Abstract
Cetaceans are a group of secondarily adapted marine mammals with an enigmatic history of transition from terrestrial to fully aquatic habitat and subsequent adaptive radiation in waters around the world. Numerous physiological and morphological cetacean characteristics have been acquired in response to this drastic habitat transition; for example, the thickened blubber is one of the most striking changes that increases their buoyancy, supports locomotion, and provides thermal insulation. However, the genetic basis underlying the blubber thickening in cetaceans remains poorly explored. Here, 88 candidate genes associated with triacylglycerol metabolism were investigated in representative cetaceans and other mammals to test whether the thickened blubber matched adaptive evolution of triacylglycerol metabolism-related genes. Positive selection was detected in 41 of the 88 candidate genes, and functional characterization of these genes indicated that these are involved mainly in triacylglycerol synthesis and lipolysis processes. In addition, some essential regulatory genes underwent significant positive selection in cetacean-specific lineages, whereas no selection signal was detected in the counterpart terrestrial mammals. The extensive occurrence of positive selection in triacylglycerol metabolism-related genes is suggestive of their essential role in secondary adaptation to an aquatic life, and further implying that ‘obesity’ might be an indicator of good health for cetaceans.
Obesity is currently considered as a major health challenge, and hundreds of thousands of individuals are dying from obesity-related chronic diseases1,2. Obesity is primarily attributed to the excess storage of triacylglycerol (TAG) once TAG synthesis (esterification) exceeds its breakdown (lipolysis)3. TAG in adipose tissue serves as the major energy storage form in mammals4, and a proper capacity for TAG storage in adipocytes is important for normal metabolic regulation. Therefore, the assessment of diversifying mechanisms and selection pressures influencing the evolution of genes closely related to TAG metabolism can potentially provide valuable information for the treatment of obesity and other related diseases.
TAG is an ester derived from glycerol and three fatty acids (FAs)5, and several genes and signaling pathways are involved in its synthesis, lipolysis, and metabolic regulation. FA arises from adipocyte using two major routes: de novo lipogenesis from non-lipid precursors or uptake of FA from the plasma6. FAs are transported through the plasma membrane by several FA transporters: fatty acid translocase (FAT/CD36), fatty acid transport protein (FATP) and fatty acid binding protein, and plasma membrane (FABPpm/GOT2)6. Glycerol and FAs are then transformed into glycerol-3P and fatty acyl-CoA, which are used for TAG synthesis. The synthesis of phosphatidic acid and its subsequent conversion to TAG are catalyzed by several enzymes, e.g., glycerol-3-phosphate acyltransferase (GPAT) and diacylglycerolacyl transferase (DGAT)4. During periods of energy demand, TAG is rapidly mobilized by the hydrolytic action of lipases [desnutrin/adipose triglyceride lipase (ATGL); hormone-sensitive lipase (HSL); and monoglyceride lipase (MGLL)] that releases free fatty acids, which in turn are utilized by other organs to meet the body’s energy requirements7,8. A series of genes [e.g., Liver X-activated receptor (LXR), perilipin (PLIN), and phosphodiesterase 3B (PDE3B)] have been demonstrated to play an important role in regulating lipogenic and lipolytic processes4,9. In addition, some genes [e.g., cell death-inducing DNA fragmentation factor-α-like effector A (CIDEA) and apolipoprotein B (APOB)] have also been associated with TAG metabolism10,11. Therefore, these genes are widely recognized as candidates for controlling TAG storage conditions by changing the expression pattern and protein sequence, and a number of genes closely related to lipid metabolism have been identified to be under adaptive evolution at the genome level12,13.
Cetaceans (whales, dolphins, and porpoises) are a highly specialized group of mammals that evolutionarily transformed from a fully terrestrial quadruped to an obligate aquatic from approximately 53–56 million years ago (Ma)14. During this evolutionary transition, energy reserves and the maintenance of body temperature were the most critical challenges that these species encountered15,16. For example, the thickness of the blubber (the specialized hypodermis) in whales is approximately 20 cm, which is 10-fold greater than that of other artiodactyls species17. The blubber of cetaceans, which comprises TAG as its most important component, is dynamic and multifunctional, and acts as a metabolic energy storage site18, contributes to positive buoyancy19,20, provides thermal insulation21, supports locomotion, and increases swimming efficiency by streamlining the body surface22,23. However, the evolutionary mechanisms and driving force for the formation and maintenance of the thickened blubber in cetaceans have not been well explored to date.
In the present study, the coding regions of 88 genes that represent nearly all of the signal pathway members involved in TAG metabolism in the various cetacean lineages, were investigated and compared to orthologous sequences from terrestrial mammals using both gene- and protein-level approaches. The goal of the present study was to test whether evolutionary changes in these TAG metabolism-related genes were associated with their transition from land to water, and to determine the molecular mechanism underlying the cetacean blubber thickening during this adaptive process.
Results
Of the 88 TAG metabolism-related genes examined in the present study, 41 key members of these genes were amplified in 5 cetacean species (Supplementary Table S1). Unfortunately, 10 genes (i.e., ACSL1, AGPAT5, AGPAT6, FABP1, FABP2, FABP6, PPARA, PPARD, PPARG, and SREBF2) were successfully amplified only in the finless porpoise (Neophocaena phocaenoides) and Omura’s baleen whale (Balaenoptera omurai), and ACSL4 was successfully amplified only in the finless porpoise (Neophocaena phocaenoides), despite various experimental optimizations.
Positive selection of TAG metabolism-related genes in cetaceans
All-mammals dataset
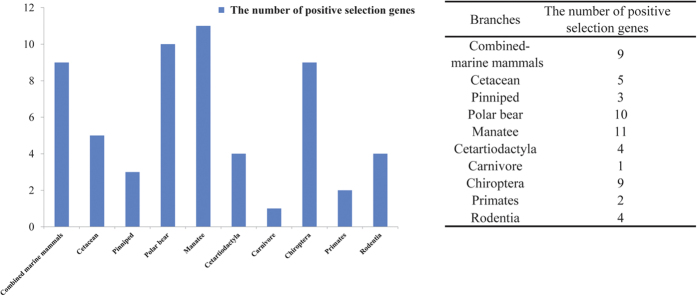
To investigate the impact of positive Darwinian selection in the 88 TAG metabolism-related genes, we used likelihood models of coding sequence evolution24 implemented in Codeml of the PAML package25. The branch-site model was used to test for positive selection in individual codons for the lineage leading to the common ancestor of each marine mammal groups (cetaceans: branch a; pinniped: branch v; polar bear: branch w; manatee: branch aa in Supplementary Fig. S1), the branch of combined marine mammals and the lineages of other groups (i.e., cetartiodactyls, carnivora, chiroptera, primates, and rodentia) across the mammalian phylogeny (Supplementary Fig. S1). Interestingly, evidence for positive selection was detected in 9 (APOB, ACSS1, AGPAT5, DGAT1, HSL1, MLXIPL, PLCB2, PLCE1, and PLIN3) of the 88 TAG metabolism-related genes examined of the combined marine mammal branches, 5 (APOB, DGAT1, MGLL, MOGAT1, and SERTAD2) genes in cetaceans, 3 (APOB, FAS, and GNPAT) genes in pinniped, 10 (APOB, DGKB, DGKZ, DGKI, FATP4, HSL, PLCB2, PLCB3, PLCH2, and PPARD) genes in polar bear and 11 (APOB, ACSL1, FAS, FATP2, MOGAT1, MOGAT2, LPL, PLCB2, PLCE1, PLIN4, and PNLIP) genes in manatee (Fig. 1 & Supplementary Table S2), which suggested the convergent evolution of TAG metabolism-related genes for the marine mammals during their adaptation to the aquatic environment. However, we also found some genes in the terrestrial groups to be positively selected, i.e. 4 genes in cetartiodactyla, 1 gene in carnivora, 9 genes in chiroptera, 2 genes in primates, and 4 genes in rodentia (Fig. 1 & Supplementary Table S2).
Figure 1. Comparison of the number of genes involved in TAG metabolism showing evidence of positive selection in mammals.
.
To further test if similar patterns of evolution occurred to the marine mammal groups, we reconstructed ancestral nodes and mapped amino acid changes along four marine mammal branches within 28 positively selected genes totally identified. Sixty-eight statistically significant (P < 0.05) parallel/convergent nonsynonymous amino acid substitutions were identified in the 14 of these 28 genes across two of the marine mammal lineages (Supplementary Table S3). In addition, 10, 21, 7, 2, 13, and 17 parallel/convergent mutations were found between branch pairs a vs v, a vs aa, w vs a, w vs aa, w vs v, and v vs aa in 4, 8, 3, 1, 2, and 8 genes, respectively. More importantly, the lineages leading to the common ancestor of pinniped, polar bear and manatee shared two amino acid changes (L316H & E331Q) in ACSL1.
Cetaceans-only dataset
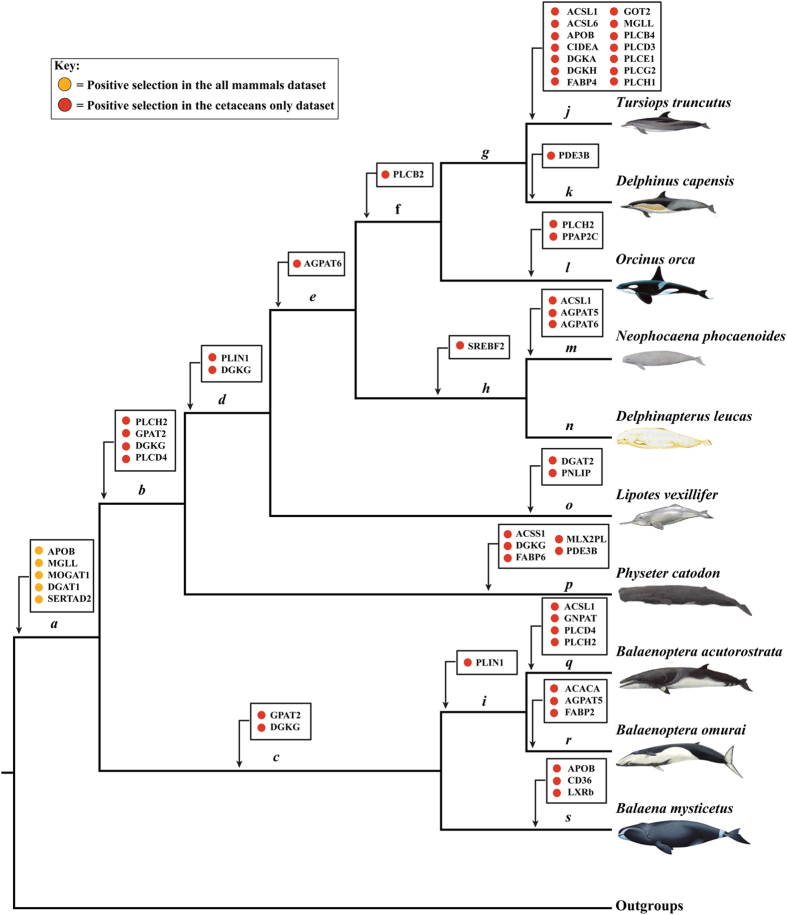
The cetaceans-only dataset consisting of 88 TAG metabolism-related genes was further used to determine the selection pattern in the interior nodes of cetaceans. Of these, 35 genes were determined to be under significant positive selection along branches b-s in Fig. 2 using the branch-site models (Supplementary Table S4). A pair of site models (M8a vs. M8)26,27 were also used to test whether specific codons in the TAG metabolism-related genes underwent positive selection, and 23 genes were determined to have undergone positive selection in the cetaceans-only dataset, where the LRTs of the site model were statistically significant (Table 1 and Supplementary Table S5). In combination with results from above branch-site and site models, it showed that 43 genes were totally detected to be under positive selection in cetaceans by PAML.
Figure 2. Positive selected genes related to TAG metabolism in cetaceans are mapped to a proposed phylogenetic tree.
Colored circles indicated the type of positive selection identified in the present study. Branches a–s in the tree are used in the branch-site models tests. Pictures of the cetacean representative members on the right of the tree are drawn by Professor Kaiya Zhou.
Table 1. M8 & M8a analysis and evidence of positive selection on 23 genes related to TAG metabolism in cetaceans.
| Gene | -InL (M8a) | -InL (M8) | P value | ω value (M8) | Positively selected sites |
|---|---|---|---|---|---|
| ACSL1 | 3880.189837 | 3877.642968 | 0.024 | 1.965 | 100 (0.800) 169 (0.842) 183 (0.829) 254 (0.877) 310 (0.863) 333 (0.852) 374 (0.936) 657 (0.849) 661 (0.838) 662 (0.877) |
| ACSL5 | 3762.751885 | 3756.161056 | <0.01 | 10.609 | 62 (0.855) 208 (0.883) 222 (0.983) 429 (0.845) 466 (0.994) 644 (0.838) 666 (0.975) 685 (0.858) |
| ACSL6 | 2974.997590 | 2972.082248 | 0.016 | 2.991 | 17 (0.872) 124 (0.908) 160 (0.923) 171 (0.917) 215 (0.899) 276 (0.931) 410 (0.986) |
| CD36 | 2970.142564 | 2958.040852 | <0.01 | 2.388 | 18 (0.964) 19 (0.953) 38 (0.874) 42 (0.886) 60 (0.991) 65 (0.939) 145 (0.935) 151 (0.934) 152 (0.911) 154 (0.874) 156 (0.998) 157 (0.892) 158 (0.964) 160 (0.999) 161 (0.916) 166 (0.916) 190 (0.857) 195 (0.948) 217 (0.899) 236 (0.927) 263 (0.969) 325 (0.949) 349 (0.919) 354 (0.963) 355 (0.951) 355 (0.951) 358 (0.855) 383 (0.961) 397 (0.946) 398 (0.899) 399 (0.981) 401 (0.905) 411 (0.997) 414 (0.855) 440 (0.915) 447 (0.846) 448 (0.934) 450 (0.961) 454 (0.945) 458 (0.858) 459 (0.942) 469 (0.957) 471 (0.894) 472 (0.897) 473 (0.999) |
| CIDEA | 1101.607445 | 1101.608921 | 0.003 | 1.337 | |
| DGAT1 | 2107.987504 | 2104.943568 | 0.014 | 14.431 | 419 (0.964) |
| DGAT2 | 1681.978411 | 1684.391038 | 0.028 | 3.305 | 41 (0.893) 42 (0.930) 67 (0.827) 117 (0.918) 177 (0.984) 227 (0.849) |
| DGKA | 3433.817808 | 3431.719676 | 0.041 | 5.593 | 451 (0.898) 572 (0.966) 720 (0.888) |
| DGKH | 5595.551465 | 5592.410358 | 0.012 | 503.770 | 1202 (0.824) |
| DGKQ | 4046.831703 | 4041.082855 | 0.001 | 31.380 | 278 (0.800) 279 (0.979) 313 (0.866) 338 (0.828) |
| FABP2 | 664.987160 | 663.011920 | 0.047 | 3.837 | 11 (0.856) 105 (0823) 26 (0.817) |
| FABP4 | 713.695341 | 705.734379 | <0.01 | 7.501 | 24 (0.957) 29 (0.951) 33 (0.943) 41 (0.959) 53 (0.961) 57 (0.955) 86 (0.954) 122 (0.996) |
| FATP2 | 3100.854796 | 3098.494072 | 0.030 | 8.292 | 176 (0.905) 242 (0.981) 251 (0.817) 356 (0.839) 392 (0.868) |
| FATP3 | 3400.109844 | 3395.996365 | 0.004 | 999.000 | 3 (0.920) 5 (0.893) 165 (0.859) 241 (0.819) 505 (0.819) 604 (0.864) 676 (0.803) |
| GNPAT | 3550.257763 | 3546.213234 | 0.004 | 2.503 | 23 (0.877) 176 (0.873) 178 (0.958) 205 (0.832) 214 (0.880) 434 (0.853) 441 (0.886) 462 (0.854) 466 (0.863) 477 (0.841) 583 (0.882) 664 (0.879) |
| GPAT2 | 4645.296344 | 4640.895446 | 0.003 | 21.420 | 439 (0.964) 445 (0.912) 479 (0.873) 572 (0.856) 677 (0.849) |
| MLXIPL | 3835.065500 | 3832.275129 | 0.018 | 346.673 | |
| MOGAT1 | 1795.790529 | 1789.419812 | <0.01 | 13.111 | 26 (0.891) 27 (0.987) 30 (0.999) 34 (0.903) 43 (0.873) 292 (0.885) |
| MOGAT2 | 2011.219495 | 2001.596828 | <0.01 | 17.931 | 74 (0.996) 238 (0.983) 255 (0.942) 295 (0.997) |
| PDE3B | 4061.866626 | 4051.580665 | <0.01 | 4.205 | 18 (0.972) 68 (0.987) 83 (0.930) 84 (0.814) 85 (0.812) 97 (0.812) 161 (0.994) 185 (0.885) 187 (0.950) 284 (0.966) 301 (0.919) 304 (0.895) 337 (0.957) 339 (0.961) 651 (0.944) 727 (0.900) |
| PLCZ1 | 2914.700506 | 2911.365281 | 0.010 | 49.708 | 340 (0.959) |
| PNLIP | 2548.952558 | 2540.199574 | <0.01 | 3.504 | 47 (0.890) 49 (0.859) 88 (0.974) 101 (0.824) 104 (0.849) 105 (0.927) 112 (0.911) 153 (0.869) 223 (0.835) 232 (0.972) 237 (0.970) 306 (0.994) 336 (0.838) 337 (0.859) 338 (0.893) 341 (0.995) 342 (0.859) 359 (0.888) 366 (0.868) 395 (0.887) 402 (0.846) |
| PPAP2C | 1454.520261 | 1447.266740 | <0.01 | 199.838 | 253 (0.823) 258 (0.847) 261 (0.855) 267 (0.863) 268 (0.931) 276 (0.924) |
Furthermore, the fixed-effects likelihood (FEL) and random-effects likelihood (REL) models were employed to confirm the selection pattern of TAG metabolism-related genes in cetaceans, and 26 genes were determined to be positively selected in cetaceans by the Datamonkey web server (Table 2 and Supplementary Table S6). The protein-level approach implemented in TreeSAAP28 identified a series of putative positively selected sites from 39 genes in cetaceans (Table 2 and Supplementary Table S6), 24 (ACSL1, ACSL5, ACSL6, ACSS1, AGPAT6, CD36, DGAT1, DGKA, DGKQ, FABP4, FATP2, GNPAT, GOT2, GPAT2, MOGAT1, MOGAT2, PDE3B, PLCB2, PLCD4, PLCH2, PLCZ1, PLIN1, PNLIP, and PPAP2C) of which were also detected by both PAML (branch-site models & site models) and Datamonkey web server (REL & FEL) methods. In addition, AGPAT5 and PLCE1 were determined to have undergone positive selection by both PAML and Datamonkey web server, whereas 15 genes (ACACA, APOB, CIDEA, DGAT2, DGKG, DGKH, FABP2, FATP3, LXRb, MGLL, PLCB4, PLCD3, PLCG2, PLCH1, and SREBF2) were detected using PAML and TreeSAAP (Table 2 and Supplementary Table S6).
Table 2. Positive selection in 41 cetacean TAG metabolism-related genes based on the analysis of the cetacean-only dataset by PAML, DATAMONKEY, or TreeSAAP.
| Gene | PAML* | DATAMONKEY* | TreeSAAP* |
|---|---|---|---|
| ACACA | Y | N | Y |
| ACSL1 | Y | Y | Y |
| ACSL5 | Y | Y | Y |
| ACSL6 | Y | Y | Y |
| ACSS1 | Y | Y | Y |
| AGPAT5 | Y | Y | N |
| AGPAT6 | Y | Y | Y |
| APOB | Y | N | Y |
| CD36 | Y | Y | Y |
| CIDEA | Y | N | Y |
| DGAT1 | Y | Y | Y |
| DGAT2 | Y | N | Y |
| DGKA | Y | Y | Y |
| DGKG | Y | N | Y |
| DGKH | Y | N | Y |
| DGKQ | Y | Y | Y |
| FABP2 | Y | N | Y |
| FABP4 | Y | Y | Y |
| FATP2 | Y | Y | Y |
| FATP3 | Y | N | Y |
| GNPAT | Y | Y | Y |
| GOT2 | Y | Y | Y |
| GPAT2 | Y | Y | Y |
| LXRb | Y | N | Y |
| MGLL | Y | N | Y |
| MOGAT1 | Y | Y | Y |
| MOGAT2 | Y | Y | Y |
| PDE3B | Y | Y | Y |
| PLCB2 | Y | Y | Y |
| PLCB4 | Y | N | Y |
| PLCD3 | Y | N | Y |
| PLCD4 | Y | Y | Y |
| PLCE1 | Y | Y | N |
| PLCG2 | Y | N | Y |
| PLCH1 | Y | N | Y |
| PLCH2 | Y | Y | Y |
| PLCZ1 | Y | Y | Y |
| PLIN1 | Y | Y | Y |
| PNLIP | Y | Y | Y |
| PPAP2C | Y | Y | Y |
| SREBF2 | Y | N | Y |
Note: Y, positive selection was detected; N, no positive selection was detected.
*Results are listed in Supplementary Tables S4,5,6.
To summarize the above results from the analyses of two datasets using different methods, 44 genes were identified to have undergone positive selection in cetaceans (Figs 2 and 3; Tables 1 and 2), and 5 (APOB, DGAT1, MGLL, MOGAT1, and SERTAD2) of these were subjected to strong positive selection in the common ancestor of the cetacean, whereas no selection was observed in terrestrial mammalian lineages such as cetartiodactyls, carnivores, and primates. It is generally accepted that a positively selected site is more reliable if it can be supported by two or more different methods. Of the positively selected genes, 41 were validated by at least two methods, thus they were used in the subsequent analyses. In terms of the functions of these 41 genes, 8 genes (ACACA, CD36, FABP2, FABP4, FATP2, FATP3, GOT2, and PNLIP) were involved in FA synthesis or transport, 26 genes (ACSL1, ACSL5, ACSL6, ACSS1, AGPAT5, AGPAT6, DGAT1, DGAT2, DGKA, DGKG, DGKH, DGKQ, GNPAT, GPAT2, MOGAT1, MOGAT2, PLCB2, PLCB4, PLCD3, PLCD4, PLCE1, PLCG2, PLCH1, PLCH2, PLCZ1, and PPAP2C) in TAG biosynthesis, and 1 gene (MGLL) in TAG lipolysis, and 6 genes (APOB, CIDEA, LXRb, PDE3B, PLIN1, and SREBF2) were associated with the regulation of TAG metabolism.
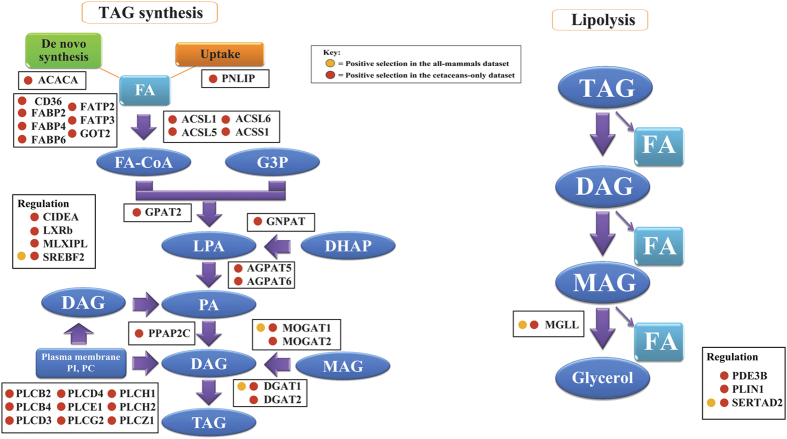
Figure 3. Mapping of the positively selected genes on the signal pathway of triacylglycerol synthesis and hydrolysis.
Colored circles indicate the type of positive selection identified in the present study. FA, fatty acid; CoA: Coenzyme A; G3P, glycerol-3-phosphate; LPA, lysophosphatidic acid; DHAP, dihydroxyacetone-P; PA, phosphatidicacid; DAG, diacylglycerol; PI, phosphatidylinositol; PC, phosphatidylcholine; MAG, monoacylglycerol; TAG, triacylglycerol.
Spatial distribution of the positively selected sites in the protein structures
Functional domains of each TAG metabolism-related gene were further examined to determine the functional significance of the putative positively selected sites, and the results showed that most identified positively selected sites were localized in or close to the functional regions in the protein structures of the 24 genes (Supplementary Fig. S2 & Supplementary Table S7). In 5 of these genes (i.e., ACSL1, CD36, DGAT2, GPAT2, and MOGAT1), 20 positively selected sites were located within the protein transmembrane domain of the corresponding genes. In addition, most of the positively selected sites were localized in the topological domain of ACSL1, ACSL5, ACSL6, CD36, DGAT1, DGAT2, FATP2, GPAT2, and SREBF2. For CD36, DGKA, DGKG, DGKH, DGKQ, LXRb, PLCD3, PLCD4, PLCE1, PLCH2, and PNLIP, some positively selected amino acids were located in the disulfide bond, glycosylation, zinc finger domain, substrate binding domain, ligand-binding domain, and so on, respectively (Supplementary Table S7). Furthermore, one positively selected site (29) was located in the nuclear localization signal motif of the FABP4 gene, and positively selected sites 440, 651, and 727 in the PDE3B gene were located in the catalytic regions, respectively (Supplementary Table S7).
Discussion
Cetaceans have a thick layer of blubber with a mean thickness of 98.4 ± 18.4 mm29. The thick blubber layer surrounding the cetacean body can comprise more than 30% of their body mass, and it is far greater than the 4–8% dissectible adipose found in the general healthy wild animal30. However, the genetic basis underlying the blubber thickening remains poorly explored. The present study therefore presents the first systematic investigation of TAG metabolism-related genes of representative cetaceans and closely related terrestrial mammals. Wide and strong signals of positive selection were detected in genes related to TAG synthesis or regulation of TAG synthesis, which could in turn provide novel insights into the evolution of blubber thickening in cetaceans.
The positive selection in cetacean DGAT1 and DGAT2 genes detected in the present study supports the morphological evidence that whales have thicker blubbers than those of other artiodactyl species17. The DGAT-catalyzed synthesis of TAG is the final and rate-limiting step in TAG formation (Fig. 3), and it is believed that DGAT is a key factor in controlling the production of triglycerides and fatty acids, as well as plays a key modulatory role in animal fat deposition31. Therefore, the observed positive selection in the DGAT1 and DGAT2 genes in cetaceans is suggestive of an enhanced capability for TAG formation. In addition, evidence of positive selection has been shown for other genes involved in the remaining pathways of TAG synthesis (Fig. 3). Effective uptake of free fatty acids can accelerate the expansion of adipocyte dimensions when lipids accumulate32. Acetyl-coA carboxylase 1 (ACC1), which play important roles in the de novo synthesis of FAs6,33, underwent positive selection in cetaceans (Table 2 and Fig. 3). Furthermore, PNLIP, the primary pancreatic TAG lipase34, was also subjected to positive selection across cetaceans, suggesting the essential role of FAs from the hydrolysis and absorption of long-chain triglyceride FAs from food during TAG synthesis in cetaceans. CD36, FABP2 (adipocytes lipid binding protein), FABP4, FATP2, FATP3, and GOT2, which are used in facilitating and regulating the transport of FAs across the plasma membrane, were determined to be under significant positive selection in cetaceans, which suggests that cetaceans might have acquired an enhanced capacity for FAs transport to maintain fat deposition. In additional, a series of essential enzymes involved in different steps of TAG biosynthesis pathways, i.e., ACSL1, ACSL5, ACSL6, ACSS1, AGPAT5, AGPAT6, DGKA, DGKG, DGKH, DGKQ, GNPAT, GPAT2, MOGAT1, MOGAT2, PLCB2, PLCB4, PLCD3, PLCD4, PLCE1, PLCG2, PLCH1, PLCH2, PLCZ1, and PPAP2C18, were determined to have undergone positive selection in cetaceans (Table 2 and Fig. 3), which in turn suggests that cetaceans have possess an effective ability to enhance their TAG synthesis during their adaptation to a fully aquatic life. Remarkably, some regulatory genes related to TAG synthesis were also determined to be under positive selection (Fig. 3), which may then imply a complex molecular mechanism of cetacean blubber thickening. Liver X-activated receptor b (LXRb), an important protein that controls the amount of cellular SREBP-1c, underwent significant positive selection in cetaceans (Table 2 and Fig. 3). The expression of FAS and mtGPAT can be greatly increased once SREBP-1c is overexpressed, which in turn results in an increase in FA synthesis and TAG deposition10,11. Furthermore, CIDEA, which enhances lipid droplet size when ectopically expressed in preadipocytes and in turn favors cellular lipid accumulation35, also presented evidence of positive selection in cetaceans. Positive selection of these genes might therefore play an important role in promoting cetacean blubber thickening.
Positive selection of different enzymes involved in cetacean TAG synthesis might directly explain the molecular basis of cetacean blubber thickening. Interestingly, besides from the TAG synthesis-related genes, MGLL, an important major lipases involved in lipolysis36, was also under strong positive selection, as indicated by the results of nucleotide- and protein- level analyses (Table 2 and Fig. 3). These findings therefore suggest that lipolysis was advanced to a certain degree in cetaceans, and the metabolic rate of cetaceans might have been increased to compensate for the energy shortage.
Cetaceans exhibit physiological and anatomical adaptations that allow them to rely on the lipids stored in their blubber as a source of energy during annual fasting periods37. During times of energy shortage, TAGs stored in lipid droplets are hydrolyzed to FAs and glycerol via lipolysis for subsequent use by other organs, and dolphins had more rapid release of non-esterified FAs than other mammals38. In addition, the water derived from metabolism, particularly lipolysis, is considered to be the primary sources of fresh water for cetaceans39. MGLL hydrolyzes MAG, thereby producing glycerol and FAs6. Therefore, positive selection of MGLL might be helpful for rapid release FAs to producing energy and to more efficiently extracting water in cetaceans, which in turn allows them to survive annual fasting periods with limited amounts of TAG stored in body tissues.
Cetacean blubber was vertically stratified and each blubber layer performs a different function, with the stable outer layer used for structural support and the more variable inner layer used for energy storage40. A suitable blubber thickness is essential for cetaceans to adapt to the aquatic environment. However, when in an emaciated state during the fasting period, the dual roles of the blubber in providing insulation and storing metabolic energy are in direct conflict20. The absence of a mechanism to regulate or control the utilization of stored TAGs in the blubber as an energy resource can result in the excessive use of TAGs during the fasting period, which in turn might greatly decrease the thickness of blubber and further weaken the activities of thermoregulation, buoyancy control, streamlining, and locomotion that are essential for aquatic life. Amazingly, evidence of positive selection in two genes involved in the regulation of lipolysis (i.e., PDE3B and PLIN1) was detected in cetacean-specific lineages, which maybe a mechanism for avoiding excessive lipolysis. PDE3B is a very powerful regulator of adipocyte lipolysis that is triggered by a decrease in cAMP levels41. The importance of PDE3B in suppressing adipocyte lipolysis has been demonstrated in PDE3B null mice42. PLIN1 is a key member of the PLIN family that encode for proteins that cover the lipid droplets in adipocytes, as well as regulate the coordination of lipid storage and utilization in various cell types36. It has been suggested that PLINs play an important role in inhibiting lipolysis when it is unphosphorylated43. Considering the function of these genes in regulating and inhibiting TAG lipolysis, positive selection of these genes suggests that cetaceans have evolved an enhanced capacity for inhibiting unrestricted lipolysis and finely control the fatty acid content of blubber, and therefore is important for the maintenance of a suitable thickness of the blubber layer.
Blubber’s thickness and lipid content extensively varies across cetaceans. For example, the lipid content of harbor porpoise blubber ranges from 76% to 88%, whereas that of minke whales ranges from 42% to 96%20. The thermal conductance capacity of the blubber is highly dependent on both its conductive quality and quantity (i.e., thickness), and the difference in the thickness and lipid content of the blubber among cetaceans might be the result of their adaptation to different aquatic habitats. Therefore, positively selected genes are distributed throughout almost the entire cetacean phylogeny, from the most common ancestral branch of cetaceans to the terminal branches (Fig. 2). A series of positively selected sites observed in 24 genes were localized in or near the functional regions on the crystal structure of the corresponding genes (Supplementary Fig. S2 and Supplementary Table S7), which indicates the ongoing adaptive evolution of cetacean TAG metabolism-related genes. It is also reasonable to assume that different cetaceans require a fine blubber layer, which has driven the relevant genes to evolve in response to continuous changes in the aquatic environment since their origin and subsequent diversification in waters across the globe.
Remarkably, APOB, the primary lipid-binding protein of chylomicrons and low-density lipoproteins (LDL)44, was determined to have undergone positive selection in the lineage leading to the common ancestor of each marine mammalian groups (i.e., cetaceans, pinnipeds, polar bear, manatee) and the branch of the combined marine mammals (Supplementary Table S2), which is consistent with the previous findings that strong adaptive selection was detected in the APOB gene of polar bears45. In cetaceans, blood-based LDL levels are much lower than humans46, and APOB may play an important role in the LDL redistribution in the blubber of cetaceans. In addition, blubber is a critical component of mammalian adaptation to the aquatic environment, and cetaceans and other aquatic animals show remarkable similarities in blubber structure and function37. Comparative genomic analyses have found that convergent amino acid substitutions were widespread throughout the genome, and PNLIP and PLIN4 genes were identified to be positively selected respective along the combined marine mammal branch and the cetacean branch in Foote et al. (2015)47. In addition, 68 parallel/convergent nonsynonymous amino acid substitutions were identified in 14 positively selected genes detected. These results further supported the adaptive evolution of cetacean TAG metabolism-related genes, and suggest that cetaceans and other marine mammals have apparently been under similar pressure for adipose tissue development and fatty acid metabolism during their adaptation to the aquatic environments. However, further investigation on the functional verification of TAG metabolism-related genes in cetaceans and other marine mammals is necessary in the future to determine their role in aquatic adaptations. Moreover, some positively selected genes were also detected in the terrestrial groups, suggesting the importance of TAG metabolism during their adaptation to various environments, and further research should be focused on this interesting phenomenon to interpret the roles of TAG metabolism in terrestrial mammals.
The present study is the first comprehensive and systematic analysis of the molecular genetic basis of blubber thickening in cetaceans. Wide and strong positive selection was detected in several genes involved in TAG synthesis, lipolysis, and regulation, which is concordant with the important functions of cetacean blubber in thermoregulation, buoyancy control, streamlining, metabolic energy storage, and locomotion. Interestingly, some regulation genes that inhibit lipolysis also showed significant evidence of positive selection, which suggests that cetaceans have evolved an enhanced capacity for inhibiting unrestricted lipolysis, particularly during fasting. This study provides novel insights into the effective and complex mechanism of maintaining a suitable blubber layer thickness in cetaceans, and also implies that ‘obesity’ might be an indicator of good health for cetaceans, compared to that determined in humans, which has been strongly associated to various chronic diseases.
Methods
Samples and DNA sequencing
Five cetacean species (two mysticetes and three odontocetes): common minke whale, Omura’s whale, Beluga, Finless porpoise, and long-beaked common dolphin were sequenced during this study. All 5 cetacean samples used in the present study were collected from dead individuals in the wild, and sampling was conducted systematically in accordance with all ethical guidelines and legal requirements in China. The protocol of this study was approved by the Institutional Review Board of Nanjing Normal University (NNU). Voucher specimens were preserved at Jiangsu Key Laboratory for Biodiversity and Biotechnology, College of Life Sciences, Nanjing Normal University (NNU), China.
Total genomic DNA was extracted from muscle with a standard phenol/chloroform procedure followed by ethanol precipitation48. The DNA integrity was checked by 1% agarose gel electrophoresis. Primers were designed for the conserved regions based on an alignment of genomic data from the cow (Bos taurus) (http://asia.ensembl.org/Bos_taurus/Info/Index) and bottlenose dolphin (Tursiops truncatus) (http://asia.ensembl.org/Tursiops_truncatus/Info/Index). All PCR amplification were conducted using a BioRAD PTC-200 with 2 × EasyTaq PCR SuperMix (TransGen Biotech) and the following profile: 34 cycles at 94 °C for 5min, 94 °C for 30 s, 53 °C−59 °C for 30 s, and 72 °C for 30 s, followed by a 10 min extension at 72 °C. The amplified PCR products were purified and sequenced in both directions using an ABI 3730 automated genetic analyzer. Three to five repeated amplification for each gene were conducted and resequenced to confirm its sequence. The specificity of these newly generated sequences was examined by comparison with the published nucleotide database at GenBank by BLAST (NCBI).
Source of data and primary treatments
Forty-one TAG metabolism-related genes (Supplementary Table S1) were sequenced in the five cetacean species mentioned above and the newly sequences were deposited in GenBank under accession numbers KR135543-KR135734. The exons of each gene were sequenced and concatenated before being analyzed together. Only high-quality and high-integrity sequences were used in the analysis. In addition, gene sequences from other mammals [including bottlenose dolphin (Tursiops truncatus)] were searched and primarily downloaded from the OrthoMaM49, and gene sequences derived from the genomes of baiji (Lipotes vexillifer), common minke whale (Balaenoptera acutorostrata scammoni), killer whale (Orcinus orca), sperm whale (Physeter macrocephalus), seal (Leptonychotes weddellii), walrus (Odobenus rosmarus divergens), manatee (Trichechus manatus latirostris), polar bear (Ursus maritimus) (ftp://ftp.ncbi.nlm.nih.gov/genomes/) and bowhead whale (Balaena mysticetus) (http://www.bowhead-whale.org/) were also used to test for positive Darwinian selection. A total of 88 TAG metabolism-related genes (Supplementary Table S1) and 23–50 species from representatives mammalian lineages (i.e., Cetaceans, Artiodactyla, Chiroptera, Rodentia, Carnivora, and Primates) were analyzed in the present study (Supplementary Table S8 and Supplementary Table S9). Nucleotide sequences of each gene examined and their deduced amino acid sequences were aligned separately using MUSCLE 3.850 and MEGA 5.051, and manually adjusted with GeneDoc.
Molecular evolutionary analyses
The codon-based maximum likelihood models implemented in CODEML program in PAML 4.725 were applied to estimate the rates of synonymous (dS) and nonsynonymous substitutions (dN), as well as dN/dS ratio (omega, ω). The non-synonymous to synonymous rate ratio ω indicates changes in selective pressures, where ω = 1, ω < 1, and ω > 1 correspond to neutral evolution, purifying, and positive selection, respectively. The well-supported phylogeny of Laurasiatheria52 and Primates53 was used as the input tree in all analyses (Tree file: Supplementary Fig. S1).
Positive selection was detected using branch-site model A, in which ω can vary among sites along specific lineages54. Modified branch-site model A (test 2) was performed for every gene in each foreground lineage, which facilitated the analysis of datasets, including all mammals (branches a, t-aa in Supplementary Fig. S1) or cetaceans only (branches b-s in Supplementary Fig. S1). To identify the probabilities of sites under positive selection in each gene for the cetacean species examined, site models were implemented where ω could vary among sites. All the positively selected sites in site models were identified by using Bayes Empirical Bayes (BEB) analysis25 with posterior probabilities of ≥0.80. The likelihood ratio test (LRT) statistic (2ΔL) approximates to a Chi-square distribution and was used to compare nested likelihood models. In addition, the improved statistical methods in Datamonkey web server55, which computed nonsynonymous and synonymous substitutions at each codon position, was used to further evaluate the selection. Sequences of each gene in cetaceans-only dataset were analyzed by using two distinct models, namely, fixed-effect likelihood (FEL) and random effect likelihood (REL).The FEL model estimates the ratio of dN/dS on a site-by-site basis, without assuming an a priori distribution across sites. The REL model first fits a distribution of rates across sites and then infers the substitution rate for individual sites. Sites with P values < 0.1 for FEL, and Bayes factor >50 for REL were considered as candidates under positive selection55.
Some studies have suggested that the ML method of evaluating positive selection might produce false-positive results even when no positively selected sites exist56 or when positively selected sites and negatively selected sites are mixed57. Further support for the PAML results were obtained using a complementary protein-level approach implemented in TreeSAAP28. TreeSAAP compares the magnitude of property changes of non-synonymous residues across a phylogeny and identifies specific amino acid properties that have likely been affected by positive destabilizing selection during evolutionary58.
Identification of parallel/convergent sites among marine mammals
The parallel/convergent sites among marine mammals were identified according to the methods previously described47. In detail, we reconstructed the ancestral sequences for 26 positive selection genes using the Codeml program in PAMLv4.7. For each of the four marine mammal groups (cetaceans, pinnipeds, polar bear and manatee) the extant sequences at each position were compared to the ancestral sequence at the node corresponding to the most recent ancestor. We used the software CONVERG 259 to test whether the number of observed parallel/convergent amino acid substitutions was significantly higher than that expected by chance, given the total numbers of amino acid replacements in the two evolutionary lineages under investigation. The positions of the parallel/convergent nonsynonymous amino acid substitutions that were found in positively selected genes are shown in Supplementary Table S3.
Mapping of positively selected sites onto protein structures
To gain insights into the functional significance of the putatively selected sites, we mapped the sites under positive selection to crystal structures. The 3D structures of genes under positive selection were predicted by using the homology modeling software provided by the I-TASSER server60. The protein sequences of positively selected genes were derived from the common bottlenose dolphin (Tursiops truncutus) genome, which were obtained from the Ensembl genome database (http://www.ensembl.org/index.html). In addition, the functional information of genes identified as being under positive selection was derived from the uniprot (http://www.uniprot.org/).
Additional Information
Accession codes: The 41 TAG metabolism-related genes (Supplementary Table S1) were sequenced in five additional cetacean species (two mysticetes and three odontocetes): common minke whale (Balaenoptera acutorostrata), Omura's whale (Balaenoptera omurai), Beluga (Delphinapterus leucas), Finless porpoise (Neophocaena phocaenoides), and long-beaked common dolphin (Delphinus capensis), and these new sequences were deposited in GenBank as accession numbers KR135543-KR135734.
How to cite this article: Wang, Z. et al .‘Obesity’ is healthy for cetaceans? Evidence from pervasive positive selection in genes related to triacylglycerol metabolism. Sci. Rep .5, 14187; doi: 10.1038/srep14187 (2015).
Supplementary Material
Acknowledgments
Financial support was provided by the National Science Fund for Distinguished Young Scholars (Grant no. 31325025) to GY, the National Natural Science Foundation of China (NSFC) (Grant no. 31172069 to GY, Grant no. 31370401 to WHR, and Grant no. U1404306 to ZC), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) to GY and SXX, and the Specialized Research Fund for the Doctoral Program of Higher Education to GY (Grant no. 20113207130001), and the Natural Science Foundation of Jiangsu Province to SXX (Grant no. BK20141449), and the Cultivation Plan for Excellent Doctorial Dissertations of Nanjing Normal University to ZFW (Grant no. 1812000002105), and the Jiangsu Provincial Innovation Project for Scientific Research of Graduate Students in Universities to ZFW (Grant no. KYLX_0708), and the Jiangsu Provincial Innovation Project for Scientific Research of Graduate Students in Universities to JL (Grant no. CXZZ13_0409). We are also grateful to Mr. Xinrong Xu, Dr. Anli Gao, and some students who have ever studied or are studying at NJNU for their assistance in sample collection. We are indebted to Yan Guo, Zhitao Niu, Kexing Du, Kui Li and Yan Liang for support and discussions. We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Footnotes
Author Contributions G.Y. and Z.W. designed the study. Z.W. and Z.C. carried out the experiments, performed the data analyses, and prepared the draft of the manuscript. S.X. and K.Z. helped to improve the manuscript. G.Y. and W.R. helped to perform the data analyses and improve the manuscript. All authors read and approved the final manuscript.
References
- Söhle J. et al. .Identification of new genes involved in human adipogenesis and fat storage. Plos One 7, e31193 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice A. M. The emerging epidemic of obesity in developing countries. Int J Epidemiol 35, 93–99 (2006). [DOI] [PubMed] [Google Scholar]
- Gregoire F. M., Smas C. M. & Sul H. S. Understanding adipocyte differentiation. Physiol Rev 78, 783–809 (1998). [DOI] [PubMed] [Google Scholar]
- Coleman R. A. & Lee D. P. Enzymes of triacylglycerol synthesis and their regulation. Prog Lipid Res 43, 134–176 (2004). [DOI] [PubMed] [Google Scholar]
- IUPAC-IUB C. O. B. N. The nomenclature of lipids. Chem Phys Lipids 21, 159–173 (1978). [PubMed] [Google Scholar]
- Large V., Peroni O., Letexier D., Ray H. & Beylot M. Metabolism of lipids in human white adipocyte. Diabetes Metab 30, 294–309 (2004). [DOI] [PubMed] [Google Scholar]
- Gibbons G. F., Islam K. & Pease R. J. Mobilisation of triacylglycerol stores. BBA-Mol Cell Biol L 1483, 37–57 (2000). [DOI] [PubMed] [Google Scholar]
- Gilham D. & Lehner R. The physiological role of triacylglycerol hydrolase in lipid metabolism. Rev Endocr Metab Dis 5, 303–309 (2004). [DOI] [PubMed] [Google Scholar]
- Ahmadian M., Wang Y. & Sul H. S. Lipolysis in adipocytes. Int J Biochem Cell B 42, 555–559 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton J. D. & Shimomura L. Sterol regulatory element-binding proteins: activators of cholesterol and fatty acid biosynthesis. Curr Opin Lipidol 10, 143–150 (1999). [DOI] [PubMed] [Google Scholar]
- Horton J. D. et al. .Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J Clin Invest 101, 2331 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. et al. .Genome-wide scans for candidate genes involved in the aquatic adaptation of dolphins. Genome Biol Evol 5, 130–139 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowen M. R., Grossman L. I. & Wildman D. E. Dolphin genome provides evidence for adaptive evolution of nervous system genes and a molecular rate slowdown. P Roy Soc B-Biol Sci 279, 3643–3651 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thewissen J. G., Cooper L. N., Clementz M. T., Bajpai S. & Tiwari B. N. Whales originated from aquatic artiodactyls in the Eocene epoch of India. Nature 450, 1190–1194 (2007). [DOI] [PubMed] [Google Scholar]
- Parry D. A. The structure of whale blubber, and a discussion of its thermal properties. Q J Microsc Sci 3, 13–25 (1949). [PubMed] [Google Scholar]
- Scholander P. F., Walters V., Hock R. & Irving L. Body insulation of some arctic and tropical mammals and birds. Biol Bull-US 99, 225–236 (1950). [DOI] [PubMed] [Google Scholar]
- Pond C. M. Morphological aspects and the ecological and mechanical consequences of fat deposition in wild vertebrates. Annu Rev Ecol Evol Syst 9, 519–570 (1978). [Google Scholar]
- Struntz D. J. et al. .Blubber development in bottlenose dolphins (Tursiops truncatus). J Morphol 259, 7–20 (2004). [DOI] [PubMed] [Google Scholar]
- Kipps E. K., McLellan W. A., Rommel S. A. & Pabst D. A. Skin density and its influence on buoyancy in the manatee (Trichechus manatus latirostris), harbor porpoise (Phocoena phocoena), and bottlenose dolphin (Tursiops truncatus). Mar Mammal Sci 18, 765–778 (2002). [Google Scholar]
- Dearolf J. L., McLellan W. A., Dillaman R. M., Frierson D. & Pabst D. A. Precocial development of axial locomotor muscle in bottlenose dolphins (Tursiops truncatus). J Morphol 244, 203–215 (2000). [DOI] [PubMed] [Google Scholar]
- Dunkin R. C., McLellan W. A., Blum J. E. & Pabst D. A. The ontogenetic changes in the thermal properties of blubber from Atlantic bottlenose dolphin Tursiops truncatus. J Exp Biol 208, 1469–1480 (2005). [DOI] [PubMed] [Google Scholar]
- Hamilton J. L., Dillaman R. M., McLellan W. A. & Pabst D. Structural fiber reinforcement of keel blubber in harbor porpoise (Phocoena phocoena). J Morphol 261, 105–117 (2004). [DOI] [PubMed] [Google Scholar]
- Pabst D. A. To bend a dolphin: convergence of force transmission designs in cetaceans and scombrid fishes. Am Zool 40, 146–155 (2000). [Google Scholar]
- Goldman N. & Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol 11, 725–736 (1994). [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- Swanson W. J., Nielsen R. & Yang Q. Pervasive adaptive evolution in mammalian fertilization proteins. Mol Biol Evol 20, 18–20 (2003). [DOI] [PubMed] [Google Scholar]
- Wong W. S., Yang Z., Goldman N. & Nielsen R. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics 168, 1041–1051 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley S., Johnson J., Smith M. J., Crandall K. A. & McClellan D. A. TreeSAAP: selection on amino acid properties using phylogenetic trees. Bioinformatics 19, 671–672 (2003). [DOI] [PubMed] [Google Scholar]
- Evans K., Hindell M. A. & Thiele D. Body fat and condition in sperm whales, Physeter macrocephalus, from southern Australian waters. Comp Biochemphys A 134, 847–862 (2003). [DOI] [PubMed] [Google Scholar]
- Koopman H. N. Phylogenetic, ecological, and ontogenetic factors influencing the biochemical structure of the blubber of odontocetes. Mar Biol 151, 277–291 (2007). [Google Scholar]
- Yu Y. & Ginsberg H. N. The role of acyl-CoA: diacylglycerol acyltransferase (DGAT) in energy metabolism. Ann Med 36, 252–261 (2004). [DOI] [PubMed] [Google Scholar]
- Hausman G. J. & Thomas G. B. The development of the inner layer of backfat in fetal and young pigs. J Anim Sci 58, 1550–1560 (1984). [DOI] [PubMed] [Google Scholar]
- Shrago E., Spennetta T. & Gordon E. Fatty acid synthesis in human adipose tissue. J Biol Chem 244, 2761–2766 (1969). [PubMed] [Google Scholar]
- Davis R. C. et al. .Assignment of human pancreatic lipase gene (PNLIP) to chromosome 10q24-q26. Genomics 11, 1164–1166 (1991). [DOI] [PubMed] [Google Scholar]
- Puri V. et al. .Cidea is associated with lipid droplets and insulin sensitivity in humans. P Natl Acad Sci USA 105, 7833–7838 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontan M. & Langin D. Lipolysis and lipid mobilization in human adipose tissue. Prog Lipid Res 48, 275–297 (2009). [DOI] [PubMed] [Google Scholar]
- Koopman H. N., Pabst D. A., McLellan W. A., Dillaman R. M. & Read A. J. Changes in blubber distribution and morphology associated with starvation in the harbor porpoise (Phocoena phocoena): evidence for regional differences in blubber structure and function. Physiol Biochemzool 75, 498–512 (2002). [DOI] [PubMed] [Google Scholar]
- Ortiz R. M., Long B., Casper D., Ortiz C. L. & Williams T. M. Biochemical and hormonal changes during acute fasting and re-feeding in bottlenose dolphins (Tursiops truncatus). Mar Mammal Sci 26, 409–419 (2010). [Google Scholar]
- Xu S. et al. .Adaptive evolution of the osmoregulation-related genes in cetaceans during secondary aquatic adaptation. BMC Evol Biol 13, 189 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel A. M. & Worthy G. A. Variability in fatty acid composition of bottlenose dolphin (Tursiops truncatus) blubber as a function of body site, season, and reproductive state. Can J Zool 82, 1933–1942 (2004). [Google Scholar]
- Degerman E. et al. .Role for phosphodiesterase 3B in regulation of lipolysis and insulin secretion. In Diabetes mellitus: a fundamental and clinical text. 3rd edition. (eds LeRoith D., Taylor S. I. & Olefsky J. M.) 373–381 (Lippincott-Raven Press, Philadelphia, 2003). [Google Scholar]
- Choi Y. H. et al. .Alterations in regulation of energy homeostasis in cyclic nucleotide phosphodiesterase 3B-null mice. J Clin Invest 116, 3240–3251 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasaemle D. L. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J Lipid Res 48, 2547–2559 (2007). [DOI] [PubMed] [Google Scholar]
- Whitfield A. J., Barrett P. H. R., Van Bockxmeer F. M. & Burnett J. R. Lipid disorders and mutations in the APOB gene. Clin Chem 50, 1725–1732 (2004). [DOI] [PubMed] [Google Scholar]
- Liu S. et al. .Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell 157, 785–794 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venn-Watson S. et al. .Blood-based indicators of insulin resistance and metabolic syndrome in bottlenose dolphins (Tursiops truncatus). Front Endocrinol (Lausanne) 4, 136 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote A. D. et al. .Convergent evolution of the genomes of marine mammals. Nat Genet 47, 272–275 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R. & Sambrook J. Molecular Cloning: A Laboratory Manual. 4th edition. (Cold Spring Harbor Laboratory Press, New York, 2012). [Google Scholar]
- Ranwez V. et al. .OrthoMaM: a database of orthologous genomic markers for placental mammal phylogenetics. BMC Evol Biol 7, 241 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. et al. .MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. et al. .Phylogenomic analysis resolves the interordinal relationships and rapid diversification of the Laurasiatherian mammals. Syst Biol 61, 150–164 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelman P. et al. .A molecular phylogeny of living primates. Plos Genet 7, e1001342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Nielsen R. & Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol 22, 2472–2479 (2005). [DOI] [PubMed] [Google Scholar]
- Pond S. L. K. & Frost S. D. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21, 2531–2533 (2005). [DOI] [PubMed] [Google Scholar]
- Suzuki Y. & Nei M. False-positive selection identified by ML-based methods: examples from the Sig1 gene of the diatom Thalassiosira weissflogii and the tax gene of a human T-cell lymphotropic virus. Mol Biol Evol 21, 914–921 (2004). [DOI] [PubMed] [Google Scholar]
- Anisimova M., Bielawski J. P. & Yang Z. Accuracy and power of Bayes prediction of amino acid sites under positive selection. Mol Biol Evol 19, 950–958 (2002). [DOI] [PubMed] [Google Scholar]
- Nery M. F., Arroyo J. I. & Opazo J. C. Accelerated Evolutionary Rate of the Myoglobin Gene in Long-Diving Whales. J Mol Evol 76, 380–387 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang J. & Kumar S. Detection of convergent and parallel evolution at the amino acid sequence level. Mol Biol Evol 14, 527–536 (1997). [DOI] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bionformatics 9, 40 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.