Abstract
Atherosclerotic cardiovascular disease is the main cause of mortality and morbidity in the world. Plasma levels of low density lipoprotein cholesterol (LDL-C) are positively correlated with the risk of atherosclerosis. High plasma LDL concentrations in patients with hypercholesterolemia lead to build-up of LDL in the inner walls of the arteries, which becomes oxidized and promotes the formation of foam cells, consequently initiating atherosclerosis. Plasma LDL is mainly cleared through the LDL receptor (LDLR) pathway. Mutations in the LDLR cause familiar hypercholesterolemia and increase the risk of premature coronary heart disease. The expression of LDLR is regulated at the transcriptional level via the sterol regulatory element binding protein 2 (SREBP-2) and at the posttranslational levels mainly through proprotein convertase subtilisin/kexin-type 9 (PCSK9) and inducible degrader of the LDLR (IDOL). In this review, we summarize the latest advances in the studies of PCSK9.
Keywords: hypercholesterolemia, low density lipoprotein receptor, statin, atherosclerosis, proprotein convertase subtilisin/kexin-type 9
Introduction
Atherosclerosis, one of the major underlying causes of cardiovascular disease, is an inflammatory response in the walls of arteries largely due to the progressive accumulation of lipid and fibrous deposits in the vessel wall[1]. When it happens in the arteries that feed the heart, it causes coronary artery disease. If it develops in the arteries that feed the brain, it results in stroke. Numerous epidemiological studies have provided unquestionable evidence that plasma levels of cholesterol, especially low density lipoprotein cholesterol (LDL-C), are positively correlated with the risk of atherosclerosis[2-3]. LDL is produced as a metabolic by-product of very low density lipoprotein (VLDL), a triglyceride-rich lipoprotein produced exclusively by the liver. LDLs are the major cholesterol transport vesicles in the blood. Approximately 65-70% of plasma cholesterol in humans is transported as LDL[2,3]. Cholesterol is carried in the highly hydrophobic core of LDL particles in its esterified form. When plasma LDL-C concentrations are too high, such as in patients with autosomal dominant/recessive hypercholesterolemia, it slowly builds up in the inner walls of the arteries, becomes oxidized and promotes the accumulation of monocyte-derived macrophages. In turn, these take up lipoprotein cholesterol and form foam cells, consequently initiating atherosclerosis[4-5].
The LDLR and cardiovascular disease
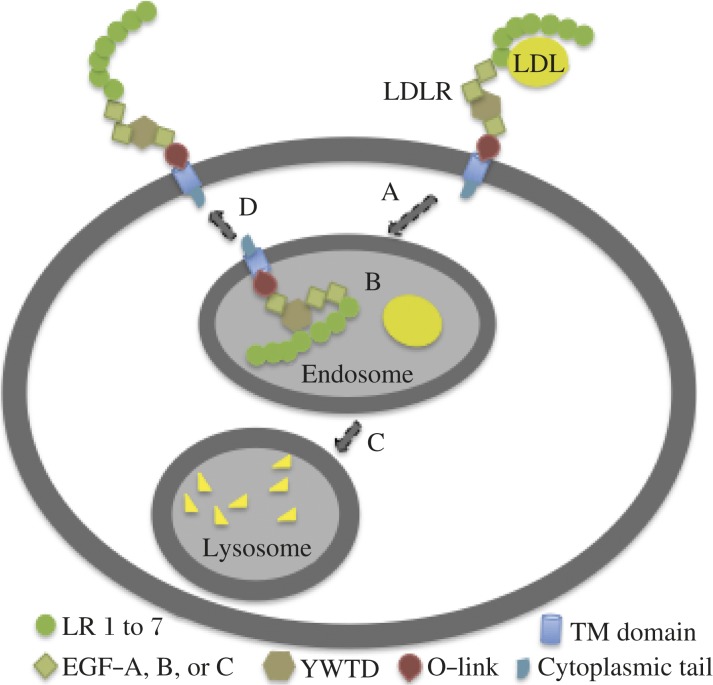
The LDLR in the liver is the protein primarily responsible for removal of LDL from the circulation[2,3]. The LDLR consists of 7 ligand-binding repeats (LR1-7) at the N terminus followed by the so-called epidermal growth factor (EGF) precursor homology domain (Fig. 1). The clustered O-linked sugar region (a threonine- and serine-rich region) is followed by the transmembrane domain and a relatively short cytoplasmic tail that are located downstream of the EGF precursor homology domain. Each LR consists of 7 individual 40-amino acid cysteine-rich tandem repeats. LR3 to LR7 are important for the binding of apolipoprotein B-100 on the surface of LDL, while LR4 and LR5 are important for binding to apolipoprotein E on VLDL surface[6]. The EGF precursor domain contains two cysteine-rich EGF-like domains (EGF-A and EGF-B) followed by a β-propeller domain/YWTD repeats and a third EGF-like domain (EGF-C)[7]. These EGF and YWTD repeats are required for lipoprotein release from the receptor in the endosome and recycling of the LDLR to the cell surface[8,9]. The cytoplasmic tail of LDLR contains all the sequences required for receptor clustering in clathrin-coated pits and for internalization of the receptor[8]. The crystallographic structure of the extracellular domain of the LDLR suggests that the extracellular domain of the cell-surface LDLR (neutral pH) adopts an extended linear conformation (open conformation) that favors interactions between the ligand binding repeats and LDL[10]. Upon ligand binding, the receptors are internalized via clathrin-coated pits and delivered to endosomes[2-3]. In the low pH environment of the endosome, the LDLR undergoes a conformational change so that LR4 and LR5 form a physical interaction with YWTD in the EGF precursor homology domain (closed conformation, Fig. 1B)[10]. The acid-dependent conformational change in the LDLR promotes the release of the bound LDL that is delivered to lysosomes for degradation (Fig. 1C) and signals recycling of the LDLR to the cell surface (Fig. 1D). Usually, each LDLR undergoes multiple rounds of internalization and recycling.
Fig. 1. LDLR-mediated LDL uptake.
A: LDL binds to the LDLR on the cell surface, enters into cells via clathrin-dependent endocytosis, and delivered to endosomes[62]. B: The conformation of the LDLR is changed to a close conformation in the low pH environment of the endosome, which promotes the release of the bound LDL[10]. C: Released LDL is delivered to lysosomes for degradation. D: The LDLR is recycled to the cell surface.
Mutations in the LDLR cause familial hypercholesterolemia (FH), an inherited disorder associated with elevated circulating levels of cholesterol, specifically LDL-C, which causes tendon and skin xanthomas, arcus corneae and/or cardiovascular deposits and leads to increased risk in coronary heart disease and mortality[2-3]. FH is the most common autosomal dominant hypercholesterolemia (ADH) with a frequency of 1 in 500 people for heterozygous FH. Ldlr-/- mice fed a chow diet display total non-fasting plasma cholesterol levels that are about two-fold higher than wild type littermates[11]. When fed a high fat/high cholesterol diet, Ldlr-/- mice show severely increased plasma levels of LDL-C, develop atherosclerosis, and are susceptible to obesity and diabetes[12-14].
LDLR is transcriptionally regulated by the sterol regulatory element binding protein 2 (SREBP-2)[2,3]. Statins, currently the most prescribed lipid-lowering drugs, act by inhibiting 3-hydroxy-3-methylglutaryl-CoA reductase that is the rate-limiting enzyme in de novo cholesterol synthesis, activating the sterol-dependent processing of SREBP-2 pathway and increasing the level of the LDLR, which results in decreased de novo cholesterol synthesis and plasma LDL-C levels[2,3]. The LDLR is also post-translationally regulated by proprotein convertase subtilisin/kexin-type 9 (PCSK9) and inducible degrader of the LDLR (IDOL)[15-17]. PCSK9 binds to LDLR and promotes lysosomal degradation of the receptor[15,16], while IDOL down-regulates LDLR via the polyubiquitination and lysosomal degradation pathway[17]. In this review, we summarize the recent findings about the important roles of PCSK9 in regulating the expression of the LDLR.
PCSK9 and the LDLR
Recently, a third form of ADH was identified that is caused by selected missense mutations in PCSK9[18,19]. Genetic studies have demonstrated that PCSK9 plays a major regulatory role in cholesterol homeostasis. Gain-of-function mutations of PCSK9 cause higher plasma LDL-C levels and lead to accelerated atherosclerosis and premature coronary heart disease[18-20]. On the other hand, loss-of-function mutations result in low concentrations of LDL-C and protection from coronary heart disease[21-23]. For example, African Americans who are heterozygous for one of two nonsense mutations in PCSK9 (Y142X and C679X) show a 30%-40% reduction in plasma LDL-C concentrations and an 88% reduction in coronary heart disease over a 15-year period[21]. PCSK9-/- mice, as well as humans with PCSK9 loss-of-function mutations show increased sensitivity to statins. Thus, inhibition of PCSK9 dramatically reduces plasma levels of LDL-C. Indeed, administration of monoclonal antibodies against PCSK9 to heterozygous FH patients significantly reduces their plasma levels of LDL-C[24-26].
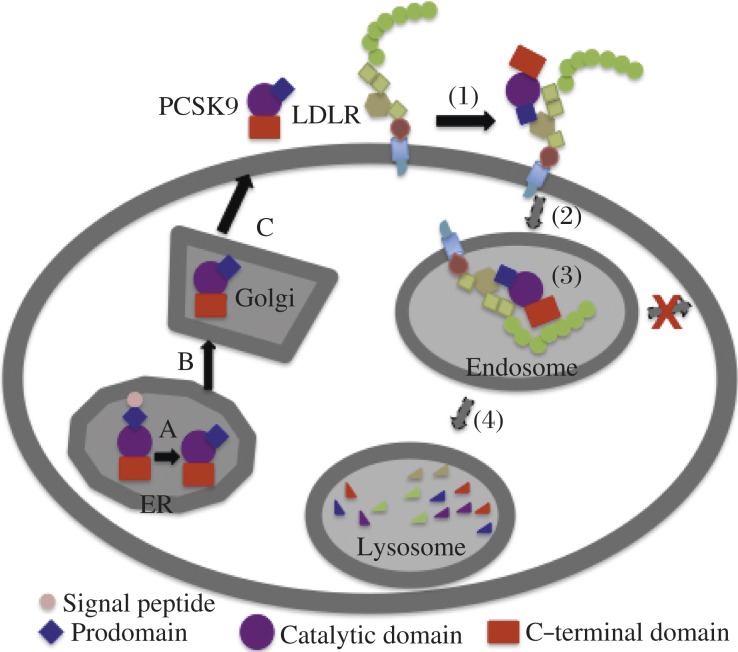
PCSK9 is a member of the subtilisin-like serine protease family that includes 7 basic amino acid-specific proprotein convertases (PC): PC1, PC2, furin, PC4, PC5/6, PACE4, and PC7, as well as two members, site-1 protease and PCSK9, that cleave at the carboxyl terminus of non-basic residues[27]. PCSK9 is a 692-amino acid secreted glycoprotein that consists of a signal sequence followed by a prodomain, a catalytic domain, and a cysteine- and histidine-rich C-terminal domain (Fig. 2). PCSK9 is synthesized as a zymogen and undergoes autocatalytic cleavage in the endoplasmic reticulum (ER) at the FAQ152SIPK site (Fig. 2A). The autocatalytic cleavage is required for protein maturation and secretion[28]. The cleaved N-terminal prodomain is tightly associated with the rest of the catalytic C-terminus and secreted together from cells, physically shielding the catalytic activity as the protein transits through the secretory pathway[27]. Sec24A is required for the transport of PCSK9 from the ER to the Golgi, while sortilin interacts with PCSK9 in the trans-Golgi network and then facilitates its secretion[29]. The expression of PCSK9 is high in the liver, intestine, kidney, and brain[30] and regulated by the SREBP2 pathway[31]. PCSK9 is present in human plasma[15,32-34], and circulating levels of PCSK9 correlate with plasma levels of LDL-C but not HDL-C[33-34]. Overexpression of recombinant PCSK9 in the liver of mice causes a significant reduction in hepatic LDLR protein levels without any effect on its mRNA levels and produces severe hypercholesterolemia[28,35-36]. On the other hand, knockdown or knockout of PCSK9 expression in mice leads to increased levels of LDLR protein in the liver and accelerated LDL clearance[37-38]. The natural gain-of-function mutation, D374Y, has a significantly increased binding affinity for the LDLR and promotes LDLR degradation much more efficiently than the wild type protein[15,39], leading to a severe form of hypercholesterolemia[18]. FH mutation, LDLR-H306Y, binds PCSK9 with a higher affinity and exhibits enhanced sensitivity to PCSK9[40]. Taken together, these findings demonstrate that the role of PCSK9 in homeostatic control of plasma LDL-C levels is dependent upon PCSK9-promoted degradation of the LDLR, thereby preventing clearance of LDL-C by the cells[15-16,28,35-37,41-44].
Fig. 2. PCSK9 secretion and PCSK9-promoted LDLR degradation.
PCSK9 contains a signal sequence followed by a prodomain, a catalytic domain, and a cysteine- and histidine-rich C-terminal domain. PCSK9 secretion: A: The protein is synthesized as a zymogen in the ER and then undergoes autocatalytic cleavage between the prodomain and the catalytic domain. The cleaved prodomain is tightly associated with the catalytic domain. B: PCSK9 is transported to the Golgi, where post-translational modifications, such as glycosylation and sulfation, occur. C: Mature PCSK9 is secreted from the cells. PCSK9-promoted LDLR degradation: (1) The catalytic domain and prodomain of PCSK9 bind to EGF-A and YWTD repeats of the LDLR, respectively. (2) PCSK9-LDLR complex enters into cells via clathrin-dependent endocytosis and is delivered to the endosome. (3) PCSK9 strongly binds to the LDLR at the acidic endosomal environment, which blocks recycling of the LDLR to the cell surface. (4) PCSK9-LDLR complex is transported to the lysosome for degradation.
Studies in cultured cells and parabiotic mice demonstrated that PCSK9 promotes degradation of the LDLR extracellularly in an adaptor protein autosomal recessive hypercholesterolemia (ARH)-dependent manner in hepatocytes and lymphocytes[15-16,41,44]. However, ARH is not required for PCSK9-promoted LDLR degradation in fibroblasts[44-45]. McNutt et al.[40] showed that PCSK9 causes degradation of the LDLR primarily through interaction with the receptor on the cell surface. However, overexpression of PCSK9 in cultured cells and mouse liver induces LDLR degradation intracellularly[36-46]. Thus, PCSK9 may be active within cells. Recently, Poirier et al.[47] reported that, upon dose and incubation period, PCSK9 could act both intracellularly and extracellularly to promote LDLR degradation in cultured cells and mouse primary hepatocytes. In addition, PCSK9's action on the LDLR is cell-type specific. Increased plasma levels of PCSK9 in mouse through infusion of purified PCSK9 or transgenic overexpression in the kidneys preferentially promote LDLR degradation in the liver but not in the adrenal glands[48-49]. Consistently, the adrenal function of a human subject with no detectable plasma PCSK9 is normal[50]. In cultured cells, expression of PCSK9 in some cell types, such as human hepatoma cells (HepG2 and HuH7), dramatically reduces LDLR levels[15-16,36,41,51]. On the other hand, PCSK9 appears to not affect LDLR expression in Chinese hamster ovarian cells (CHO-K1), monkey kidney cells (COS-7) and rat liver cells (McArdle RH7777)[15-16,36,52]. Recently, studies from Mayer et al.[53] showed that the C-terminal cysteine- and histidine-rich domain of PCSK9 directly interacts with annexin A2, which subsequently inhibits PCSK9-promoted LDLR degradation. The high expression of annexin 2 in fibroblasts and COS-7 cells may account for PCSK9-resistance in these cells. On the other hand, the dissociation of PCSK9 for the LDLR after endocytosis may be responsible for the inability of PCSK9 to promote LDLR degradation in human skin fibroblasts SV-589[54].
Secreted PCSK9-promoted degradation of the LDLR requires binding of PCSK9 to the LDLR and internalization of the receptor but does not require the proteolytic activity of PCSK9[15-16,55]. We have shown that PCSK9 interacts with the EGF-A of the LDLR at the cell surface and binds the receptor with a much higher affinity at the acidic environment of the endosome (Fig. 2). Consequently, the receptor traffics from the endosome to the lysosome for degradation, rather than being recycled[16]. Binding of PCSK9 to LDLR interferes with the acid-dependent conformational change of the receptor but disrupting the pH-dependent conformational change in the LDLR is not sufficient to trigger LDLR degradation[41]. In addition, we demonstrated that YWTD repeats and a minimum of three ligand-binding repeats in the LDLR that are not required for PCSK9 binding at neutral pH are essential for efficient LDLR degradation induced by PCSK9[41-42]. We also reported that the C-terminal domain of PCSK9 is essential for PCSK9-promoted degradation of the LDLR but is not required for binding to the LDLR at the neutral pH values[41]. The X-ray crystallographic structure of PCSK9-LDLR complex shows that YWTD repeats of the LDLR interact with the prodomain of PCSK9[56]. Several biochemical studies indicate that the negatively charged ligand binding repeats of the LDLR may interact with the positively charged C-terminal domain of PCSK9 in the acidic endosomal environment, to enhance PCSK9 binding[57-59] (Fig. 2, step 3). Neither PCSK9 or the LDLR contains a lysosomal targeting signal. Presently, the mechanism by which binding of PCSK9 to the LDLR reroutes the receptor to the lysosomes for degradation is not understood and is believed to be complex.
Conclusion
Phase III clinical trials of the monoclonal antibodies against PCSK9 show impressive LDL-C-lowering effect of PCSK9 inhibition in heterozygous and homozygous FH patients, high-risk patients intolerant to statins, and patients with poor LDL-C-lowering response even with maximally tolerated statin dosages[24-26]. Furthermore, when adding to the statin therapy, PCSK9 inhibitors can markedly reduce cardiovascular events, such as myocardial infarction and ischemic stroke, with no significant adverse side effects[24,26]. However, low plasma cholesterol levels have been linked to increased cancer incidence or mortality and higher hemorrhagic stroke risk[60]. Sattar et al. reported that statin treatments are associated with a 9% increased risk for incident diabetes[61]. With approval of PCSK9 inhibitors imminent, the potential for harm of PCSK9 inhibition needs to be evaluated. In addition, all PCSK9 inhibitors currently under phase III clinical trials are monoclonal anti-PCSK9 antibodies. Given that the treatment of patients with hypercholesterolemia is lifelong and the antibody therapy requires injections of large amounts of antibodies to achieve clinical efficacy with extremely high production costs, the wide use of these drugs may be limited. Thus, the need for more effective and more cost-efficient therapies to lower LDL-C is urgent. However, the molecular mechanisms of PCSK9-promoted LDLR degradation are not completely understood, which impedes the future design of PCSK9 specific small molecule inhibitors. Crystallographic studies of PCSK9-EGF-AB complex show that the interaction face between the catalytic domain of PCSK9 and the EGF-A of the LDLR is relatively flat and big, making it impossible to design a specific inhibitor to block the interaction between PCSK9 and the EGF-A of the LDLR. Therefore, mechanistic studies of PCSK9-promoted LDLR degradation, such as identification of new interaction regions between PCSK9 and the LDLR and potential cofactors important for PCSK9-promoted LDLR degradation, are necessary.
Acknowledgments
D.W.Z. is a Scholar of the Alberta Heritage Foundation for Medical Research and is supported in part by a Canadian Institutes of Health Research New Investigator Award. Zhang laboratory is supported by Canadian Foundation for Innovation, grants from a Grant-in-Aid for Heart and Stroke Foundation of Canada, Pfizer Canada, the Canadian Institutes of Health Research (MOP 93794), and Mazankowski Alberta Heart Institute.
References
- [1].Libby P. Inflammation in atherosclerosis[J] Nature. 2002;420(6917):868–874. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- [2].Brown MS, Goldstein JL. Biomedicine. Lowering LDL–not only how low, but how long?[J] Science. 2006;311(5768):1721–1723. doi: 10.1126/science.1125884. [DOI] [PubMed] [Google Scholar]
- [3].Goldstein JL, Brown MS. The LDL receptor[J] Arterioscler Thromb Vasc Biol. 2009;29(4):431–438. doi: 10.1161/ATVBAHA.108.179564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Tiwari RL, Singh V, Barthwal MK. Macrophages: an elusive yet emerging therapeutic target of atherosclerosis[J] Med Res Rev. 2008;28(4):483–544. doi: 10.1002/med.20118. [DOI] [PubMed] [Google Scholar]
- [5].Platt N, Gordon S. Is the class A macrophage scavenger receptor (SR-A) multifunctional? - The mouse's tale[J] J Clin Invest. 2001;108(5):649–654. doi: 10.1172/JCI13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Russell DW, Brown MS, Goldstein JL. Different combinations of cysteine-rich repeats mediate binding of low density lipoprotein receptor to two different proteins[J] J Biol Chem. 1989;264(36):21682–21688. [PubMed] [Google Scholar]
- [7].Jeon H, Meng W, Takagi J, et al. Implications for familial hypercholesterolemia from the structure of the LDL receptor YWTD-EGF domain pair[J] Nat Struct Biol. 2001;8(6):499–504. doi: 10.1038/88556. [DOI] [PubMed] [Google Scholar]
- [8].Davis CG, Goldstein JL, Sudhof TC, et al. Acid-dependent ligand dissociation and recycling of LDL receptor mediated by growth factor homology region[J] Nature. 1987;326(6115):760–765. doi: 10.1038/326760a0. [DOI] [PubMed] [Google Scholar]
- [9].Beglova N, Jeon H, Fisher C, et al. Cooperation between fixed and low pH-inducible interfaces controls lipoprotein release by the LDL receptor[J] Mol Cell. 2004;16(2):281–292. doi: 10.1016/j.molcel.2004.09.038. [DOI] [PubMed] [Google Scholar]
- [10].Rudenko G, Henry L, Henderson K, et al. Structure of the LDL receptor extracellular domain at endosomal pH[J] Science. 2002;298(5602):2353–2358. doi: 10.1126/science.1078124. [DOI] [PubMed] [Google Scholar]
- [11].Ishibashi S, Brown MS, Goldstein JL, et al. Hypercholesterolemia in low density lipoprotein receptor knockout mice and its reversal by adenovirus-mediated gene delivery[J] J Clin Invest. 1993;92(2):883–893. doi: 10.1172/JCI116663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Schreyer SA, Vick C, Lystig TC, et al. LDL receptor but not apolipoprotein E deficiency increases diet-induced obesity and diabetes in mice[J] Am J Physiol Endocrinol Metab. 2002;282(1):E207–E214. doi: 10.1152/ajpendo.2002.282.1.E207. [DOI] [PubMed] [Google Scholar]
- [13].Merat S, Casanada F, Sutphin M, et al. Western-type diets induce insulin resistance and hyperinsulinemia in LDL receptor-deficient mice but do not increase aortic atherosclerosis compared with normoinsulinemic mice in which similar plasma cholesterol levels are achieved by a fructose-rich diet[J] Arterioscler Thromb Vasc Biol. 1999;19(5):1223–1230. doi: 10.1161/01.atv.19.5.1223. [DOI] [PubMed] [Google Scholar]
- [14].Bonfleur ML, Vanzela EC, Ribeiro RA, et al. Primary hypercholesterolaemia impairs glucose homeostasis and insulin secretion in low-density lipoprotein receptor knockout mice independently of high-fat diet and obesity[J] Biochim Biophys Acta. 2010;1801(2):183–190. doi: 10.1016/j.bbalip.2009.10.012. [DOI] [PubMed] [Google Scholar]
- [15].Lagace TA, Curtis DE, Garuti R, et al. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice[J] J Clin Invest. 2006;116(11):2995–3005. doi: 10.1172/JCI29383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhang DW, Lagace TA, Garuti R, et al. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation[J] J Biol Chem. 2007;282(25):18602–18612. doi: 10.1074/jbc.M702027200. [DOI] [PubMed] [Google Scholar]
- [17].Zelcer N, Hong C, Boyadjian R, et al. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor[J] Science. 2009;325(5936):100–104. doi: 10.1126/science.1168974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Abifadel M, Varret M, Rabes JP, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia[J] Nat Genet. 2003;34(2):154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- [19].Leren TP. Mutations in the PCSK9 gene in Norwegian subjects with autosomal dominant hypercholesterolemia[J] Clin Genet. 2004;65(5):419–422. doi: 10.1111/j.0009-9163.2004.0238.x. [DOI] [PubMed] [Google Scholar]
- [20].Abifadel M, Rabes JP, Devillers M, et al. Mutations and polymorphisms in the proprotein convertase subtilisin kexin 9 (PCSK9) gene in cholesterol metabolism and disease[J] Hum Mutat. 2009;30(4):520–529. doi: 10.1002/humu.20882. [DOI] [PubMed] [Google Scholar]
- [21].Cohen JC, Boerwinkle E, Mosley TH, et al. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease[J] N Engl J Med. 2006;354(12):1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- [22].Seidah NG, Khatib AM, Prat A. The proprotein convertases and their implication in sterol and/or lipid metabolism[J] Biol Chem. 2006;387(7):871–877. doi: 10.1515/BC.2006.110. [DOI] [PubMed] [Google Scholar]
- [23].Abifadel M, Rabes JP, Jambart S, et al. The molecular basis of familial hypercholesterolemia in Lebanon: spectrum of LDLR mutations and role of PCSK9 as a modifier gene[J] Hum Mutat. 2009;30(7):E682–E691. doi: 10.1002/humu.21002. [DOI] [PubMed] [Google Scholar]
- [24].Sabatine MS, Giugliano RP, Wiviott SD, et al. Efficacy and safety of evolocumab in reducing lipids and cardiovascular events[J] N Engl J Med. 2015;372(16):1500–1509. doi: 10.1056/NEJMoa1500858. [DOI] [PubMed] [Google Scholar]
- [25].Cannon CP, Cariou B, Blom D, et al. Efficacy and safety of alirocumab in high cardiovascular risk patients with inadequately controlled hypercholesterolaemia on maximally tolerated doses of statins: the ODYSSEY COMBO II randomized controlled trial[J] Eur Heart J. 2015;36(19):1186–1194. doi: 10.1093/eurheartj/ehv028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Robinson JG, Farnier M, Krempf M, et al. Efficacy and safety of alirocumab in reducing lipids and cardiovascular events[J] N Engl J Med. 2015;372(16):1489–1499. doi: 10.1056/NEJMoa1501031. [DOI] [PubMed] [Google Scholar]
- [27].Seidah NG, Mayer G, Zaid A, et al. The activation and physiological functions of the proprotein convertases[J] Int J Biochem Cell Biol. 2008;40(6–7):1111–1125. doi: 10.1016/j.biocel.2008.01.030. [DOI] [PubMed] [Google Scholar]
- [28].Benjannet S, Rhainds D, Essalmani R, et al. NARC–1/PCSK9 and its natural mutants: zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol[J] J Biol Chem. 2004;279(47):48865–48875. doi: 10.1074/jbc.M409699200. [DOI] [PubMed] [Google Scholar]
- [29].Gustafsen C, Kjolby M, Nyegaard M, et al. The Hypercholesterolemia-Risk Gene SORT1 Facilitates PCSK9 Secretion[J] Cell Metab. 2014;19(2):310–318. doi: 10.1016/j.cmet.2013.12.006. [DOI] [PubMed] [Google Scholar]
- [30].Seidah NG, Benjannet S, Wickham L, et al. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation[J] Proc Natl Acad Sci U S A. 2003;100(3):928–933. doi: 10.1073/pnas.0335507100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Horton JD, Shah NA, Warrington JA, et al. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes[J] Proc Natl Acad Sci U S A. 2003;100(21):12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mayne J, Dewpura T, Raymond A, et al. Plasma PCSK9 levels are significantly modified by statins and fibrates in humans[J] Lipids Health Dis. 2008;7(22) doi: 10.1186/1476-511X-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lambert G, Ancellin N, Charlton F, et al. Plasma PCSK9 concentrations correlate with LDL and total cholesterol in diabetic patients and are decreased by fenofibrate treatment[J] Clin Chem. 2008;54(6):1038–1045. doi: 10.1373/clinchem.2007.099747. [DOI] [PubMed] [Google Scholar]
- [34].Alborn WE, Cao G, Careskey HE, et al. Serum proprotein convertase subtilisin kexin type 9 is correlated directly with serum LDL cholesterol[J] Clin Chem. 2007;53(10):1814–1819. doi: 10.1373/clinchem.2007.091280. [DOI] [PubMed] [Google Scholar]
- [35].Maxwell KN, Breslow JL. Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype[J] Proc Natl Acad Sci U S A. 2004;101(18):7100–7105. doi: 10.1073/pnas.0402133101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Park SW, Moon YA, Horton JD. Post-transcriptional regulation of low density lipoprotein receptor protein by proprotein convertase subtilisin/kexin type 9a in mouse liver[J] J Biol Chem. 2004;279(48):50630–50638. doi: 10.1074/jbc.M410077200. [DOI] [PubMed] [Google Scholar]
- [37].Rashid S, Curtis DE, Garuti R, et al. Decreased plasma cholesterol and hypersensitivity to statins in mice lacking Pcsk9[J] Proc Natl Acad Sci U S A. 2005;102(15):5374–5379. doi: 10.1073/pnas.0501652102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Frank-Kamenetsky M, Grefhorst A, Anderson NN, et al. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates[J] Proc Natl Acad Sci U S A. 2008;105(33):11915–11920. doi: 10.1073/pnas.0805434105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Cunningham D, Danley DE, Geoghegan KF, et al. Structural and biophysical studies of PCSK9 and its mutants linked to familial hypercholesterolemia[J] Nat Struct Mol Biol. 2007;14(5):413–419. doi: 10.1038/nsmb1235. [DOI] [PubMed] [Google Scholar]
- [40].McNutt MC, Kwon HJ, Chen C, et al. Antagonism of secreted PCSK9 increases low-density lipoprotein receptor expression in HEPG2 cells[J] J Biol Chem. 2009;284(16):10561–10570. doi: 10.1074/jbc.M808802200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhang DW, Garuti R, Tang WJ, et al. Structural requirements for PCSK9-mediated degradation of the low-density lipoprotein receptor[J] Proc Natl Acad Sci U S A. 2008;105(35):13045–13050. doi: 10.1073/pnas.0806312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gu HM, Adijiang A, Mah M, et al. Characterization of the role of EGF-A of low-density lipoprotein receptor in PCSK9 binding[J] J Lipid Res. 2013;54(12):3345–3357. doi: 10.1194/jlr.M041129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lalanne F, Lambert G, Amar MJ, et al. Wild-type PCSK9 inhibits LDL clearance but does not affect apoB-containing lipoprotein production in mouse and cultured cells[J] J Lipid Res. 2005;46(6):1312–1319. doi: 10.1194/jlr.M400396-JLR200. [DOI] [PubMed] [Google Scholar]
- [44].Wang Y, Huang Y, Hobbs HH, et al. Molecular characterization of proprotein convertase subtilisin/kexin type 9-mediated degradation of the LDLR[J] J Lipid Res. 2012;53(9):1932–1943. doi: 10.1194/jlr.M028563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Fasano T, Sun XM, Patel DD, et al. Degradation of LDLR protein mediated by 'gain of function' PCSK9 mutants in normal and ARH cells[J] Atherosclerosis. 2009;203(1):166–171. doi: 10.1016/j.atherosclerosis.2008.10.027. [DOI] [PubMed] [Google Scholar]
- [46].Maxwell KN, Fisher EA, Breslow JL. Overexpression of PCSK9 accelerates the degradation of the LDLR in a post-endoplasmic reticulum compartment[J] Proc Natl Acad Sci U S A. 2005;102(6):2069–2074. doi: 10.1073/pnas.0409736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Poirier S, Mayer G, Poupon V, et al. Dissection of the endogenous cellular pathways of PCSK9-induced LDLR degradation: Evidence for an intracellular route[J] J Biol Chem. 2009;284(42):28856–28864. doi: 10.1074/jbc.M109.037085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Grefhorst A, McNutt MC, Lagace TA, et al. Plasma PCSK9 preferentially reduces liver LDL receptors in mice[J] J Lipid Res. 2008;49(6):1303–1311. doi: 10.1194/jlr.M800027-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Luo Y, Warren L, Xia D, et al. Function and distribution of circulating human PCSK9 expressed extrahepatically in transgenic mice[J] J Lipid Res. 2009;50(8):1581–1588. doi: 10.1194/jlr.M800542-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Cariou B, Benoit I, Le May C. Preserved adrenal function in fully PCSK9-deficient subject[J] Int J Cardiol. 2014;176(2):499–500. doi: 10.1016/j.ijcard.2014.07.057. [DOI] [PubMed] [Google Scholar]
- [51].Benjannet S, Rhainds D, Hamelin J, et al. The proprotein convertase (PC) PCSK9 is inactivated by furin and/or PC5/6A: functional consequences of natural mutations and post-translational modifications[J] J Biol Chem. 2006;281(41):30561–30572. doi: 10.1074/jbc.M606495200. [DOI] [PubMed] [Google Scholar]
- [52].Maxwell KN, Breslow JL. Proprotein convertase subtilisin kexin 9: the third locus implicated in autosomal dominant hypercholesterolemia[J] Curr Opin Lipidol. 2005;16(2):167–172. doi: 10.1097/01.mol.0000162321.31925.a3. [DOI] [PubMed] [Google Scholar]
- [53].Mayer G, Poirier S, Seidah NG. Annexin A2 is a C-terminal PCSK9 binding protein that regulates endogenous LDL receptor levels[J] J Biol Chem. 2008;283(46):31791–31780. doi: 10.1074/jbc.M805971200. [DOI] [PubMed] [Google Scholar]
- [54].Nguyen MA, Kosenko T, Lagace TA. Internalized PCSK9 dissociates from recycling LDL receptors in PCSK9-resistant SV-589 fibroblasts[J] J Lipid Res. 2014;55(2):266–275. doi: 10.1194/jlr.M044156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].McNutt MC, Lagace TA, Horton JD. Catalytic activity is not required for secreted PCSK9 to reduce low density lipoprotein receptors in HepG2 cells[J] J Biol Chem. 2007;282(29):20799–20803. doi: 10.1074/jbc.C700095200. [DOI] [PubMed] [Google Scholar]
- [56].Surdo PL, Bottomley MJ, Calzetta A, et al. Mechanistic implications for LDL receptor degradation from the PCSK9/LDLR structure at neutral pH[J] EMBO Rep. 2011;12(12):1300–1305. doi: 10.1038/embor.2011.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Tveten K, Holla OL, Cameron J, et al. Interaction between the ligand-binding domain of the LDL receptor and the C-terminal domain of PCSK9 is required for PCSK9 to remain bound to the LDL receptor during endosomal acidification[J] Hum Mol Genet. 2012;21(6):1402–1409. doi: 10.1093/hmg/ddr578. [DOI] [PubMed] [Google Scholar]
- [58].Holla OL, Cameron J, Tveten K, et al. Role of the C-terminal domain of PCSK9 in degradation of the LDL receptors[J] J Lipid Res. 2011;52(10):1787–1794. doi: 10.1194/jlr.M018093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Yamamoto T, Lu C, Ryan RO. A two-step binding model of PCSK9 interaction with the low density lipoprotein receptor[J] J Biol Chem. 2011;286(7):5464–5470. doi: 10.1074/jbc.M110.199042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Stein EA, Raal FJ. Targeting LDL: is lower better and is it safe?[J] Best Pract Res Clin Endocrinol Metab. 2014;28(3):309–324. doi: 10.1016/j.beem.2013.10.010. [DOI] [PubMed] [Google Scholar]
- [61].Sattar N, Preiss D, Murray HM, et al. Statins and risk of incident diabetes: a collaborative meta-analysis of randomised statin trials[J] Lancet. 2010;375(9716):735–742. doi: 10.1016/S0140-6736(09)61965-6. [DOI] [PubMed] [Google Scholar]
- [62].Goldstein JL, Hobbs HH, Brown MS. Scriver CR. The Metabolic & Molecular Bases of Inherited Disease. eighth edition. New York: McGraw-Hill; 2001. Familial Hypercholesterol[J] pp. 2863–2913. [Google Scholar]