of miRNAs in H. annuus root and leaf tissues based on their expression patterns
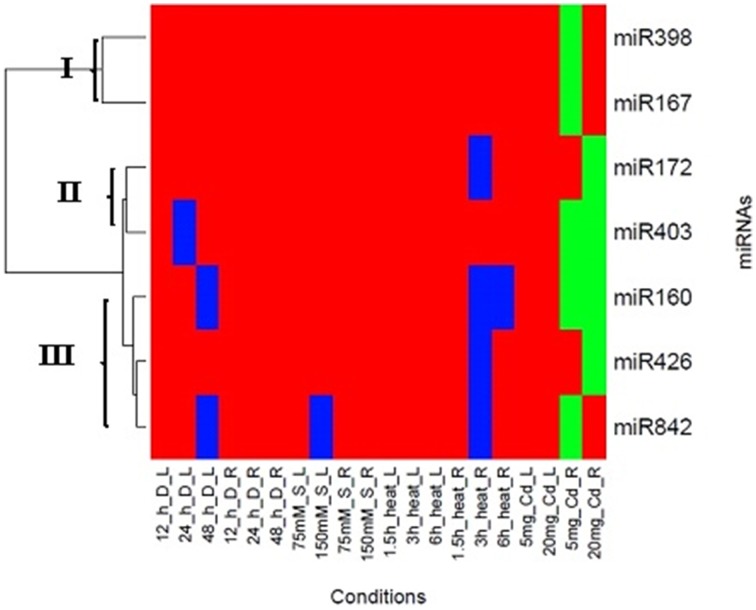
Cluster analysis was performed to further analyze the pattern of expression of these miRNAs under different stress conditions. A comparison of expression profiles of these seven miRNAs in both tissues was conducted to find their tissue-specific expression patterns, regardless of their response to different stress conditions. Three clusters were formed, when the expression pattern of the seven miRNAs were analyzed in response to stress (Figure 5). Cluster I included miR167 and miR398. The expression pattern of leaf and root was opposite, but their trend was similar in each tissue over time and stress. In root tissue, these miRNAs revealed slight alteration in three treatments; the highest peak was in 5 mg/L cadmium stress. Cluster II contained miR172 and miR403 families and showed a fluctuated pattern in leaves and roots in all of stress conditions. Median value of expression of these miRNAs exhibited increasing pattern in four stresses. Due to similarity in expression patterns of miR160, miR426 and miR842, they grouped in the cluster III. They displayed similar trend in each tissue during stress. The lowest peak of miR842 was in severe stress through drought and salt condition in the leaf tissue, while it was at 3 h after heat treatment in the root tissue.
Figure 5.
Clustering of miRNAs expression profiles. Heat map diagram of miRNA expression prepared with two-way unsupervised hierarchical clustering of miRNAs expression under different abiotic stress. miRNAs are given in the rows and each columns represent a sample. The miRNA clustering tree is shown on the left (cluster I, II and III). Abbreviations: L, Leaf; R, Root; h, Hour; D, Drought stress; S, Salt Stress; Cd, Cadmium stress.
Relationship of miRNAs and target gene in stress response
Analysis of Pearson correlation showed positive and negative correlation between expression of miRNAs and target genes against stress in both tissues. The results indicated significant negative correlation between miR172, miR398 and miR403 and their putative targets in leaf tissue under cadmium stress. On the contrary, miR398 presented significant negative correlation in root tissue after heat treatment. Weak correlation was observed between some miRNAs and their candidate targets in both tissues against specific stress such as miR160 in drought stress and miR167 after heat treatment (Table S4). Some miRNAs displayed significantly positive correlation, which indicated other mechanisms are involved in target gene regulation.
Interaction between miRNAs and TFs-mediated gene regulatory subnetworks
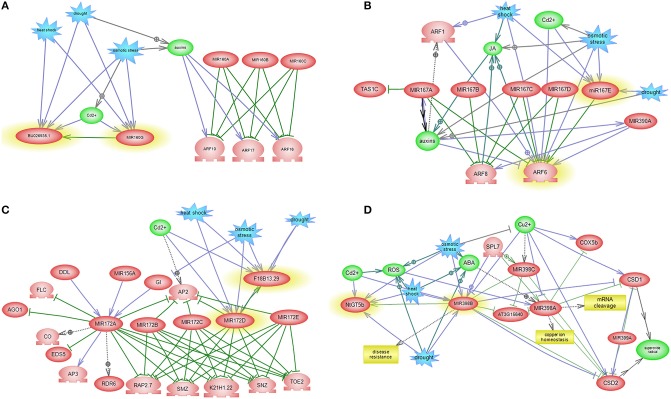
The regulatory subnetworks that are constructed for each miRNA elucidated some of the intermingled miRNA and TF relationships as well as miRNA-miRNA relationships and the involvement of other miRNA families in miRNA specific post-transcriptional regulation pathways (Figures 6, 7). References of interaction relations between miRNAs and genes in subnetworks and their correlated references are listed in Table S5.
Figure 6.
miRNA-mediated gene regulatory sub networks in response to abiotic stress. miRNA-target-abiotic stress interaction are shown. (A) Feedback loop between miR160, its target and abiotic stress. (B) Feedback loop between miR167, its target and abiotic stress and abiotic stress. (C) Feedback loop between miR172, its target and abiotic stress. (D) Feedback loop between miR398, its target and abiotic stress.
Figure 7.

miRNA-mediated gene regulatory sub networks in response to abiotic stress. miRNA-target-abiotic stress interaction are shown. (A) Feedback loop between miR403, its target and abiotic stress. (B) Feedback loop between miR842, its target and abiotic stress.
GO analysis is a robust approach in understanding underlying molecular mechanisms of different cellular events and offer a reliable tool for GO based gene selection (Fruzangohar et al., 2013). GO classification showed that the target genes are involved in auxin signaling, RNA mediated silencing, ethylene signaling, DNA methylation and response to abiotic stress.
Subnetwork of miR426 did not display any connectivity with other participants in the network. As miR426 exhibits stress and tissue specific pattern, this miRNA is a suitable candidate for further studies in the context of miRNA mediated gene regulatory pathway. MiR842 showed connectivity with MBB18_8 and (R)-mandelonitrile lyase that drought, heat, salt and cadmium affected its expressions (Figure 7B).
MiR160 centered network exposed interaction between QHG18J04.yg.ab1 and three TFs belonging to the ARF family (ARF16, ARF17, and ARF18), where four abiotic stress affect expression of miR160 and QHG18J04.yg.ab1 (Figure 6A). Interestingly, in this network, two other ARF family members; ARF6 and ARF8 regulate JA (Jasmonic acid) pathway and are in turn regulated by miR167. In addition, sub network of miR167 includes regulation of TAS1C, which encodes a ta-siRNA (Figure 6B). Expression of TFs involved in ethylene signaling pathway and flower and seed development is regulated by the members of the miR172 family. But miR172 itself is regulated by miR156 and DDL (a gene silencer) (Figure 6C). MiR398 revealed cross talk with SPL, with central roles in many cellular processes such as auxin metabolism, root growth and cytokinesis; where SPL is regulated by miR156, miR157 and miR159. Therefore, these three miRNAs are speculated to regulate the expression of miR398 indirectly. MiR156 have excelled as a key regulator in the regulation of other miRNAs. MiR398 causes mRNA cleavage and disease resistance, which regulates expression of CSDs gene and NtGT5b (Figure 6D). Subnetwork of miR403 revealed that SE and miR402 regulate AGO2. It is known that SE has role in miRNA biogenesis, RNA splicing and chromatin modification. MiR402 displayed connectivity with DML1 and DML3, which are involved in DNA methylation and transcriptional control (Figure 7A).
Discussion
The role of seven miRNAs in response to several abiotic stresses was studied in H. annuus leaf and root tissues by RT-qPCR. Changes in RWC, expression of HSP70-related protein, sodium, potassium, and cadmium concentration revealed obviously various mechanisms stimulation to cope with stress. The measured alterations of metabolic and physiological status in H. annuus provide additional support for modification reactions toward stress conditions.
The temporal and spatial expression profiles of seven miRNAs in both tissues at altered time points were in agreement with previous studies (Sunkar et al., 2012; Zandkarimi et al., 2015; Zhang, 2015). For instance, high-throughput sequencing of Brassica napus showed up-regulation of miR172 in the root tissue under cadmium excess, up-regulation of miR167 from 2 to 24 h after exposure in 300 mM NaCl in Arabidopsis and down-regulation of miR398 under drought stress in cotton which treated with PEG.
The different expression patterns of miR167 observed in photosynthetic and non-photosynthetic tissues suggest that miR167 may have a role in tissue-specific adaptation to stress. MiR160 and miR167 regulate ARF families and play roles in the auxin signaling pathway (Khraiwesh et al., 2012), and were differentially expressed under stresses compared to control conditions. In addition, ARF domain was found in the protein sequence of QHG18J04.yg.ab1 (target of miR160). Therefore, this gene may be involved in auxin signaling pathway and have a role in promoting plant tolerance to stress. The alteration of miR160 and miR167 in H. annuus at early stage of treatments suggests that they are responsive to early stress response. Reduced expression of miR160 and miR167 in leaves under severe stress may alter basal level of auxin and consequently restrict plant growth through the antagonism between abscisic acid and auxin because of escape of tension. In turn, this could lead to attenuation of plant growth and development under stress to cope with the imposed stress (Sunkar et al., 2012). Even though they have common targets, in our study, miR160 and miR167 did not group together in one cluster. On the other hand, these miRNAs showed various expression patterns in other plant species under different stress (Khraiwesh et al., 2012; Sunkar et al., 2012). Besides, under normal conditions, their expression pattern was also variable in different plant genus and species (Zeng et al., 2010).
Some studies have suggested roles for miR172 during cadmium, drought, cold and heat stress conditions (Sunkar et al., 2012; Zhou et al., 2012). In line with the previous studies, miR172 revealed temporal up- or down-regulation in leaf and root in response to stress conditions except for the salt stress. Expression of miR172 showed inverse correlation with its target COX5b, only in leaf tissue under cadmium stress. The post-transcriptional silencing of COX5b by miR172 may reflect the providence of Helianthus plant from energy loss via avoiding respiration under excessive cadmium concentrations (Sunkar et al., 2012).
We found that miR398 is especially up-regulated in leaf tissues during heat stress in line with the previous reports (Guan et al., 2013). Interestingly, miR398 was down-regulated in root tissue while expression of HSP70-related was up-regulated, which may indicate that miR398 has a tissue specific mode of action and localization during heat stress condition. Differential expression of miR398 in response to drought, salt, heat and cadmium stress have been shown in several species such as wild emmer wheat, Medicago, Nicotiana, Brassica and Arabidopsis (Trindade et al., 2010; Frazier et al., 2011; Kantar et al., 2011; Zhou et al., 2012; Guan et al., 2013). In contrast, under severe drought and cadmium stress conditions, miR398 was down-regulated in leaf tissue indicating a tissue-specific and stress-specific response orchestrated by this miRNA. In this study, the probable target of miR398 was NtGT5b which is a microsomal enzyme responsible for glucuronidation reactions with a role in the storage of secondary metabolites and plants defense against stress (Miners et al., 2002; Ko et al., 2006). On the other hand, new target genes were predicted for miR398 and miR167 in Phaseolus vulgaris and Malus hupehensis in vegetative phase which involved in monogalactosyl diacylglycerol synthase, acyltransferase and dioxygenase, gluconeogenesis pathway and glycolytic process (Heyndrickx and Vandepoele, 2012; Han et al., 2014; Xing et al., 2014). Furthermore, in Nicotiana tabacum in response to TiO2 nanoparticles, these miRNAs were grouped in one cluster (Frazier et al., 2014). All of these results have led to the conclusion that miR398 may be involved in the sugar biosynthesis pathway, associated with reduction in energy consumption for photosynthesis, and increase the tolerance of plant in abiotic stress conditions.
Surprisingly, the pattern of miR403 was varying in each stress condition. Its expression was declined and was induced in both tissues in drought and cadmium stress, respectively. In contrast, miR403 displayed increased expression in the leaf and decreased expression in the root during heat and salt stress. In the previous study, abundance of miR403 was high in heat and salt libraries in Raphanus sativus (Wang et al., 2014; Sun et al., 2015). As a result, this discrepancy suggested that miR403 was potentially expressed in stress-, tissue-, and species- specific manner during abiotic stress. Also, the expression of AGO2 at all stages of treatment in both tissues was aberrant. In plants, AGOs are involved in various small RNA pathways from post-transcriptional gene silencing to epigenetic silencing phenomena such as RNA-directed DNA methylation (RdDM) pathway in Arabidopsis (Schraivogel and Meister, 2014). AGO1 and AGO2 proteins were regulated by miR403 (Figure 7A). In addition, AGO2 has been shown to have an antiviral role (Harvey et al., 2011). Therefore, it is possible that this miRNA is involved in the regulation of miRNA-mediated RNA cleavage carried out by other miRNAs during stress conditions. Furthermore, AGO1 is also regulated by miR172 family (Ronemus et al., 2006), which may designate a crucial role for miR172 in general miRNA mediated gene silencing pathways (Figure 6C). This result, based on altered expression of miR403 and its target under different abiotic stress and its subnetwork (Figure 7A) which showed DML1 and DML3, involved in DNA methylation, may pave the ways for intricate control mechanisms for drought, heat, salt and cadmium stress tolerance in sunflower. Interestingly, miR172 and miR403 were grouped in one cluster (Figure 5) and they showed common targets; AGO1 and AGO2, which may suggest a general role for these miRNAs in small RNA pathway and DNA methylation.
In this study, miR426 and miR842 show differential expression under abiotic stress. However, their expression exhibited stress-dependent manner during stress which was declined in root tissue under heat and drought stress and was reversed in salt and cadmium treatment. Also, inverse correlation of these miRNAs with their target was temporary in some stages. Their possible targets, TPP2 and (R)-mandelonitrile lyase, were induced in response to some abiotic stresses and are reported to be involved in defense mechanism, oxidation-reduction process, cyanide biosynthetic process and alcohol metabolic process, which is indicated as a biocatalyst in organic chemistry (Hu and Poulton, 1999; Shima et al., 2007). MiR842 revealed no significant change after waterlogging conditions (Moldovan et al., 2010), whereas it was repressed after ABA treatment of roots in Arabidopsis (Jia and Rock, 2013) which was similar with the expression of miR842 under drought and heat stress in both tissues. In the earlier studies, MBB18_8 a member of Jacalin lectin family (Gustafson et al., 2005) and a kinase-like protein (Barozai et al., 2012) were predicted as targets of miR842. As a result, miR842 might have a role not only in sugar biosynthesis and sugar mediated signaling pathways but may also have a role as an osmoprotectant. Surprisingly, miR842 gene was post-transcriptionally regulated by alternative splicing (Jia and Rock, 2013). Consequently, temporal expression in response to stress and regulation of its gene revealed that miR842 might have a complicated function in plants. Our results suggest a novel function for miR426 and miR842 in the regulation of sunflower tolerance to abiotic stress. Interestingly, miR160, miR426 and miR842 showed similar pattern and were grouped in one cluster and according to their target, they are probably involved in carbohydrate signaling pathway.
In this study, expressions of miRNA targets are not consistent with the expression pattern of their related miRNAs at all times. The pattern of miRNAs and their target genes were semi-coherent, coherent or non-coherent type during stresses in leaf and root tissues. For example, expression of miR842 and its target in leaf tissue showed inverse correlation during mild drought and cadmium stress. A recent study has revealed, the expression pattern of four miRNAs and their target have a semi-coherent fashion under salt stress in the halophyte smooth cordgrass (Zandkarimi et al., 2015). In addition, we did not analyze protein levels of target genes during stress, therefore we can neither confirm nor reject that these genes are direct targets for these miRNAs. It is possible that these miRNAs and their targets are expressed in a non-overlapping manner and regulate their targets in different cells, as reported for miR395 and AST68 in Arabidopsis (Kawashima et al., 2009). The coherent correlation between miRNA and mRNA is still under debate (Li et al., 2012), while the non-coherent type implicated the post transcriptional regulation mechanism by miRNA-directed cleavage for target mRNAs (Zandkarimi et al., 2015). Taken together, this information suggests that miRNAs play a versatile role for plant's acclimation to stress conditions. The kinetics of miRNAs and target regulation over time and in different tissues against tension and compression stresses imply complicated physiological and genetic mechanisms in H. annuus in order to deal with and adapt to harsh environment. Indeed, aberrant expression of many miRNAs during stress revealed that they respond to environmental stresses in a miRNA-, stress-, and tissue-dependent manner. Nevertheless, differential expression of certain miRNA rely on the specific stress condition, even in the same plant species (Zhang, 2015). As a consequence, aberrant expression of these miRNAs may reflect synergistic activities at the biochemical, physiological and molecular levels such as auxin signaling and sugar response, and finally at the organismal level to attenuate plant growth and development under stress. Interaction between miRNA and target gene is more flexible because of regulation of a mRNA target gene by multiple miRNAs or on the contrary regulation of numerous mRNA target by individual miRNA (Zandkarimi et al., 2015). As well, alternative splicing regulates miRNA biogenesis and expression of target genes to make different isoforms (Jia and Rock, 2013). This indicates that plants employ unrecognized regulatory loops to achieve tolerance via these regulatory small RNAs and suggests that they selectively regulate the expression of specific target genes under each condition. In conclusion this study adds to the growing body of literature on stress-responsive miRNAs in plants.
Author contributions
BS conceived and designed the research. BS, EE, HF, FR, and HB conducted experiment. RE, SM, FK, and ZF carried out experiment and analyzed data. RE, BS, HF, and EE wrote the manuscript. All authors read and approved the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Shahrekord University for partial financial assistance. We would like also to express our gratitude to Parisa Shiran (PhD candidate at the University of Melbourne) for editing the manuscript.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00741
References
- Alanazi I., Ebrahimie E., Hoffmann P., Adelson D. L. (2013). Combined gene expression and proteomic analysis of EGF induced apoptosis in A431 cells suggests multiple pathways trigger apoptosis. Apoptosis 18, 1291–1305. 10.1007/s10495-013-0887-6 [DOI] [PubMed] [Google Scholar]
- Alimohammadi A., Shiran B., Martinez-Gomez P., Ebrahimie E. (2013). Identification of water-deficit resistance genes in wild almond Prunus scoparia using cDNA-AFLP. Sci. Hortic. 159, 19–28. 10.1016/j.scienta.2013.04.023 [DOI] [Google Scholar]
- Allen E., Xie Z., Gustafson A. M., Carrington J. C. (2005). microRNA-directed phasing during Trans-Acting siRNA biogenesis in plants. Cell 121, 207–221. 10.1016/j.cell.2005.04.004 [DOI] [PubMed] [Google Scholar]
- Barozai M. Y. K., Baloch I. A., Din M. (2012). Identification of MicroRNAs and their targets in Helianthus. Mol. Biol. Rep. 39, 2523–2532. 10.1007/s11033-011-1004-y [DOI] [PubMed] [Google Scholar]
- Čatský J. (1960). Determination of water deficit in disks cut out from leaf blades. Biol. Plant 2, 76–78. 10.1007/BF02920701 [DOI] [Google Scholar]
- Chiasson D. M., Loughlin P. C., Mazurkiewicz D., Mohammadidehcheshmeh M., Fedorova E. E., Okamoto M., et al. (2014). Soybean SAT1 (Symbiotic Ammonium Transporter 1) encodes a bHLH transcription factor involved in nodule growth and NH4+ transport. Proc. Natl. Acad. Sci. U.S.A. 111, 4814–4819. 10.1073/pnas.1312801111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deihimi T., Niazi A., Ebrahimi M., Kajbaf K., Fanaee S., Bakhtiarizadeh M. R., et al. (2012). Finding the undiscovered roles of genes: an approach using mutual ranking of coexpressed genes and promoter architecture-case study: dual roles of thaumatin like proteins in biotic and abiotic stresses. Springerplus 1, 1–10. 10.1186/2193-1801-1-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Caterina R., Giuliani M., Rotunno T., De Caro A., Flagella Z. (2007). Influence of salt stress on seed yield and oil quality of two sunflower hybrids. Ann. Appl. Biol. 151, 145–154. 10.1111/j.1744-7348.2007.00165.x [DOI] [Google Scholar]
- Ebrahimie M., Esmaeili F., Cheraghi S., Houshmand F., Shabani L., Ebrahimie E. (2014). Efficient and simple production of insulin-producing cells from embryonal carcinoma stem cells using mouse neonate pancreas extract, as a natural inducer. PLoS ONE 9:e90885. 10.1371/journal.pone.0090885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier T. P., Burklew C. E., Zhang B. (2014). Titanium dioxide nanoparticles affect the growth and microRNA expression of tobacco (Nicotiana tabacum). Funct. Integr. Genomics 14, 75–83. 10.1007/s10142-013-0341-4 [DOI] [PubMed] [Google Scholar]
- Frazier T. P., Sun G., Burklew C. E., Zhang B. (2011). Salt and drought stresses induce the aberrant expression of microRNA genes in tobacco. Mol. Biotechnol. 49, 159–165. 10.1007/s12033-011-9387-5 [DOI] [PubMed] [Google Scholar]
- Fruzangohar M., Ebrahimie E., Ogunniyi A. D., Mahdi L. K., Paton J. C., Adelson D. L. (2013). Comparative GO: A web application for comparative gene ontology and gene ontology-based gene selection in bacteria. PLoS ONE 8:e58759. 10.1371/journal.pone.0058759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomelli J. I., Weigel D., Chan R. L., Manavella P. A. (2012). Role of recently evolved miRNA regulation of sunflower HaWRKY6 in response to temperature damage. New Phytol. 195, 766–773. 10.1111/j.1469-8137.2012.04259.x [DOI] [PubMed] [Google Scholar]
- Guan Q., Lu X., Zeng H., Zhang Y., Zhu J. (2013). Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 74, 840–851. 10.1111/tpj.12169 [DOI] [PubMed] [Google Scholar]
- Gustafson A. M., Allen E., Givan S., Smith D., Carrington J. C., Kasschau K. D. (2005). ASRP: the Arabidopsis small RNA project database. Nucleic Acids Res. 33, D637–D640. 10.1093/nar/gki127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L., Bussell J. D., Păcurar D. I., Schwambach J., Pãcurar M., Bellini C. (2009). Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21, 3119–3132. 10.1105/tpc.108.064758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Xie H., Kong M. L., Sun Q. P., Li R. Z., Pan J. B. (2014). Computational identification of miRNAs and their targets in Phaseolus vulgaris. Genet. Mol. Res. 13, 310–322. 10.4238/2014.January.17.16 [DOI] [PubMed] [Google Scholar]
- Harvey J. J., Lewsey M. G., Patel K., Westwood J., Heimstädt S., Carr J. P., et al. (2011). An antiviral defense role of AGO2 in plants. PLoS ONE 6:e14639. 10.1371/journal.pone.0014639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyndrickx K. S., Vandepoele K. (2012). Systematic identification of functional plant modules through the integration of complementary data sources. Plant Physiol. 159, 884–901. 10.1104/pp.112.196725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland D. R., Arnon D. I. (1950). The water-culture method for growing plants without soil. Circ. Calif. Agric. Exp. Stn. 347. [Google Scholar]
- Hu Z., Poulton J. E. (1999). Molecular analysis of (R)-(+)-mandelonitrile lyase microheterogeneity in black cherry. Plant Physiol. 119, 1535–1546. 10.1104/pp.119.4.1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia F., Rock C. D. (2013). MIR846 and MIR842 comprise a cistronic MIRNA pair that is regulated by abscisic acid by alternative splicing in roots of Arabidopsis. Plant Mol. Biol. 81, 447–460. 10.1007/s11103-013-0015-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D., Wang Y., Zhao Y., Chen M. (2013). MicroRNAs and their cross-talks in plant development. J. Genet. Genomics 40, 161–170. 10.1016/j.jgg.2013.02.003 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M. W., Bartel D. P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14, 787–799. 10.1016/j.molcel.2004.05.027 [DOI] [PubMed] [Google Scholar]
- Kantar M., Lucas S. J., Budak H. (2011). miRNA expression patterns of Triticum dicoccoides in response to shock drought stress. Planta 233, 471–484. 10.1007/s00425-010-1309-4 [DOI] [PubMed] [Google Scholar]
- Kawashima C. G., Yoshimoto N., Maruyama−Nakashita A., Tsuchiya Y. N., Saito K., Takahashi H., et al. (2009). Sulphur starvation induces the expression of microRNA−395 and one of its target genes but in different cell types. Plant J. 57, 313–321. 10.1111/j.1365-313X.2008.03690.x [DOI] [PubMed] [Google Scholar]
- Khraiwesh B., Zhu J. K., Zhu J. (2012). Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta 1819, 137–148. 10.1016/j.bbagrm.2011.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J. H., Kim B. G., Hur H. G., Lim Y., Ahn J. H. (2006). Molecular cloning, expression and characterization of a glycosyltransferase from rice. Plant Cell Rep. 25, 741–746. 10.1007/s00299-006-0119-4 [DOI] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. (2011). miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39, D152–D157. 10.1093/nar/gkq1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Sun Q., Jia Y., Cong R., Ni Y., Yang X., et al. (2012). Coordinated miRNA/mRNA expression profiles for understanding breed-specific metabolic characters of liver between Erhualian and large white pigs. PLoS ONE 7:e38716. 10.1371/journal.pone.0038716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdi L. K., Ebrahimie E., Adelson D. L., Paton J. C., Ogunniyi A. D. (2013). A transcription factor contributes to pathogenesis and virulence in Streptococcus pneumoniae. PLoS ONE 8:e70862. 10.1371/journal.pone.0070862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsen E., Kilian J., Harter K., Jansson C., Wanke D., Sundberg E. (2011). Transcriptome analysis of high-temperature stress in developing barley caryopses: early stress responses and effects on storage compound biosynthesis. Mol. Plant 4, 97–115. 10.1093/mp/ssq058 [DOI] [PubMed] [Google Scholar]
- Mashkina E. V., Usatov A. V., Skorina M. V. (2010). Comparative analysis of thermotolerance of sunflower chlorophyll mutants. Russ. J. Genet. 46, 178–184. 10.1134/S1022795410020079 [DOI] [PubMed] [Google Scholar]
- Merah O., Langlade N., Alignan M., Roche J., Pouilly N., Lippi Y., et al. (2012). Genetic analysis of phytosterol content in sunflower seeds. Theor. Appl. Genet. 125, 1589–1601. 10.1007/s00122-012-1937-0 [DOI] [PubMed] [Google Scholar]
- Miners J. O., McKinnon R. A., Mackenzie P. I. (2002). Genetic polymorphisms of UDP-glucuronosyltransferases and their functional significance. Toxicology 181, 453–456. 10.1016/S0300-483X(02)00449-3 [DOI] [PubMed] [Google Scholar]
- Moldovan D., Spriggs A., Yang J., Pogson B. J., Dennis E. S., Wilson I. W. (2010). Hypoxia-responsive microRNAs and trans-acting small interfering RNAs in Arabidopsis. J. Exp. Bot. 61, 165–177. 10.1093/jxb/erp296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monavar Feshani A., Mohammadi S., Frazier T. P., Abbasi A., Abedini R., Karimi Farsad L., et al. (2012). Identification and validation of Asteraceae miRNAs by the expressed sequence tag analysis. Gene 493, 253–259. 10.1016/j.gene.2011.11.024 [DOI] [PubMed] [Google Scholar]
- Nikitin A., Egorov S., Daraselia N., Mazo I. (2003). Pathway studio—the analysis and navigation of molecular networks. Bioinformatics 19, 2155–2157. 10.1093/bioinformatics/btg290 [DOI] [PubMed] [Google Scholar]
- Niu Z. X., Sun L. N., Sun T. H., Li Y. S., Wang H. (2007). Evaluation of phytoextracting cadmium and lead by sunflower, ricinus, alfalfa and mustard in hydroponic culture. J. Environ. Sci. 19, 961–967. 10.1016/S1001-0742(07)60158-2 [DOI] [PubMed] [Google Scholar]
- Panahi B., Mohammadi S. A., Ebrahimie E. (2013). Identification of miRNAs and their potential targets in halophyte plant Thellungiella halophila. Biotechnologia 94, 285–290. 10.5114/bta.2013.46422 [DOI] [Google Scholar]
- Ronemus M., Vaughn M. W., Martienssen R. A. (2006). MicroRNA-targeted and small interfering RNA–mediated mRNA degradation is regulated by Argonaute, Dicer, and RNA-dependent RNA polymerase in Arabidopsis. Plant Cell 18, 1559–1574. 10.1105/tpc.106.042127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Schraivogel D., Meister G. (2014). Import routes and nuclear functions of Argonaute and other small RNA-silencing proteins. Trends Biochem. Sci. 39, 420–431. 10.1016/j.tibs.2014.07.004 [DOI] [PubMed] [Google Scholar]
- Senthil-Kumar M., Srikanthbabu V., Mohan Raju B., Ganeshkumar Shivaprakash, N., Udayakumar M. (2003). Screening of inbred lines to develop a thermotolerant sunflower hybrid using the temperature induction response (TIR) technique: a novel approach by exploiting residual variability. J. Exp. Bot. 54, 2569–2578. 10.1093/jxb/erg278 [DOI] [PubMed] [Google Scholar]
- Shima S., Matsui H., Tahara S., Imai R. (2007). Biochemical characterization of rice trehalose−6−phosphate phosphatases supports distinctive functions of these plant enzymes. FEBS J. 274, 1192–1201. 10.1111/j.1742-4658.2007.05658.x [DOI] [PubMed] [Google Scholar]
- Srivastava S., Srivastava A. K., Suprasanna P., D'souza S. (2013). Identification and profiling of arsenic stress-induced microRNAs in Brassica juncea. J. Exp. Bot. 64, 303–315. 10.1093/jxb/ers333 [DOI] [PubMed] [Google Scholar]
- Sun X., Xu L., Wang Y., Yu R., Zhu X., Luo X., et al. (2015). Identification of novel and salt-responsive miRNAs to explore miRNA-mediated regulatory network of salt stress response in radish (Raphanus sativus L.). BMC Genomics 16:197. 10.1186/s12864-015-1416-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Li Y. F., Jagadeeswaran G. (2012). Functions of microRNAs in plant stress responses. Trends Plant Sci. 17, 196–203. 10.1016/j.tplants.2012.01.010 [DOI] [PubMed] [Google Scholar]
- Trindade I., Capitão C., Dalmay T., Fevereiro M. P., dos Santos D. M. (2010). miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta 231, 705–716. 10.1007/s00425-009-1078-0 [DOI] [PubMed] [Google Scholar]
- Varkonyi-Gasic E., Wu R., Wood M., Walton E. F., Hellens R. P. (2007). Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3:12. 10.1186/1746-4811-3-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687. 10.1016/j.cell.2009.01.046 [DOI] [PubMed] [Google Scholar]
- Wang R., Xu L., Zhu X., Zhai L., Wang Y., Yu R., et al. (2014). Transcriptome-wide characterization of novel and heat-stress-responsive micrornas in radish (Raphanus Sativus L.) using next-generation sequencing. Plant Mol. Biol. Rep. 33 10.1007/s11105-014-0786-1 [DOI] [Google Scholar]
- Xing L., Zhang D., Li Y., Zhao C., Zhang S., Shen Y., et al. (2014). Genome-wide identification of vegetative phase transition-associated microRNAs and target predictions using degradome sequencing in Malus hupehensis. BMC Genomics 15:1125. 10.1186/1471-2164-15-1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandkarimi H., Bedre R., Solis J., Mangu V., Baisakh N. (2015). Sequencing and expression analysis of salt-responsive miRNAs and target genes in the halophyte smooth cordgrass (Spartina alternifolia Loisel). Mol. Biol. Rep. 42, 1341–1350. 10.1007/s11033-015-3880-z [DOI] [PubMed] [Google Scholar]
- Zeng C., Wang W., Zheng Y., Chen X., Bo W., Song S., et al. (2010). Conservation and divergence of microRNAs and their functions in Euphorbiaceous plants. Nucleic Acids Res. 38, 981–995. 10.1093/nar/gkp1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. (2015). MicroRNA: a new target for improving plant tolerance to abiotic stress. J. Exp. Bot. 66, 1749–1761. 10.1093/jxb/erv013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z. S., Song J. B., Yang Z. M. (2012). Genome-wide identification of Brassica napus microRNAs and their targets in response to cadmium. J. Exp. Bot. 63, 4597–4613. 10.1093/jxb/ers136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Ding Y., Liu H. (2011). MiR398 and plant stress responses. Physiol. Plant 143, 1–9. 10.1111/j.1399-3054.2011.01477.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.