Highlight
We describe new quantitative trait loci for growth and transpiration in wheat under two water regimes using an imaging platform, and co-location with loci for yield components in the field.
Key words: Drought, leaf expansion, Lemnatec, QTL, Triticum aestivum, water-use efficiency.
Abstract
Crop yield in low-rainfall environments is a complex trait under multigenic control that shows significant genotype×environment (G×E) interaction. One way to understand and track this trait is to link physiological studies to genetics by using imaging platforms to phenotype large segregating populations. A wheat population developed from parental lines contrasting in their mechanisms of yield maintenance under water deficit was studied in both an imaging platform and in the field. We combined phenotyping methods in a common analysis pipeline to estimate biomass and leaf area from images and then inferred growth and relative growth rate, transpiration, and water-use efficiency, and applied these to genetic analysis. From the 20 quantitative trait loci (QTLs) found for several traits in the platform, some showed strong effects, accounting for between 26 and 43% of the variation on chromosomes 1A and 1B, indicating that the G×E interaction could be reduced in a controlled environment and by using dynamic variables. Co-location of QTLs identified in the platform and in the field showed a possible common genetic basis at some loci. Co-located QTLs were found for average growth rate, leaf expansion rate, transpiration rate, and water-use efficiency from the platform with yield, spike number, grain weight, grain number, and harvest index in the field. These results demonstrated that imaging platforms are a suitable alternative to field-based screening and may be used to phenotype recombinant lines for positional cloning.
Introduction
Drought is a major cause of decreased crop production worldwide. In Australian dryland agriculture, grain crop yields are approximately 50% of their potential and are highly unpredictable. The 2006 drought reduced the total Australian wheat (Triticum aestivum L.) yield by 46% (FAO, 2013). During the 1990s, the rate of productivity increase in Australian broadacre cropping improved by 3.4% annually but has since slowed to about 1.4%. Yield is the end product of a grain crop, integrating the genetic ability of the plant to grow, assimilate carbon, and transfer it to the grain, and the effects of environmental conditions on these different plant processes. Yield is therefore a complex trait under multigenic control and is highly influenced by genotype×environment (G×E) interactions (Tardieu, 2010).
Although many quantitative trait loci (QTLs) have been identified in wheat for yield and yield components in low-yielding, rain-fed environments (reviewed by Fleury and Langridge, 2014), the underlying genes have yet to be cloned. Many QTLs called ‘unstable’ across environments are observed under specific environmental conditions only (G×E interactions). For example, the allele carried by the RAC875 parental line at the QTL for yield on chromosome 3B in wheat is only positive in hot and dry environments where the soil is deep, such as in northern Australia and Mexico, but not in southern Australia (Bonneau et al., 2013). In addition, gene cloning in wheat is still a difficult task due to the size (17 Gb) and complexity of the genome: the bread wheat genome is hexaploid with three homeologous A, B, and D genomes, and contains about 80% repeat sequences (Smith and Flavell, 1975; Choulet et al., 2010). The availability of genomic resources has substantially increased in recent years, significantly enhancing progress in genetic mapping and gene identification (Periyannan et al., 2013; Saintenac et al., 2013). As the availability of these resources becomes commonplace, phenotypic screening has become a bottleneck in the understanding and tracking of complex phenotypes such as yield in dry environments.
While plant physiologists have made progress in understanding the mechanisms of drought tolerance, this has not translated into tools that can be effectively utilized by breeding programmes. Tardieu and Tuberosa (2010) suggested that dissecting complex traits into simple components independent of many confounding environmental effects could be undertaken in highly controlled phenotyping platforms. The development of high-throughput phenotyping platforms over the last 10 years has improved evaluation of large genetic populations (Berger et al., 2010). It is now possible to apply these technical advances to the fine-mapping and possible cloning of genetic loci underlying QTLs.
The imaging platform of The Plant Accelerator at the Australian Plant Phenomics Facility and University of Adelaide, Australia, allowed us to image, weigh, and water thousands of plants every 2 d (Honsdorf et al., 2014). We developed routines to convert pixels from red/green/blue images into biomass and leaf area, to infer growth and relative growth rate, transpiration, and water-use efficiency (WUE) and applied these techniques to the analyses of a recombinant inbred line (RIL) population under different watering regimes.
The selected genetic population was a set of RILs derived from a cross between Drysdale and Gladius, two modern bread wheat varieties adapted to the southern region of the Australian wheat belt, characterized by a Mediterranean-type climate. As a consequence of the limited water storage in these soils, crops rely on season rainfall, which becomes sporadic in spring, leading to cyclic drought from the heading stage until the end of grain filling. Both Gladius and Drysdale perform well in a low-to-medium-rainfall environment but show different mechanisms of response to drought (Fleury et al., 2010). Gladius is an erect and waxy leaf variety selected for yield under severe drought in South Australia. Drysdale is a transpirationally efficient variety, which was selected for high carbon isotope discrimination, a surrogate for transpiration efficiency (Rebetzke et al., 2002; Condon et al., 2006). Segregation of these traits in Drysdale×Gladius progeny makes this population an interesting resource for dissecting the genetics of physiological mechanisms of adaptation to drought.
In this study, we used a genetic population with parental lines contrasting in their mechanisms of yield maintenance under water deficit. We analysed plant growth, transpiration, and WUE from imaging and watering data from a phenotyping platform for plants grown under two different watering regimes. The results were compared with QTLs identified for yield components in a field environment controlled for temperature and the watering regime. The comparison led to the discovery of co-located QTLs for plant growth, transpiration, and yield components, indicating that imaging platforms can be used to phenotype recombinant lines.
Materials and methods
Plant material
Genetic analysis was undertaken in a RIL population of 5000 lines from a cross between Drysdale and Gladius, two spring wheat varieties with the following respective pedigrees: Hartog×3 Quarrion for Drysdale, and RAC875/Krichauff//Excalibur/Kukri/3/RAC875/Krichauff/4/RAC875//Excalibur/Kukri for Gladius. Four sets of lines were used for different purposes. Set 1 consisting of 250 randomly chosen RILs was used for the simple sequence repeat (SSR) and diversity arrays technology (DArT) genetic map construction and phenotyping under semi-controlled field conditions in polyurethane tunnels (polytunnels) in 2010. A subset of 150 lines (set 2) flowering in a 6 d window was chosen from set 1 in order to apply the drought stress at a similar stage of development. This set was complemented with 100 randomly chosen RILs to construct the single nucleotide polymorphism (SNP) genetic map (set 3). The 115 RILs (set 4) common between sets 1 and 3 were genotyped to construct an integrated SSR, DArT, and SNP map.
Field trials
Two experiments were carried out under semi-controlled field conditions in the polytunnel facility of the University of Adelaide (Urrbrae, South Australia, Australia, 35° S 139° E). This facility includes bird nets and polytunnels, equipped with automatic watering systems (drippers) and weather stations (MEA, Adelaide, Australia) recording air temperature and humidity at 2 m height and at the plant canopy level, as well as soil temperature and wind. Gypsum blocks (MEA) were used for measuring soil water tension at three different soil depth (15, 30, and 40cm from the soil surface, eight sensors per watering regime). All climatic data were averaged and stored every 10min in a data logger (MEA).
Plant growth conditions
In 2010, 250 RILs of set 1 were grown under well-watered and water-deficit treatments. This trial was sown on 9 June, later than the normal commercial sowing time (April/May) for wheat in South Australia, in order to expose plants to drought stress during flowering and grain filling. In both treatments, micro-plots (10×120cm, 16 plants, density of 133 plants m–2) were randomly distributed and partially replicated (two replicates for 60 RILs and three replicates for the parental lines). On both sides of each lane of micro-plots, a micro-plot was sown with genotype Gladius (not analysed) to prevent any border effect. Both treatments were sown under bird nets and maintained as well watered (soil water tension > –0.1MPa) during vegetative growth using a dripper system. In the drought treatment, watering was stopped at stem elongation, and a polytunnel was installed as a rain shelter. When the soil dried below –0.6MPa, a light watering of 15mm was applied three times before harvesting the plants. In the well-watered treatment, watering was maintained until 15 d after flowering. Plants were fertilized (Aquasol, Hortico, Clayton, VIC, Australia) twice in both treatments: at stem elongation stage and at flowering stage. A fungicide (Bayfidan; Bayer Australia, Pymble, NSW, Australia) was applied at booting and flowering stages.
Plants experienced similar shoot micro-environments in both water regimes, with air temperature from 3.8 to 31 °C (average 12.4 °C) and a maximum vapour pressure deficit (VPD) of 3.5 kPa. Soil water tension in the well-watered treatment stayed above –0.1MPa until 15 d20°C (equivalent day at 20 °C, explained in ‘Thermal compensation of time and rates’) after flowering when plants experienced a moderate water deficit (> –0.4MPa) for 10 d. In the water-deficit treatment, the soil started to dry when irrigation was stopped and reached –0.6MPa around 5 d20°C after flowering. The soil water tension was then kept around –0.6MPa with light watering.
In 2011, 148 lines of the Drysdale/Gladius RIL set 2 were grown in the same platform under two water regimes: well watered and water deficit. This experiment was sown on 14 July, with micro-plots of 40×60cm (32 plants, 133 plants m–2). Both treatments were well watered until stem elongation. Then, watering was stopped in the drought treatment and lightly watered as in 2010. Urea (Manutec, Cavan, SA, Australia) was mixed with the top soil before sowing (N rate 225kg ha–1); the fertilizer and fungicide regime during plant growth was the same as in 2010.
In both water regimes, air conditions were warmer and dryer than in 2010, due to the later sowing, with air temperature from 4.8 to 35.2 °C (average 15.2 °C) and a maximum VPD of 4.9 kPa. Soil water potential stayed above –0.1MPa until 10 d20°C before flowering in both treatments and then started to dry at different speeds depending on treatment. Soil water potential reached –0.6MPa around flowering in the water-deficit treatment and around 20 d20°C after flowering in the well-watered treatment.
Plant measurements
Heading time (d; six first spikes appeared in the plot), and Flowering time (d; six spikes flowering on one-third of the spike length) were scored every day from the first spike appearance. All tillers were manually harvested, and the number of tillers per plant (Tiller number) and spikes per plant (Spike number) were counted. After drying (10 % moisture content), Stem weight (g), Grain weight (g), and total plant weight (Biomass, g) were measured and calculated per plant. A sample of 100ml of seed was weighed, and the seeds were counted with a seed counter (Pfueffer GmBH, Germany) to estimate Single grain weight (g) and total Seed number per plant. Grain weight per plot was converted to Yield (t ha–1) for an easier comparison with other trials. Harvest index was calculated as the ratio of Grain weight/Biomass.
Imaging platform experiment
Plant growth conditions
An experiment with 150 RILs of set 2 was performed in The Plant Accelerator (Honsdorf et al., 2014) greenhouse facilities in Urrbrae, South Australia, Australia (34°58′16.18″ S; 138°38′23.88″ E). Two watering regimes were applied (well watered and stable drought), and each line was replicated twice. Well-watered and drought-stressed plants of the same line and replicate number were placed next to each other. Single plants were grown in 2.5 l plastic pots filled with 3kg of potting mix (50% coco/peat mix, 50% clay/loam). Three seeds per pot were sown and thinned to one plant per pot at the three-leaf stage. Two pots per treatment contained artificial plants and were placed in the middle of the greenhouse, under the same watering regimes as pots with plants in order to estimate the evaporation from the soil surface.
During the first 3 weeks, plants were manually well- watered (> –0.02MPa) in both treatments. Plants were then transferred to the imaging platform of The Plant Accelerator, where each pot was placed onto a cart on a conveyor belt until flowering. Plants were imaged using a LemnaTec 3D Scanalyzer (LemnaTec, GmbH, Aachen, Germany). Three red/green/blue images (2056×2454 pixels) were taken with top and two side views with a 90° horizontal rotation (Honsdorf et al., 2014). Background–foreground separation was then applied to separate the plant tissue area from the background, and pixel numbers per image were counted after noise removal.
Environmental conditions fluctuated naturally in the greenhouse as plants experienced natural lighting and temperature ranking from 17 °C (night) to 25 °C (day). Every second day, pots were automatically weighed and watered to –0.02 and –0.05MPa (well-watered and drought treatments, respectively). Soil water content was measured by automatically weighing the pots. Differences in weight were attributed to changes in soil water content, after correction for the increase in plant Biomass mean (see below). A water-release curve of the soil was obtained on additional pots. Five pots containing three plants were dried from soil water retention capacity to –1.6MPa. After long nights (>12h) in a growth chamber with air saturated with water, pre-dawn leaf water potential was measured on non-expanding leaves using a Scholander-type pressure chamber (Soil Moisture Equipment Corp., Santa Barbara, USA). A Van Genucheten curve (Van Genuchten, 1980) was fitted to these data (soil water potential vs soil water content), thereby allowing calculation of the mean soil water potential in each pot at each weighing time (Supplementary Fig. S1, available at JXB online). Water loss per pot between two watering events was considered as plant Transpiration after correcting for soil evaporation measured using pots with artificial plants.
Calibration by plant destructive measurements
A separate experiment with similar conditions of culture was carried out on the two parental lines in order to convert pixel values obtained by image analysis into biological variables. From 2 weeks after sowing to flowering, three plants per genotype and per treatment were harvested twice a week. Plants were directly weighed (Plant weight, g). Leaf area (mm2) was measured with a planimeter (PATON electronic belt driven planimeter, CSIRO, Canberra, Australia). Biomass (g) was measured after 1 week at 65 °C.
Data analysis
Analysis of variance (ANOVA) and correlation analysis was performed for data obtained from different experiments using PROC GLM and PROC CORR, respectively, in SAS Software v.8.1 (SAS Institute, 2000). Broad-sense heritability (h 2) was calculated from variance components according to Kearsey and Pooni (1996). Data obtained from the imaging platform was analysed using R Software (R Development Core Team, 2013). Non-linear models were fitted with the nls function of the R package.
Thermal compensation of time and rates
Time and rates were expressed as thermal time as described by Parent et al. (2010) and Parent and Tardieu (2012). Briefly, the temperature responses of development processes were described by the equation of Johnson et al. (1942), modified by Parent and Tardieu (2012), and applied in different studies of developmental processes (Parent et al., 2009, 2010; Louarn et al., 2010):
| (1) |
where F(T) is the considered rate, T is the temperature (K), ∆HA ‡ (J mol–1) is the enthalpy of activation of the process and determines the curvature at low temperature, α (dimensionless) determines how sharp the rate decrease is at high temperature and is fixed at 3.5 for developmental processes (Parent and Tardieu, 2012), T0 (K) determines the temperature at which the rate is maximum, and A is the trait scaling coefficient.
For any measured rate J(T) at temperature T, a temperature compensated rate was calculated as the equivalent rate at 20 °C.
| (2) |
with F(T) being the response of development to temperature. As developmental time (or thermal time t 20°C) is the reciprocal of development rate, it results in:
| (3) |
with t 20°C being expressed either as equivalent hour at 20 °C (h20°C) or equivalent day at 20 °C (d20°C), depending on the unit of t.
Analysis pipeline for platform data
An analysis pipeline was developed to convert pixels from platform images into variables of biological interest such as Biomass, Plant weight, Leaf area, Average growth rate or WUE.
Converting pixels into projected areas by using simple trigonometric equations
A length measured in pixels (side.distance) from a side-view image can be converted to a distance in mm with a single coefficient (side.coeff), the size of a pixel:
| (4) |
with side.coeff=0.5156mm pixel–1 for the camera settings used.
For a top-view image, this coefficient (top.coeff, mm pixel–1) depends on plant height. First, the size of a pixel is calculated depending on the distance between the object and the camera:
| (5) |
with top.slope (0.0001097mm pixel–2) and top.offset (0.2899mm pixel–1) as the linear parameters of the relationship and cm.pix (pixel) as the y-coordinate of the centre of mass of the plant obtained from the side view images.
The projected areas can then be calculated as:
| (6) |
| (7) |
Linear models converting projected areas to Biomass, Plant weight, and Leaf area For each biological variable (Biomass, Plant weight, or Leaf area), the complete linear model with projected areas at the second order was tested, as well as all derived simpler models on data from the second experiment with destructive measurements. The complete model is:
| (8) |
These models were compared using the Bayesian information criterion (BIC) and the model with the lowest value was selected.
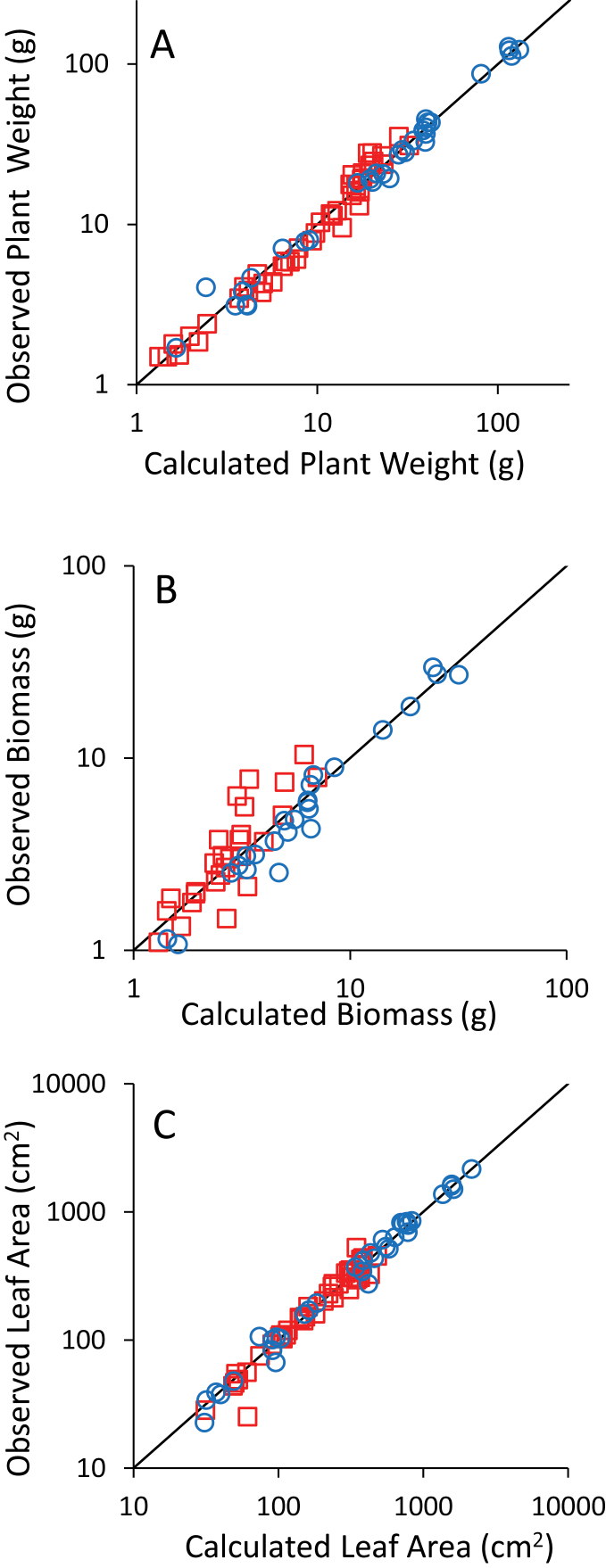
This procedure produced similar results for Biomass and Leaf area but different results for Plant weight (Table 1). The models were used to infer biological variables with the same parameters for well-watered plants and drought-stressed plants, but the error was higher for Biomass than for Plant weight and Leaf area (Fig. 1). It was decided to keep the same model for plants under well-watered and water-deficit conditions to better compare the treatments and to allow the calculation of variable responses to soil water potential.
Table 1.
Models selected for inferring Leaf Area, Plant Weight and Biomass from projected areas
Side and top are the average projected shoot area (mm2) on the side view and the projected shoot area on the top view (mm2), respectively. Models have been selected with BIC. ***P<0.001, **P <0.01, and (.), P<0.1 in an ANOVA test. Where no sign is given, this predictor was not selected by the BIC test.
| Intercept | Leaf area | Plant weight | Biomass |
|---|---|---|---|
| Side | *** | *** | *** |
| Top | (.) | *** | |
| Side2 | ** | *** | ** |
| Top2 | *** | *** | |
| Side:top | *** | (.) | *** |
Fig. 1.
Plots of observed/calculated variables for Plant weight (A), Biomass (B), and Leaf area (C). Circles are data for well-watered plants and squares are for drought-stressed plants. The line is the 1:1 line. The scale is logarithmic for better data visualization but models were selected on raw data. (This figure is available in colour at JXB online.)
Fitting growth curves to experimental data
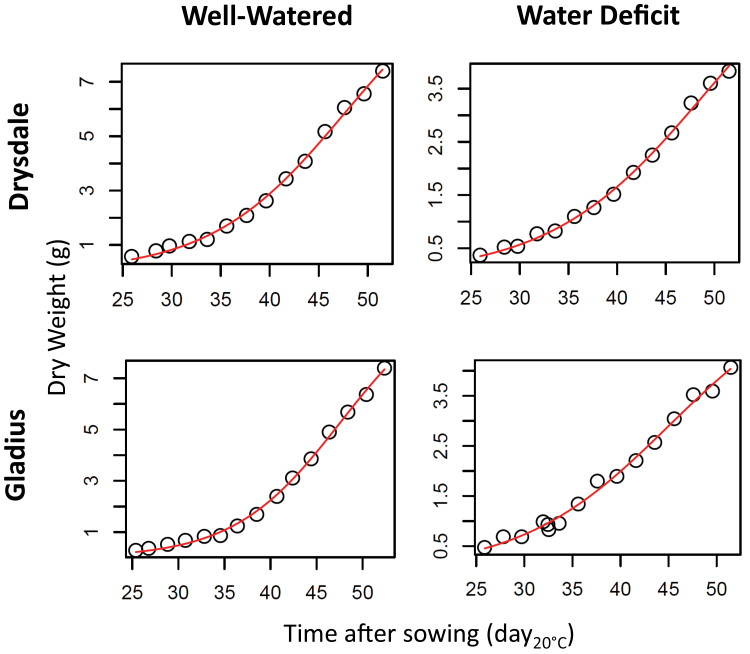
Different models for fitting Biomass, Plant weight, or Leaf area over time have been tested with the R function for non-linear regression (nls(), with our own self-start functions) on the two parental lines Gladius and Drysdale: exponential, linear, logistic with three (Chen et al., 2014) or four parameters, thye equation of Richards with four or five parameters, Gompertz with four parameters (Chen et al., 2014), and Weibull with three or four parameters. Some models were not adapted to all datasets. Only the linear, exponential, and logistic three parameters converged for all plants, but the logistic equation fitted best (BIC tests; results not shown) and was therefore applied to all data (Fig. 2).
Fig. 2.
Growth curves for calculated Biomass over thermal time in parental lines. Growth curves were calculated on single plants and these plots are examples of single plants. Circles indicate the calculated data. The solid line indicates the logistic (three-parameter) models. (This figure is available in colour at JXB online.)
| (9) |
In this equation, t 0 is the inflexion point and is considered as the transition from the vegetative to the reproductive stage. K is the Maximum relative growth rate (KRGR).
A logistic curve was fitted for each plant for Biomass, Plant weight, and Leaf area with time expressed as d20°C. This model for Biomass, Plant weight, and Leaf area fitted well with experimental data, even for Biomass (Fig. 2).
Water-use calculations
Soil water potential was calculated at each date using a water-release curve (Supplementary Fig. S1), measuring pot weight and calculating Plant weight using the logistic equation applied to plant images.
Average transpiration rate (TR, g d20°C –1) was calculated from water loss between days 35 and 50 after sowing (vegetative stage in all genotypes), taking into account the soil evaporation and plant weight. Average transpiration rate per unit of leaf area (TRarea, g mm–2 d20°C –1) was calculated from the leaf area inferred by the growth model:
| (10) |
Derived variables such as Growth, RGR, and WUE
For the three variables (Biomass, Plant weight, or Leaf area), their growth rate and Average relative growth rate (RGR) were derived from their growth curve:
| (11) |
| (12) |
In this analysis, Growth (g d20°C –1) is the increase of biomass with time, and Average leaf expansion rate (LER, mm2 d20°C –1) is the increase of leaf area. RGR (d20°C –1) is the relative increase of biomass and Average relative leaf expansion rate (RER, d20°C –1) is the relative increase of Leaf area. Average growth rate (Growth.AVE, g d20°C –1) and Average leaf expansion rate (LERAVE, mm2 d20°C –1) were calculated from day 35 to day 50.
WUE (g g–1) was calculated as the Average growth rate divided by transpiration from day 35 to day 50.
For all of these variables, their relative response to soil water potential (Ψ) was calculated as:
| (13) |
with Variable WW and Variable D, respectively, being the value of the considered biological variable in well-watered and drought conditions and Ψ WW and Ψ D, respectively, being the value of Ψ in well-watered and drought conditions.
Genetic map construction
DNA was extracted from 2.0g of bulked leaf tissue (three to six plants per line) of 8-week-old plants using a mini prep ball bearing extraction method with minor modifications (Pallotta et al., 2000). The RIL sets 1 and 3 were genotyped with markers for genes that control phenology in wheat: vernalization genes Vrn-A1 and Vrn-D1, and photoperiod genes Ppd-B1 and Ppd-D1, as described by Maphosa et al. (2014). For each line to be genotyped with SNP markers (set 3), approximately 100ng μl–1 of DNA (30 μl) was sent to the Department of Primary Industries, Victoria, to be assayed on an 9k SNP iSelect BeadChip array as described by Akhunov et al. (2009).
Three linkage maps were used. The first was based on DArT, SSR, and gene-based markers on RIL set 1 (Maphosa et al., 2014). The second map was constructed using SNP and gene-based markers on RIL set 3, which consisted of a mixture of random and a selection of mid-maturing lines. The third map was based on DArT, SSR, and SNP using RIL set 4. The method of genetic map construction was described by Maphosa et al. (2014).
QTL mapping
QTL analysis was performed for the mean spatial adjusted value of traits using PROC GLM, SAS software (v.9). Initially, single marker analysis was used for each trait to identify markers associated with variation for traits. Further evaluation was carried out by composite interval mapping with a 15 cM window and a maximum of 15 marker co-factors per model using Windows QTL Cartographer version 2.0 (Wang et al., 2004). As the 150 lines of set 2 were still segregating for the Vrn-A1, Vrn-D1, Ppd-B1, and Ppd-D1 genes (Beales et al., 2007; Zhang et al., 2008), the model was adjusted to remove their effect on growth and yield. Tests were performed at 1 cM intervals, and effect of these genes was selected as the co-factor by forward–backward stepwise regression (Model 6). Genome-wide, trait-specific threshold values (α= 0.05) of the likelihood ratio test statistic for declaring the presence of a QTL were estimated from 1000 permutation tests by random sampling of phenotypic data (Churchill and Doerge, 1994; Doerge and Churchill, 1996). The phenotypic variation explained by a QTL (R 2) conditioned by the composite interval mapping co-factors included in the model was calculated at the most likely QTL position. The additive effect of an allelic substitution at each QTL was also obtained. The log of odds (LOD) peak of each significant QTL was considered as the QTL location on the linkage map.
The QTL analysis of the imaging platform experiment on RIL set 2 was done using the SNP map. All QTL intervals were located on the SSR–DArT–SNP map and the SNP map for comparing the position of QTLs for different traits and platforms.
Results
Analysis of the imaging platform data
Biomass, Plant weight, Leaf area, Average growth rate, and WUE were calculated using the new analysis pipeline. The same pipeline was adapted to plants for all genotypes and treatments (Fig. 2). These data were then used for genetic analysis. Transgressive segregation and significant genetic variation among the lines was observed for most of these traits (Table 2). As expected, large differences were observed between the two watering regimes, so each treatment was analysed separately. The broad-sense heritability (h 2) values ranged from 0 to 54%, indicating that the proportion of genetic-to-environmental variation differed among traits.
Table 2.
Descriptive statistics and heritability (h2, %) for the traits measured on the 150 Drysdale/Gladius RIL phenotyped in the imaging platform
SD, standard deviation; Min, minimum, Max, maximum; ***P<0.001, **P<0.01, *P <0.05 and NS, P >0.05 (not significant) in an ANOVA test.
| Variable | Well watered | Drought | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Name | Acronym | Mean±SD | (Min – Max) | h 2 | P | Mean±SD | (Min – Max) | h 2 | P |
| Average growth rate | GrowthAVE (g d20°C –1) | 0.332±0.076 | (0.108–0.533) | 40.8 | *** | 0.121±0.047 | (0.032–0.27) | 51.1 | *** |
| Average relative growth rate | RGR (d20°C –1) | 0.102±0.008 | (0.076–0.126) | 5.3 | NS | 0.075±0.014 | (0.038–0.117) | 46.1 | *** |
| Inflexion point in growth curves | Txgrowth (d20°C) | 56.11±2.83 | (50.43–65.14) | 6.6 | NS | 51.41±7.75 | (38.84–97.06) | 43.3 | *** |
| Maximum relative growth rate | KRGR (d20°C –1) | 0.143±0.014 | (0.113–0.188) | 3.2 | NS | 0.130±0.023 | (0.079–0.215) | 30.0 | *** |
| Average leaf expansion rate | LERAVE (mm2 d20°C –1) | 3062±725 | (997–4842) | 37.4 | *** | 999±364 | (329–2137) | 41.1 | *** |
| Average relative leaf expansion rate | RER (d20°C –1) | 0.092±0.007 | (0.072–0.110) | 35.7 | *** | 0.056±0.010 | (0.031–0.084) | 47.6 | *** |
| Maximum relative leaf expansion rate | KRER (d20°C –1) | 0.122±0.010 | (0.086–0.146) | 24.4 | *** | 0.102±0.014 | (0.070–0.139) | 53.8 | *** |
| Average transpiration rate | TR (g d–1) | 88.9±16.0 | (46.6–123.2) | 14.1 | * | 38.2±7.1 | (25.1–78.7) | 22.2 | ** |
| Average transpiration rate per unit leaf area | TRarea (g mm–2 d20°C –1) | 2.98±0.47 | (1.24–4.68) | 11.9 | * | 2.08±0.37 | (1.40–3.61) | 25.7 | ** |
| WUE | WUE (g g–1) | 0.003±0.001 | (0.002–0.006) | 21.7 | *** | 0.003±0.001 | (0.001–0.005) | 44.6 | *** |
We compared the data from the 2011 experiments in semi-controlled field conditions (polytunnel) and from the platform experiment, both performed on the same RIL. No significant correlations were found between traits measured in the imaging platform in pots (growth, transpiration, and WUE) and traits measured in the polytunnel (yield, yield components, and phenology), with r values ranging from –0.3 to 0.2 (Supplementary Fig. S2, available at JXB online).
QTLs for plant growth using the imaging platform
Using a genetic map of 3200 SNP markers and 783 loci, a total of 21 QTLs were identified for the platform variables in the Drysdale/Gladius population (Table 3). We identified a total of 14 QTLs for traits related to plant growth in both water treatments. Four QTLs showed strong effects ranging from 16 to 43% of the genetic variation of the trait on chromosomes 1A and 1B. GrowthAVE was controlled by four QTLs, which altogether explained 61% of the variation. Six QTLs for traits related to transpiration were identified.
Table 3.
QTLs for traits measured using the imaging platform
The additive effect is expressed in specific trait units. A positive value means that the trait increase is due to the Drysdale allele, while a negative value indicates the Gladius allele. Chr, chromosome. R 2 is the percentage of the genetic variation of the trait explained by the QTL.
| Trait | QTL | Chr | LOD thresholda | Marker interval | QTL (cM) | LOD | Additive effect | R 2 (%) |
|---|---|---|---|---|---|---|---|---|
| Well-watered | ||||||||
| GrowthAVE | QGRO.atw-1B | 1BL | 3.3 | Ex_c5296_9365847 | 68.2 | 6.2 | +0.030 | 43 |
| CAP7_c4778_2155754 | ||||||||
| QGRO.atw-2A.1 | 2AS | 3.3 | JD_c18695_17091254 | 52.1 | 4.3 | +0.025 | 9 | |
| Ex_rep_c66709_65042923 | ||||||||
| QGRO.atw-2A.2 | 2AL | 3.3 | BF475068A_Ta_2_1 | 63.2 | 3.8 | –0.010 | 5 | |
| Ex_rep_c69799_68761171 | ||||||||
| QGRO.atw-5A | 5AL | 3.3 | Ex_c32414_41076471 | 8 | 5.3 | +0.060 | 4 | |
| Ex_c2505_4679749 | ||||||||
| LERAVE | QLERAVE.atw-1B.1 | 1BL | 3.5 | Ex_c5296_9365847 | 68.8 | 4.4 | +242 | 26 |
| CAP7_c4778_2155754 | ||||||||
| QLERAVE.atw-1B.2 | 1BL | 3.4 | Ex_c5296_9365847 | 60.6 | 6.4 | –304 | 16 | |
| CAP11_c1902_1022590 | ||||||||
| RER | QRER.atw-1A | 1AL | 3.6 | Ra_c2227_4304970 | 50.3 | 3.6 | +0.002 | 30 |
| Ex_c15377_23637176 | ||||||||
| TR | QTR.atw-1B | 1BL | 3.1 | Ex_c5296_9365847 | 72.8 | 4.9 | +6.2 | 15 |
| CAP7_c4778_2155754 | ||||||||
| TRarea | QTRarea.atw-2D | 2DL | 2.9 | Ex_rep_c69782_68740893 | 38.1 | 3 | +0.036 | 3 |
| Ra_c3057_5773026 | ||||||||
| Drought | ||||||||
| GrowthAVE | QGRO.atd-5B | 5BL | 3.3 | Ex_c35398_43558614 | 111.1 | 4.8 | –0.020 | 9 |
| Ex_c11951_19164786 | ||||||||
| RGR | QRGR.atd-4A | 4AL | 3.4 | Ex_c5487_9686018 | 120.8 | 3.4 | –0.001 | 10 |
| Ex_c14478_22481430 | ||||||||
| Txgrowth | QTxgrowth.atd-7D | 7DS | 2.8 | Ex_c17914_26681837 | 3.5 | 3.2 | +2.75 | 10 |
| Ex_c11813_18968198 | ||||||||
| LERAVE | QLERAVE.atd-5B | 5BL | 3.3 | Ex_c35398_43558614 | 112.4 | 4.4 | –130 | 10 |
| Ex_c11951_19164786 | ||||||||
| TxLER | QTxLER.atd-5B | 5BL | 3.3 | Ex_c35398_43558614 | 113.6 | 3.6 | –2.7 | 6 |
| Ex_c11951_19164786 | ||||||||
| TR | QTR.atd-3A | 3AL | 2.8 | Ex_c11877_19055556 | 97 | 31 | +2.1 | 2 |
| Ex_c15674_24004810 | ||||||||
| QTR.atd-4B | 4BL | 2.8 | Ex_c28687_37791888 | 61.6 | 4.6 | –2.6 | 3 | |
| Ex_c17211_25859780 | ||||||||
| WUE | QWUE.atd-2A | 2AL | 3.0 | BE406351A_Ta_2_3 | 66.5 | 4 | –0.0003 | 3 |
| Ex_rep_c69799_68761171 | ||||||||
| Relative response to soil water potential | ||||||||
| KRGR | QKRGR.atr-5A | 5AS | 3.3 | JD_c5795_6955031 | 32.9 | 3.9 | –0.2 | 3 |
| Ra_c8898_14972290 | ||||||||
| KRER | QKRER.atr-3A | 3AL | 3.1 | Ex_c11910_19101291 | 67.2 | 4.1 | –0.1 | 2 |
| Ku_c38911_47455674 | ||||||||
| TRarea | QTRarea.atr-6A | 6AS | 1.8 | CAP12_c1663_836753 | 6.7 | 2 | +0.2 | 10 |
| Ex_c965_1846161 | ||||||||
a The LOD threshold was empirically estimated at α= 0.05 from 1000 permutation tests by random sampling of phenotypic data for each trait.
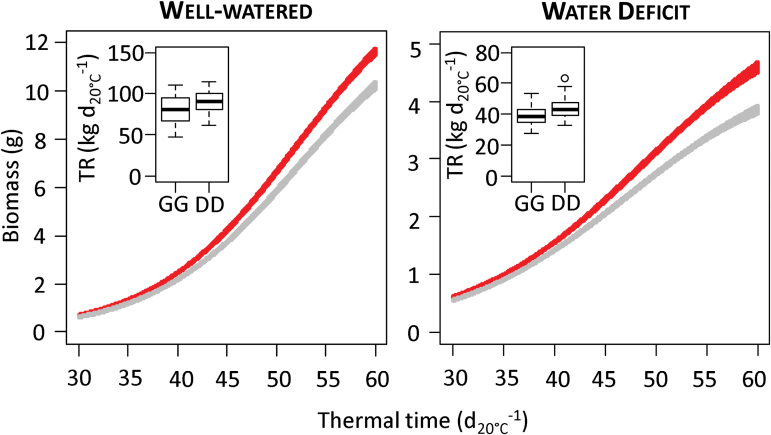
We found overlaps between QTLs for different variables measured in the platform on three regions of chromosomes 1B, 2A, and 5B (Table 4). On chromosome 1B, the QTL QGRO.atw-1B for GrowthAVE and QLERAVE.atw-1B.1 for Average leaf expansion rate coincided with the QTL peak of QTR.atw-1B for Average transpiration rate. These three QTLs carried Drysdale as a positive allele. When looking at pools of RILs carrying Gladius or Drysdale alleles at the marker wsnp_CAP11_c1902_1022590 (Fig. 3), it appeared that the higher transpiration rate and higher growth rate for plants carrying the Drysdale allele were observed in both treatments. This QTL effect seemed intrinsic rather than specific to well-watered conditions.
Table 4.
Co-localization of QTLs for traits studied in the imaging platform and the polytunnel facility
Position on the SSR–DArT–SNP map (1) and SNP map (2). Apd, ACPFG polytunnel drought; apw, ACPFG polytunnel well-watered; 10, year 2010; 11, year 2011; atd, The Plant Accelerator drought; atw, The Plant Accelerator well-watered.. Light-grey shading indicates a positive additive allelic effect (Drysdale); dark grey shading indicates a negative additive allelic effect (Gladius).
| Genetic map | Imaging platform QTL | Polytunnel QTL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | cM (1) | cM (2) | LERAVE | Growth.AVE | LER.Tx | TR | TR | WUE | Yield | Grain number | Grain weight | Harvest index | Spike number | Tiller number |
| 1BL | 74.3 | QLERAVE. atw-1B.2 | QTR. atw-1B | |||||||||||
| 86.7 | QLERAVE. atw-1B.1 | QGRO. atw-1B | QSnp.apw11-1B | |||||||||||
| 112.9 | ||||||||||||||
| 2AS | 88.2 | QYie.apw11-2A.2 | ||||||||||||
| 90.9 | QGRO. atw-2A.1 | |||||||||||||
| 91.4 | QGns.apd10-2A | |||||||||||||
| 2AL | 93.4 | QWUE.atd-2A | QYie.apw11-2A.1 | QGra.apw11-2A | ||||||||||
| 109.8 | QGRO. atw-2A.2 | |||||||||||||
| 4BL | 43.3 | QTR. atd-4B | ||||||||||||
| 46.2 | ||||||||||||||
| 47.1 | QTil.apd10-4B.2 | |||||||||||||
| 5BL | 106.9 | QLERAVE. atd-5B | QGRO. atd-5B | QTxLER. atd-5B | QHar.apw11-5B.1 | |||||||||
| 133.1 | ||||||||||||||
Fig. 3.
Difference between lines with the Drysdale or Gladius alleles at marker wsnp_CAP11_c1902_1022590 (position 74.3 on chromosome 1B) for their growth curve and Average transpiration rate (TR) in the well-watered treatment of the experiment in the imaging platform. The graphs show growth curves for lines with the Gladius allele (lower curve) or the Drysdale allele (upper curve) at this locus. Curves are the logistic inference±standard deviation obtained with 1000 bootstrap replicates (function boot in R) on all lines having the considered allele at this locus. Insets show boxplots of TR per unit leaf area for lines with the Gladius (G) or Drysdale (D) allele at this locus. (This figure is available in colour at JXB online.)
Overlap between QTLs detected with the imaging platform and field trials
In two experiments run in semi-controlled field conditions that included two watering treatments, a total of 84 QTLs (Supplementary Table S1, available at JXB online) were found for traits related to biomass, yield and phenology. All QTL identified using the imaging platform, the polytunnel experiments and the field data from Maphosa et al. (2014) were positioned on the SSR–DArT–SNP map and the SNP map. This enabled us to identify seven regions with co-located QTLs (Tables 4 and 5).
Table 5.
Co-localization of QTLs for traits studied in the imaging platform and in the field (Maphosa et al., 2014)
Position on the SSR–DArT–SNP map (1) and SNP map (2). Light-grey shading indicates a positive additive allelic effect (Drysdale); dark grey shading indicates a negative additive allelic effect (Gladius).
| Genetic map | Imaging platform QTL | Field QTL | |||||
|---|---|---|---|---|---|---|---|
| Chr | cM (1) | cM (2) | TR | KRGR | Yield | Grain number | Screening |
| 3AL | 60.5 | 116.2 | |||||
| 62.0 | NSW09 | ||||||
| 67.2 | QTR.atd-3A | NSW10 SAU10 SAU10D | |||||
| 67.0 | 102.4 | ||||||
| 69.4 | SAB09 NSW09 SAU10D | ||||||
| 74.7 | 94.4 | ||||||
| 5AS | 21.6 | SAR08 SAR09 MEX11 | NSW09 | ||||
| NSW09 | SAU10, SAU10D | ||||||
| 26.2 | QKRGR.atr-5A | ||||||
| 27.6 | |||||||
A region on chromosome 1B showed QTLs for Spike number in the polytunnel (QSnp.apw11-1B) in 2011 and for plant growth-related traits (QGRO.atw-1B and QLERAVE.atw-1B.1) and transpiration (QTR.atw-1B) in the platform (Table 4 and Fig. 3). All these QTLs were found in plants grown under well-watered conditions, but some effects could be seen under water deficit. It is worth noting that QGRO.atw-1B, which explained 43% of the genetic variation of Growth under well-watered conditions, overlapped with QSnp.apw11-1B, which explained 20% of the variation of Spike number.
Two regions of chromosome 2A showed overlapping QTLs between the platform and the polytunnel experiments (Table 4). QGRO.atw-2A.2 for Average growth rate in the imaging platform overlapped with three QTLs for Yield, Grain number, and Grain weight in the polytunnel. The second overlap was between two polytunnel QTLs for Yield and Grain weight and a QTL that explained 9% of the genetic variation of Average growth rate in well-watered conditions. It also overlapped with a QTL for WUE in the drought treatment in the platform.
Two QTLs for platform and polytunnel traits were found in close proximity on chromosome 4B (Table 4). The QTL QTR.atd-4B.1 for Average transpiration rate was mapped near the QTL for Tiller number in the polytunnel, QTil.apd10-4B.2.
Chromosome 5B showed an overlap of loci found in the platform under drought and a QTL controlling Harvest index in the polytunnel 2011 experiment (Table 4). Four QTLs associated with Average leaf expansion rate, for T 0, for leaf expansion curves, and for Harvest index were co-mapped on the interval 106.9–133.1 cM of the long arm of this chromosome. Although the Harvest index QTL was found for the irrigated treatment, the environmental conditions were hot and dry in 2011 with a maximum VPD of 4.9 kPa in 2011 (vs 3.5 kPa in 2010).
Two regions showed co-location of QTLs from the imaging platform and field when compared with the results of Maphosa et al. (2014) (Table 5). A QTL for Average transpiration rate in the platform overlapped on chromosome 3A with seven QTLs for yield, grain number, and screenings in the field previously found by Maphosa et al. (2014) (Table 5). The transpiration QTL QTR.atd-3A was identified under drought, which matched with the conditions where the yield-related QTLs were expressed: in rain-fed conditions of New South Wales and South Australia, and under cyclic drought in South Australia semi-irrigated conditions.
Discussion
Using imaging platforms for analyses of drought responses
One role of imaging platforms, such as The Plant Accelerator, is to enable phenotyping of large numbers of plants using stable environmental conditions and consistent methods across experiments. Previous reports have shown the potential of imaging platforms for QTL mapping and the identification of heritable traits in barley (Chen et al., 2014; Honsdorf et al., 2014). However, for the last 10 years, most studies using such platforms have focused on platform development and analysis procedures (see examples in this special issue), more than application to specific biological questions related to crop improvement. In this study, known procedures of phenotyping have been linked through an operational analysis pipeline to support the genetic analysis of quantitative traits from non-destructive measurements. This allowed not only calculation of dynamic profiles but also derived traits such as soil water potential and transpiration, taking into account the plant weight, and the transpiration per unit leaf area or dynamic WUE.
Although the heritability values were generally low, QTLs of strong effect were found on chromosomes 1A and 1B using the imaging platform that explained between 26 and 43% of the genetic variation (Fig. 3). The G×E interaction could be reduced to some extent by using dynamic variables such as biomass increase and leaf expansion over time in response to the water treatment.
It is worthwhile noting that different sets of QTLs were identified in well-watered and drought conditions. Four QTLs on chromosomes 1A, 2D, and 6B were specifically expressed under well-watered conditions. Five QTLs were found only in the drought treatment of the platform on chromosomes 3A, 4A, 4B, 5B, and 7D and are therefore drought responsive. For fine-mapping and positional cloning, only one watering condition would be required, allowing phenotyping of a large number of lines.
Genetics of growth, transpiration, and WUE in wheat
To the best of our knowledge, this study is the first to describe QTLs for leaf expansion and plant growth in wheat. Fourteen QTLs were found for growth-related traits. Since several of the variables measured are mathematically dependent, some QTLs are likely to represent the same loci identified using different equations. These QTLs are readily identified since they are located in the same genetic position and show the same additivity. Using these criteria, the total number of loci controlling growth traits in our study is 11.
QTLs for transpiration efficiency have been found previously in wheat (Rebetzke et al., 2008; Wu et al., 2011) using the carbon isotope discrimination method. By comparing our results with those of Rebetzke et al. (2008), the QTL QTR.atw-1B of Drysdale/Gladius may be collocated with a QTL for carbon isotope discrimination found on the long arm of chromosome 1B, but more markers will be needed to compare the two map positions. The five QTLs of Drysdale/Gladius for transpiration and WUE on chromosomes 2D, 6A, 3A, 4Bs and 2A were not detected previously by Rebetzke et al. (2008) or Wu et al. (2011).
Two QTLs for average transpiration rate per unit leaf area (QTRarea.atw-2D and QTR.atd-4B) were found on chromosomes 2D and 4B. These chromosomes also carry the Reduced height (Rht) semi-dwarfing genes known to control plant development in wheat (Ellis et al., 2002, 2005). Since TR is calculated from plant biomass, developmental genes, such as the Rht genes, are likely to influence TR. However, the locations of Rht8 (Gasperini et al., 2012) and QTRarea.atw-2D do not match, and the Rht1 diagnostic marker (Ellis et al., 2002) was monomorphic in Drysdale/Gladius. Therefore, Rht genes do not explain the transpiration QTLs. Further investigations will be needed to identify candidate genes underneath these QTLs.
Common loci for growth and transpiration rates, and yield components
There were no significant genetic correlations between growth, transpiration, or WUE data from experiments in the platform and yield component traits in the polytunnel. Low correlation is often observed when comparing very different traits (vegetative growth and transpiration vs grain yield) under different growth conditions (glasshouse vs field, and pot vs plot). However, we identified some common QTLs between the platform and the field experiments, which indicate a common genetic basis for both traits. For example, the QTL for biomass increase in pots on chromosomes 1B and 2A under well-watered conditions overlapped with QTLs for yield components in the polytunnel. This indicates that these QTLs might control plant growth across environments, which could be translated into grain yield.
Co-location of QTLs for transpiration traits and yield were also found by Maphosa et al. (2014) by comparing their data with the studies of Rebetzke et al. (2008) and Wu et al. (2011). However, the only co-locations found were in the Ppd-B1 and Ppd-D1 regions, which control phenology. This is a well-known drought escape mechanism where the plant cycle is accelerated so that plants flower before the onset of severe drought late in the cropping season. Since these loci are well known, our study aimed to identify loci that were not related to phenology. Consequently, the effects of these two genes were excluded by including their genotypes in the QTL model so that the QTLs we identified are likely to control yield and growth-related traits per se.
A locus on chromosome 1B (86.7–112.9 cM) showed three overlapping QTLs controlling LERAVE and GrowthAVE in the platform under well-watered conditions, and spike number in the polytunnel in 2011 (Table 4). The higher transpiration rate for plants carrying the Drysdale allele was mainly driven by a higher leaf area and biomass. This QTL seems to be constitutive, as some effects have been found in well-watered as well as under drought conditions (Fig. 3). Several other QTLs for yield and yield components have been detected on chromosome 1B under rain-fed, well-watered, and drought-stressed environments in different wheat mapping populations in Australia, China, India, Mexico, and Spain (Quarrie et al., 2005; Kuchel et al., 2007; Kumar et al., 2007; Maccaferri et al., 2008; Wang et al., 2009; Pinto et al., 2010; Edwards, 2011; Bennett et al., 2012). There is a strong association between the number of stems per plant and relative growth rate in wheat (Dreccer et al., 2013). Our result suggests that the QTL on 1B could control plant growth rate during the vegetative phase, which could affect the number of spikes and grain yield. This QTL would be an interesting target for cloning to identify genes controlling yield across environments in wheat.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Water-release curve of experiment in the imaging platform.
Supplementary Fig. S2. Correlation between the traits from the platform and the polytunnel experiments.
Supplementary Table S1. QTLs identified for phenology, biomass, and yield-related traits in the polytunnel experiments run in 2010 and 2011 in Urrbrae (South Australia).
Acknowledgements
This work was supported by the Australian Research Council, The Grains Research and Development Corporation (ACP00002-Q), The Government of South Australia, and the European Union Framework Program 7 ‘Drought-tolerant yielding plants’ (DROPS) project (FP7-KBBE-244374). The Plant Accelerator, Australian Plant Phenomics Facility, is funded under the National Cooperative Research Infrastructure Strategy of the Australian Commonwealth.
Glossary
Abbreviations:
- BIC
Bayesian information criterion
- DArT
diversity arrays technology
- GrowthAVE
Average growth rate
- G×E
genotype by environment
- h2
heritability
- KRER
Maximum relative leaf expansion rate
- KRGR
Maximum relative growth rate
- LER
leaf expansion rate
- LERAVE
Average leaf expansion rate
- LOD
log of odds
- QTL
quantitative trait locus
- RER
Average relative leaf expansion rate
- RGR
Average relative growth rate
- RIL
recombinant inbred line
- SNP
single nucleotide polymorphism
- SSR
simple sequence repeat
- TR
Average transpiration rate
- TRarea
Average transpiration rate per unit leaf area
- Txgrowth
Inflexion point in growth curves
- TxLER
Inflexion point in leaf expansion curves
- VPD
vapour pressure deficit
- WUE
water-use efficiency.
References
- Akhunov E, Nicolet C, Dvorak J. 2009. Single nucleotide polymorphism genotyping in polyploid wheat with the Illumina GoldenGate assay. Theoretical and Applied Genetics 119, 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beales J, Turner A, Griffiths S, Snape JW, Laurie DA. 2007. A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theoretical and Applied Genetics 115, 721–733. [DOI] [PubMed] [Google Scholar]
- Bennett D, Izanloo A, Reynolds M, Kuchel H, Langridge P, Schnurbusch T. 2012. Genetic dissection of grain yield and physical grain quality in bread wheat (Triticum aestivum L.) under water-limited environments. Theoretical and Applied Genetics 125, 255–271. [DOI] [PubMed] [Google Scholar]
- Berger B, Parent B, Tester M. 2010. High-throughput shoot imaging to study drought responses. Journal of Experimental Botany 61, 3519–3528. [DOI] [PubMed] [Google Scholar]
- Bonneau J, Taylor J, Parent B, Bennett D, Reynolds M, Feuillet C, Langridge P, Mather D. 2013. Multi-environment analysis and improved mapping of a yield-related QTL on chromosome 3B of wheat. Theoretical and Applied Genetics 126, 747–761. [DOI] [PubMed] [Google Scholar]
- Chen D, Neumann K, Friedel S, Kilian B, Chen M, Altmann T, Klukas C. 2014. Dissecting the phenotypic components of crop plant growth and drought responses based on high-throughput image analysis. The Plant Cell 26, 4636–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choulet F, Wicker T, Rustenholz C, et al. 2010. Megabase level sequencing reveals contrasted organization and evolution patterns of the wheat gene and transposable element spaces. The Plant Cell 22, 1686–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. 1994. Empirical threshold values for quantitative trait mapping. Genetics 138, 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon A, Farquhar G, Rebetzke G, Richards R. 2006. The application of carbon isotope discrimination in cereal improvement for water-limited environments. In: Ribaut J, ed. Drought adaptation in cereals. New York: Haworth Press, 171–211. [Google Scholar]
- Doerge RW, Churchill GA. 1996. Permutation tests for multiple loci affecting a quantitative character. Genetics 142, 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreccer MF, Chapman SC, Rattey AR, Neal J, Song Y, Christopher JJ, Reynolds M. 2013. Developmental and growth controls of tillering and water-soluble carbohydrate accumulation in contrasting wheat (Triticum aestivum L.) genotypes: can we dissect them? Journal of Experimental Botany 64, 143–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J. 2011. A genetic analysis of drought related traits in hexaploid wheat. PhD thesis, University of Adelaide, Adelaide, Australia. [Google Scholar]
- Ellis H, Spielmeyer W, Gale R, Rebetzke J, Richards A. 2002. “Perfect” markers for the Rht-B1b and Rht-D1b dwarfing genes in wheat. Theoretical and Applied Genetics 105, 1038–1042. [DOI] [PubMed] [Google Scholar]
- Ellis MH, Rebetzke GJ, Azanza F, Richards RA, Spielmeyer W. 2005. Molecular mapping of gibberellin-responsive dwarfing genes in bread wheat. Theoretical and Applied Genetics 111, 423–430. [DOI] [PubMed] [Google Scholar]
- FAO. 2013. Drought. http://www.fao.org/docrep/017/aq191e/aq191e.pdf.
- Fleury D, Jefferies S, Kuchel H, Langridge P. 2010. Genetic and genomic tools to improve drought tolerance in wheat. Journal of Experimental Botany 61, 3211–3222. [DOI] [PubMed] [Google Scholar]
- Fleury D, Langridge P. 2014. Quantitative trait loci and association mapping for plant abiotic stress tolerance: trait characterization and introgression for crop improvement and breeding. In: Jenks MA, Hasegawa PM, eds. Plant abiotic stress, 2nd edn John Wiley & Sons, 257–288. [Google Scholar]
- Gasperini D, Greenland A, Hedden P, Dreos R, Harwood W, Griffiths S. 2012. Genetic and physiological analysis of Rht8 in bread wheat: an alternative source of semi-dwarfism with a reduced sensitivity to brassinosteroids. Journal of Experimental Botany 63, 4419–4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honsdorf N, March TJ, Berger B, Tester M, Pillen K. 2014. High-throughput phenotyping to detect drought tolerance QTL in wild barley introgression lines. PloS One 9, e97047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson FH, Eyring H, Williams RW. 1942. The nature of enzyme inhibitions in bacterial luminescence: Sulfanilamide, urethane, temperature and pressure. Journal of Cellular and Comparative Physiology 20, 247–268. [Google Scholar]
- Kearsey M, Pooni H. 1996. The genetical analysis of quantitative traits. London: Chapman & Hall. [Google Scholar]
- Kuchel H, Williams KJ, Langridge P, Eagles HA, Jefferies SP. 2007. Genetic dissection of grain yield in bread wheat. I. QTL analysis. Theoretical and Applied Genetics 115, 1029–1041. [DOI] [PubMed] [Google Scholar]
- Kumar N, Kulwal PL, Balyan HS, Gupta PK. 2007. QTL mapping for yield and yield contributing traits in two mapping populations of bread wheat. Molecular Breeding 19, 163–177. [Google Scholar]
- Louarn G, Andrieu B, Giauffret C. 2010. A size-mediated effect can compensate for transient chilling stress affecting maize (Zea mays) leaf extension. New Phytologist 187, 106–118. [DOI] [PubMed] [Google Scholar]
- Maccaferri M, Sanguineti MC, Corneti S, et al. 2008. Quantitative trait loci for grain yield and adaptation of durum wheat (Triticum durum Desf.) across a wide range of water availability. Genetics 178, 489–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maphosa L, Langridge P, Taylor H, et al. 2014. Genetic control of grain yield and grain physical characteristics in a bread wheat population grown under a range of environmental conditions. Theoretical and Applied Genetics 127, 1607–1624. [DOI] [PubMed] [Google Scholar]
- Pallotta M, Graham R, Langridge P, Sparrow D, Barker S. 2000. RFLP mapping of manganese efficiency in barley. Theoretical and Applied Genetics 101, 1100–1108. [Google Scholar]
- Parent B, Conejero G, Tardieu F. 2009. Spatial and temporal analysis of non-steady elongation of rice leaves. Plant, Cell and Environment 32, 1561–1572. [DOI] [PubMed] [Google Scholar]
- Parent B, Suard B, Serraj R, Tardieu F. 2010. Rice leaf growth and water potential are resilient to evaporative demand and soil water deficit once the effects of root system are neutralized. Plant, Cell and Environment 33, 1256–1267. [DOI] [PubMed] [Google Scholar]
- Parent B, Tardieu F. 2012. Temperature responses of developmental processes have not been affected by breeding in different ecological areas for 17 crop species. New Phytologist 194, 760–774. [DOI] [PubMed] [Google Scholar]
- Periyannan S, Moore J, Ayliffe M, et al. 2013. The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science 341, 786–788. [DOI] [PubMed] [Google Scholar]
- Pinto RS, Reynolds MP, Mathews KL, McIntyre CL, Olivares-Villegas JJ, Chapman SC. 2010. Heat and drought adaptive QTL in a wheat population designed to minimize confounding agronomic effects. Theoretical and Applied Genetics 121, 1001–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarrie SA, Steed A, Calestani C, et al. 2005. A high-density genetic map of hexaploid wheat (Triticum aestivum L.) from the cross Chinese Spring X SQ1 and its use to compare QTLs for grain yield across a range of environments. Theoretical and Applied Genetics 110, 865–880. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. 2013. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Rebetzke G, Condon A, Richards R, Farquhar G. 2002. Selection for reduced carbon isotope discrimination increases aerial biomass and grain yield of rainfed bread wheat. Crop Science 42, 739–745. [Google Scholar]
- Rebetzke GJ, Condon AG, Farquhar GD, Appels R, Richards RA. 2008. Quantitative trait loci for carbon isotope discrimination are repeatable across environments and wheat mapping populations. Theoretical and Applied Genetics 118, 123–137. [DOI] [PubMed] [Google Scholar]
- Saintenac C, Zhang W, Salcedo A, Rouse MN, Trick HN, Akhunov E, Dubcovsky J. 2013. Identification of wheat gene Sr35 that confers resistance to Ug99 stem rust race group. Science 341, 783–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute. 2000. The SAS system for Windows. Cary, NC: SAS Institute. [Google Scholar]
- Smith D, Flavell R. 1975. Characterisation of the wheat genome by renaturation kinetics. Chromosoma 50, 223–242. [Google Scholar]
- Tardieu F, Tuberosa R. 2010. Dissection and modelling of abiotic stress tolerance in plants. Current Opinion in Plant Biology 13, 206–212. [DOI] [PubMed] [Google Scholar]
- Tardieu F. 2010. Why work and discuss the basic principles of plant modelling 50 years after the first plant models? Journal of Experimental Botany 61, 2039–2041. [DOI] [PubMed] [Google Scholar]
- Van Genuchten M. 1980. A closed form equation for predicting the hydraulic conductivity of unsatured soils. Soil Science Society of America Journal 4, 892–898. [Google Scholar]
- Wang RX, Hai L, Zhang XY, You GX, Yan CS, Xiao SH. 2009. QTL mapping for grain filling rate and yield-related traits in RILs of the Chinese winter wheat population Heshangmai X Yu8679. Theoretical and Applied Genetics 118, 313–325. [DOI] [PubMed] [Google Scholar]
- Wang S, Basten C, Gaffney P, Zeng Z. 2004. Windows QTL Cartographer 2.0 User Manual. NC, USA: Bioinformatics Research Center. [Google Scholar]
- Wu X, Chang X, Jing R. 2011. Genetic analysis of carbon isotope discrimination and its relation to yield in a wheat doubled haploid population. Journal of Integrative Plant Biology 53, 719–730. [DOI] [PubMed] [Google Scholar]
- Zhang XK, Xiao YG, Zhang Y, Xia XC, Dubcovsky J, He ZH. 2008. Allelic variation at the vernalization genes Vrn-A1, Vrn-B1, Vrn-D1, and Vrn-B3 in Chinese wheat cultivars and their association with growth habit. Crop Science 48, 458–470. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.