Abstract
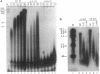
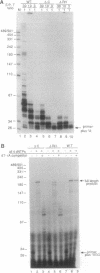
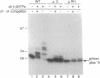
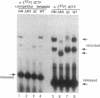
The active sites for the polymerase and nuclease activities of Moloney murine leukemia virus (M-MuLV) reverse transcriptase (RT) reside in separate domains of a single polypeptide. We have studied the effects of RNase H domain mutations on DNA polymerase activity. These mutant RTs displayed decreased processivity of DNA synthesis. We also compared complexes formed between primer-templates and mutant and wild-type reverse transcriptase (RT). Although M-MuLV RT is monomeric in solution, two molecules of RT bound DNA cooperatively, suggesting that M-MuLV RT binds primer-template as a dimer. Some mutant RTs with decreased processivity failed to form the putative dimer.
Full text
PDF
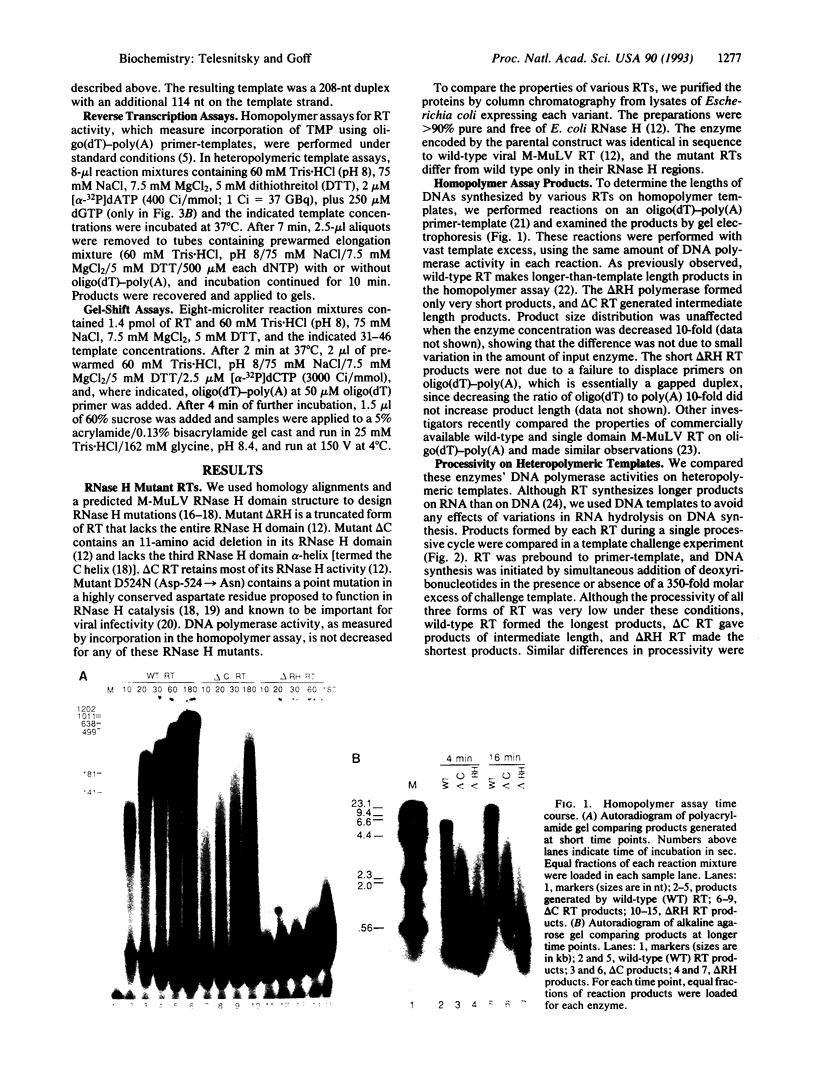

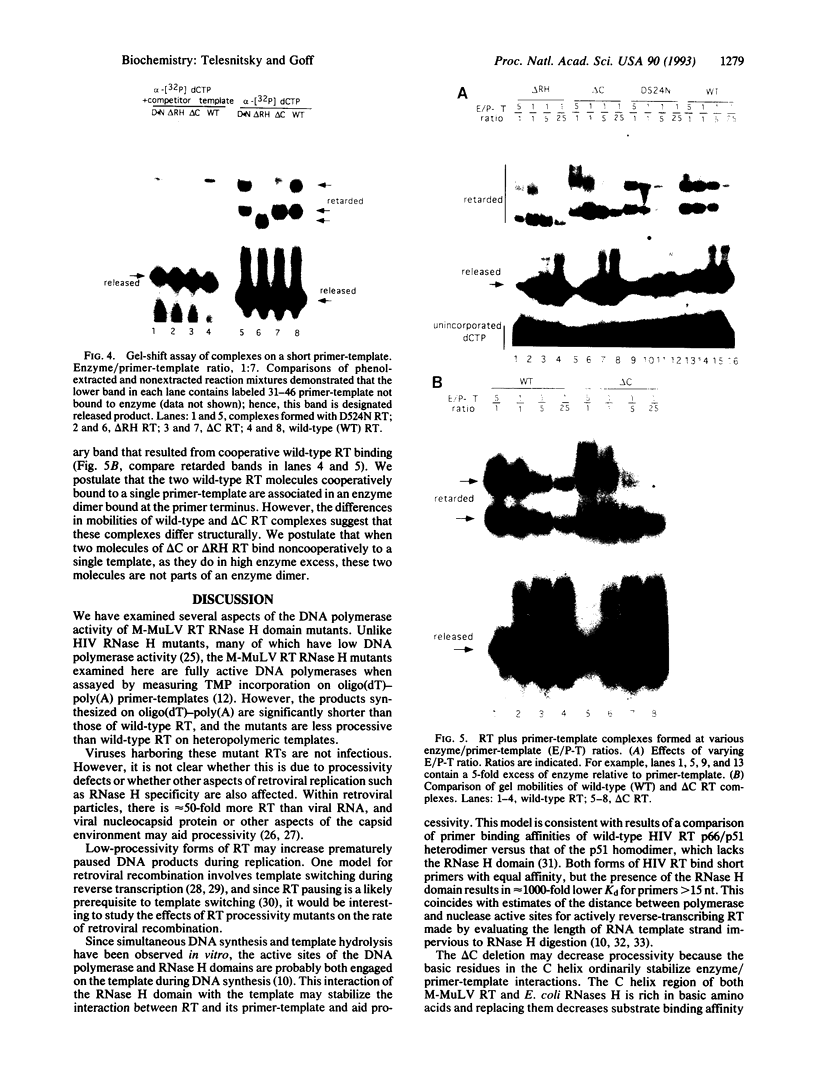

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Buiser R. G., DeStefano J. J., Mallaber L. M., Fay P. J., Bambara R. A. Requirements for the catalysis of strand transfer synthesis by retroviral DNA polymerases. J Biol Chem. 1991 Jul 15;266(20):13103–13109. [PubMed] [Google Scholar]
- Chao K. L., Lohman T. M. DNA-induced dimerization of the Escherichia coli Rep helicase. J Mol Biol. 1991 Oct 20;221(4):1165–1181. doi: 10.1016/0022-2836(91)90926-w. [DOI] [PubMed] [Google Scholar]
- Cheng N., Painter G. R., Furman P. A. Crosslinking of substrates occurs exclusively to the p66 subunit of heterodimeric HIV-1 reverse transcriptase. Biochem Biophys Res Commun. 1991 Jan 31;174(2):785–789. doi: 10.1016/0006-291x(91)91486-v. [DOI] [PubMed] [Google Scholar]
- Colicelli J., Goff S. P. Sequence and spacing requirements of a retrovirus integration site. J Mol Biol. 1988 Jan 5;199(1):47–59. doi: 10.1016/0022-2836(88)90378-6. [DOI] [PubMed] [Google Scholar]
- DeStefano J. J., Buiser R. G., Mallaber L. M., Myers T. W., Bambara R. A., Fay P. J. Polymerization and RNase H activities of the reverse transcriptases from avian myeloblastosis, human immunodeficiency, and Moloney murine leukemia viruses are functionally uncoupled. J Biol Chem. 1991 Apr 25;266(12):7423–7431. [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Johnson M. S., McClure M. A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989 Mar;64(1):1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- Dudding L. R., Nkabinde N. C., Mizrahi V. Analysis of the RNA- and DNA-dependent DNA polymerase activities of point mutants of HIV-1 reverse transcriptase lacking ribonuclease H activity. Biochemistry. 1991 Oct 29;30(43):10498–10506. doi: 10.1021/bi00107a019. [DOI] [PubMed] [Google Scholar]
- Furfine E. S., Reardon J. E. Reverse transcriptase.RNase H from the human immunodeficiency virus. Relationship of the DNA polymerase and RNA hydrolysis activities. J Biol Chem. 1991 Jan 5;266(1):406–412. [PubMed] [Google Scholar]
- Gilboa E., Mitra S. W., Goff S., Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979 Sep;18(1):93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- Goff S. P. Retroviral reverse transcriptase: synthesis, structure, and function. J Acquir Immune Defic Syndr. 1990;3(8):817–831. [PubMed] [Google Scholar]
- Goff S., Traktman P., Baltimore D. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J Virol. 1981 Apr;38(1):239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hizi A., Hughes S. H., Shaharabany M. Mutational analysis of the ribonuclease H activity of human immunodeficiency virus 1 reverse transcriptase. Virology. 1990 Apr;175(2):575–580. doi: 10.1016/0042-6822(90)90444-v. [DOI] [PubMed] [Google Scholar]
- Hostomsky Z., Hostomska Z., Fu T. B., Taylor J. Reverse transcriptase of human immunodeficiency virus type 1: functionality of subunits of the heterodimer in DNA synthesis. J Virol. 1992 May;66(5):3179–3182. doi: 10.1128/jvi.66.5.3179-3182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W. S., Temin H. M. Retroviral recombination and reverse transcription. Science. 1990 Nov 30;250(4985):1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- Huber H. E., McCoy J. M., Seehra J. S., Richardson C. C. Human immunodeficiency virus 1 reverse transcriptase. Template binding, processivity, strand displacement synthesis, and template switching. J Biol Chem. 1989 Mar 15;264(8):4669–4678. [PubMed] [Google Scholar]
- Johnson M. S., McClure M. A., Feng D. F., Gray J., Doolittle R. F. Computer analysis of retroviral pol genes: assignment of enzymatic functions to specific sequences and homologies with nonviral enzymes. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya S., Katsuda-Nakai C., Ikehara M. Importance of the positive charge cluster in Escherichia coli ribonuclease HI for the effective binding of the substrate. J Biol Chem. 1991 Jun 25;266(18):11621–11627. [PubMed] [Google Scholar]
- Katz R. A., Skalka A. M. Generation of diversity in retroviruses. Annu Rev Genet. 1990;24:409–445. doi: 10.1146/annurev.ge.24.120190.002205. [DOI] [PubMed] [Google Scholar]
- Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992 Jun 26;256(5065):1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- Kotewicz M. L., Sampson C. M., D'Alessio J. M., Gerard G. F. Isolation of cloned Moloney murine leukemia virus reverse transcriptase lacking ribonuclease H activity. Nucleic Acids Res. 1988 Jan 11;16(1):265–277. doi: 10.1093/nar/16.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Grice S. F., Naas T., Wohlgensinger B., Schatz O. Subunit-selective mutagenesis indicates minimal polymerase activity in heterodimer-associated p51 HIV-1 reverse transcriptase. EMBO J. 1991 Dec;10(12):3905–3911. doi: 10.1002/j.1460-2075.1991.tb04960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin J. G., Crouch R. J., Post K., Hu S. C., McKelvin D., Zweig M., Court D. L., Gerwin B. I. Functional organization of the murine leukemia virus reverse transcriptase: characterization of a bacterially expressed AKR DNA polymerase deficient in RNase H activity. J Virol. 1988 Nov;62(11):4376–4380. doi: 10.1128/jvi.62.11.4376-4380.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G. X., Taylor J. Template switching by reverse transcriptase during DNA synthesis. J Virol. 1990 Sep;64(9):4321–4328. doi: 10.1128/jvi.64.9.4321-4328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrahi V., Usdin M. T., Harington A., Dudding L. R. Site-directed mutagenesis of the conserved Asp-443 and Asp-498 carboxy-terminal residues of HIV-1 reverse transcriptase. Nucleic Acids Res. 1990 Sep 25;18(18):5359–5363. doi: 10.1093/nar/18.18.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama F., Kikuchi R., Crouch R. J., Uchida T. Intrinsic properties of reverse transcriptase in reverse transcription. Associated RNase H is essentially regarded as an endonuclease. J Biol Chem. 1989 Nov 5;264(31):18808–18817. [PubMed] [Google Scholar]
- Prasad V. R., Goff S. P. Linker insertion mutagenesis of the human immunodeficiency virus reverse transcriptase expressed in bacteria: definition of the minimal polymerase domain. Proc Natl Acad Sci U S A. 1989 May;86(9):3104–3108. doi: 10.1073/pnas.86.9.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon J. E., Furfine E. S., Cheng N. Human immunodeficiency virus reverse transcriptase. Effect of primer length on template-primer binding. J Biol Chem. 1991 Jul 25;266(21):14128–14134. [PubMed] [Google Scholar]
- Repaske R., Hartley J. W., Kavlick M. F., O'Neill R. R., Austin J. B. Inhibition of RNase H activity and viral replication by single mutations in the 3' region of Moloney murine leukemia virus reverse transcriptase. J Virol. 1989 Mar;63(3):1460–1464. doi: 10.1128/jvi.63.3.1460-1464.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth M. J., Tanese N., Goff S. P. Purification and characterization of murine retroviral reverse transcriptase expressed in Escherichia coli. J Biol Chem. 1985 Aug 5;260(16):9326–9335. [PubMed] [Google Scholar]
- Tanese N., Goff S. P. Domain structure of the Moloney murine leukemia virus reverse transcriptase: mutational analysis and separate expression of the DNA polymerase and RNase H activities. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1777–1781. doi: 10.1073/pnas.85.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesnitsky A., Blain S. W., Goff S. P. Defects in Moloney murine leukemia virus replication caused by a reverse transcriptase mutation modeled on the structure of Escherichia coli RNase H. J Virol. 1992 Feb;66(2):615–622. doi: 10.1128/jvi.66.2.615-622.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma I. M. The reverse transcriptase. Biochim Biophys Acta. 1977 Mar 21;473(1):1–38. doi: 10.1016/0304-419x(77)90005-1. [DOI] [PubMed] [Google Scholar]
- Yang W., Hendrickson W. A., Crouch R. J., Satow Y. Structure of ribonuclease H phased at 2 A resolution by MAD analysis of the selenomethionyl protein. Science. 1990 Sep 21;249(4975):1398–1405. doi: 10.1126/science.2169648. [DOI] [PubMed] [Google Scholar]