Abstract
Major porins are among the most abundant proteins embedded in the outer membrane (OM) of Gram-negative bacteria, playing crucial roles in maintenance of membrane structural integrity and OM permeability. Although many OM proteins (especially c-type cytochromes) in Shewanella oneidensis, a research model for respiratory versatility, have been extensively studied, physiological significance of major porins remains largely unexplored. In this study, we show that OmpS38 and OmpA are two major porins, neither of which is responsive to changes in osmolarity or contributes to the intrinsic resistance to β-lactam antibiotics. However, OmpS38 but not OmpA is largely involved in respiration of non-oxygen electron acceptors. We then provide evidence that expression of ompS38 is transcribed from two promoters, the major of which is favored under anaerobic conditions while the other appears constitutive. The major promoter is under the direct control of Crp, the master regulator dictating respiration. As a result, the increase in the level of OmpS38 correlates with an elevated activity in Crp under anaerobic conditions. In addition, we show that the activity of the major promoter is also affected by Fur, presumably indirectly, the transcription factor for iron-dependent gene expression.
The outer membrane (OM) of Gram-negative bacteria is a highly asymmetric barrier that hinders the permeability of both hydrophilic and hydrophobic compounds1. To facilitate uptake of nutrients and other molecules for growth and diverse functions of the cell, the OM is embedded with a large number of channels formed by β-barrel proteins, including porins, substrate-specific transporters, and active transporters2. Porins are among most abundant proteins and important constituents of the OM, comprising up to 2% of the entire protein content of the cell1. Classic porins (~40 kDa), exemplified by Escherichia coli OmpC and OmpF, consist of 12–22 transmembrane segments while small porins (<25 kDa), such as E. coli OmpW, have 8–103,4. It has been well established that porins are water-filled open channels allowing the passive penetration of hydrophilic molecules, which are discriminated generally by their gross physiochemical properties, such as size, hydrophobicity, and charge5,6. As bacterial antibiotic resistance is strongly correlated with permeability changes of the OM, porins have been a research focus for their role in transporting antibiotics7. Additionally, porins have been implicated in respiration, electron transfer, and pathogenesis by acting as receptors for phages, adhesins, and even as virulence factors8,9,10,11,12.
Most biological functions with which porins are associated are attributed to major porins as they are significantly more abundant than others. Given the diverse processes in which major porins are involved, their expression is tightly regulated, often at different levels by various regulators. A well-known osmotic response regulatory system is E. coli EnvZ-OmpR two-component system (TCS), controlling transcription of ompF and ompC as well as a diverse set of other genes13,14,15. Other regulators that converge to transcriptionally regulate the differential expression of major porin genes in response to environmental disturbances including Fnr (fumarate nitrate regulator), Crp (cyclic-AMP receptor protein), Lrp (amino acid metabolism regulator), Fur (iron homeostasis regulator), and CpxA-CpxR (cell-envelope stress regulator, to name a few16. In addition, two small regulatory RNAs, MicF and MicC, significantly affect OmpF and OmpC production at the translational level17.
Shewanella, a group of facultative anaerobic γ-proteobacteria widely distributed in nature have received intensive attentions owing to their remarkably diverse respiratory capacities and the potential for environmental remediation and microbial fuel cell, which largely rely on extracellular electron transfer18,19,20. Two mechanisms are adopted by Shewanella to deliver electrons generated intracellularly to electron acceptors (EA) in the extracellular space: direct transfer by redox c-type cytochromes on the OM and indirect means mediated by electron shuttles, such as riboflavin21,22,23. While the OM integrity could be critical to the former, the OM permeability is important to the latter. In addition, the genus is regarded as emerging human pathogens and a reservoir for antibiotics resistance given that a number of β-lactamases and Qnr-type determinants have been isolated24,25,26,27,28. Hence, insights into major porins are needed because of their abundance in the OM and role in transport of hydrophilic molecules, such as electron shuttles and some antibiotics.
In S. oneidensis MR-1, the most extensively studied Shewanella strain, a large number of porins have been predicted in the genome annotation29. No known biological function has been demonstrated for these putative porins except for SO_3896, which is found to have a role in anaerobic respiration and its promoter is one of the most robust12,30. For regulation, the bacterium contains an E. coli EnvZ-OmpR analogue, but it has diverged considerably from its E. coli counterpart despite its connection with osmoregulation13,31. In this study we first made efforts to identify and functionally characterize the major porins in S. oneidensis. Our results demonstrate that two porins, SO_3896 and SO_3545, named as OmpS38 and OmpA respectively, stand out as the most abundant ones. Interestingly, neither is notably responsive to changes in osmolarity or required for transport of β-lactam antibiotics into the periplasm. Further investigation reveals that OmpS38 is generally important in respiration of non-oxygen EAs and that the enhanced expression during anaerobic respiration largely relies on Crp activation.
Materials and Methods
Bacterial strains, plasmids, and culture conditions
A list of all bacterial strains and plasmids used in this study is given in Table 1. All chemicals were acquired from Sigma Co. (Shanghai, China) unless otherwise noted. Information for primers used in this study is available upon request. For genetic manipulation, E. coli and S. oneidensis strains under aerobic conditions were grown in Lysogeny Broth (LB, Difco, Detroit, MI) medium (tryptone, 10 g/L; yeast extract, 5 g/L; and NaCl, 5 g/L) at 37 and 30 °C, respectively. When needed, the growth medium was supplemented with chemicals at the following concentrations: 2, 6-diaminopimelic acid (DAP), 0.3 mM; ampicillin sodium, 50 μg/ml; kanamycin sulfate, 50 μg/ml; and gentamycin sulfate; 15 μg/ml.
Table 1. Strains and plasmids used in this study.
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | Host for cloning | Lab stock |
| WM3064 | ΔdapA, donor strain for conjugation | W. Metcalf, UIUC |
| XL1-Blue MRF’Kan | Recipient strain for one-hybrid system | Stratagene |
| S. oneidensis strains | ||
| MR-1 | Wild type | ATCC 700550 |
| HG0624 | Δcrp derived from MR-1 | (35) |
| HG1215 | ΔSO1215 derived from MR-1 | This study |
| HG1821 | ΔSO1821 derived from MR-1 | This study |
| HG1937 | ∆fur derived from MR-1 | This study |
| HG2356 | Δfnr derived from MR-1 | (35) |
| HG3099 | ΔfadL derived from MR-1 | This study |
| HG3284 | ΔcydX derived from MR-1 | (59) |
| HG3545 | ΔompA derived from MR-1 | This study |
| HG3896 | ΔompS38 derived from MR-1 | This study |
| HG3988 | ΔarcA derived from MR-1 | (44) |
| HG3545-3896 | ΔompAΔompS38 (Δdouble) derived from MR-1 | This study |
| Plasmid | ||
| pHGM01 | Apr, Gmr, Cmr, att-based suicide vector | (33) |
| pHG101 | Kmr, promoterless broad-host vector | (34) |
| pHGEI01 | Kmr, integrative lacZ reporter vector | (48) |
| pBBR-Cre | Spr, helper plasmid for antibiotic cassette removal | (38) |
| pBXcmT | B1H bait vector | (45) |
| pTRG | B1H target vector | Stratagene |
| pHGE-PompS38-lacZ | pHGEI01 containing the ompS38 promoter | This study |
Physiological characterization of S. oneidensis strains
MS defined medium with 30 mM sodium lactate as the carbon source was used for growth and respiration characterization as described previously32. For anaerobic respiration, electron acceptors studied in this study included nitrate (3 mM), fumarate (20 mM), trimethylamine N-oxide (TMAO, 20 mM), ferric citrate (10 mM), and goethite (FeO(OH), 10 mM). Growth of S. oneidensis strains under aerobic or anaerobic conditions was determined by recording optical densities of cultures at 600 nm (OD600). For aerobic growth, fresh medium was inoculated to ~0.01 of OD600 with overnight cultures grown from a single colony. For anaerobic growth, cultures of mid-log phase (~0.2 of OD600, the same through the study) grown aerobically were centrifuged, washed with fresh medium twice, purged in nitrogen and suspended in fresh medium to ~0.01 of OD600 in an anaerobic glove box. To cultivate ∆crp under anaerobic conditions, TMAO was used as EA.
Mutagenesis and genetic complementation
S. oneidensis in-frame deletion strains were constructed using the att-based Fusion PCR method33. In brief, two fragments flanking the target gene were generated by PCR with primers containing attB and the gene-specific sequence, and then joined by the second round PCR. The fusion fragments were introduced into pHGM01 by site-specific recombination using the BP Clonase (Invitrogen) according to the manufacturer’s instruction. The resulting mutagenesis vectors were transformed into E. coli WM3064 and then transferred into relevant S. oneidensis strains via conjugation. Integration of the mutagenesis constructs into the chromosome were selected by gentamycin resistance and confirmed by PCR. Verified transconjugants were grown in NaCl-less LB broth and plated on LB supplemented with 10% sucrose. Gentamycin-sensitive and sucrose-resistant colonies were screened by PCR for the intended deletion. The mutants were then verified by sequencing.
Mutants from previous studies had been successfully complemented by using either multiple-copy or single-copy (integrative) vectors (Table 1). In this study, the same complementation constructs were used and similar results were obtained as indicated in the relevant figure legends. For newly constructed mutants, plasmids pHG101 and pHG102 were utilized for genetic complementation34. For genes adjacent to their promoter, a fragment containing the coding sequence and its native promoter was generated by PCR and cloned into pHG101. For others, the coding sequence was amplified and inserted into the multiple-cloning site (MCS) of pHG102 under the control of the arcA promoter, which is constitutively active35. After verified by sequencing, the resulting complementation vector was transferred into relevant mutant strains via conjugation.
Extraction and analysis of OM proteins
Envelope fractions were prepared according to an established method with modifications13,36. Briefly, cultures of log phase were normalized to the lowest OD to allow for comparison of OM protein quantities across strain backgrounds. Ten ml normalized culture was pelleted, washed once in 30 mM Tris-HCl (pH 8.1), and pelleted again. Cell pellets were then resuspended in 500 μl 20% sucrose in 30 mM Tris-HCl buffer (pH 8.1), followed by the addition of 20 μL of 20 mg/ml lysozyme in 0.1 mM Ethylenediaminetetraacetic acid (EDTA) (pH 7.3), and incubated on ice for 30 min. Following lysozyme treatment, 1 ml of 3 mM ETDA (pH 7.3) was added and the resulting extract was disrupted with a single 20 s pulse using a microtip sonicator (Haishu Instrument, Ningbo, China). A 1.5 ml fraction of the extract was then centrifuged at 16,000 g for 60 min. Envelope fractions were collected as centrifuged precipitate and resuspended in 30 μl of Laemmli SDS sample buffer. After boiled for 5 min, 15 ul samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE, 10% acrylamide, 4 M urea, 1% SDS). Tandem mass spectrometry (MS/MS) analysis of major OM proteins was carried out essentially the same as described before37.
Antibiotic and nitrite susceptibility assays
The assays were carried out as previously described25,38. In brief, cells of mid-log phase were adjusted to approximately 109 cfu/ml with fresh LB and followed by 10-fold serial dilutions. Five μl of each dilution was spotted onto LB plates containing antibiotics or nitrite of varying concentrations. The plates were incubated for 24 h or longer before being read. For each strain, experiment was performed in triplicate and repeated independently at least three times.
Chemical assays
Mid-log phase cultures were used to inoculate Bis-Tris propane (BTP) buffer (pH 5.8) with goethite as sole EA to ~0.1 of OD600 and incubated under anaerobic condition. Concentrations of Fe (II) of cultures were quantified using ferrozine reagent spectrophotometrically at 562 nm39. Standard curves were made using ferrous sulphate dissolved in 0.5 mol/L hydrochloric acid. Concentrations of nitrite in culture supernatants were measured by a modified Griess assay40.
Identification of the SO_3896 transcriptional start sites
S. oneidensis cells were grown in LB to the mid-log phase, collected by centrifugation, and applied to RNA extraction using the RNeasy minikit (Qiagen, Shanghai) as described before41. RNA was quantified by using a NanoVue spectrophotometry (GE healthcare). The transcriptional start sites of SO_3896 were determined using 5′ rapid amplification of cDNA ends (RACE) according to the manufacturer’s instruction (Invitrogen, Shanghai).
Electrophoretic motility shift assay (EMSA)
Expression and purification of S. oneidensis Crp protein as His-tagged fusion converted from the entire cloneset of S. oneidensis ORFs was described before42,43. The probes used for EMSA were prepared by PCR with 33P end-labeled primers. The binding reaction was performed with ~25–50 fmol (~2–5 nM) of labeled probes and various amounts of protein with or without 10 μM cAMP in 12 μl binding buffer containing 100 mM Tris/HCl (pH 7.4), 20 mM KCl, 10 mM MgCl2, 2 mM DTT, 0.2 μg/μL poly (dI ·dC), and 10% glycerol at 15 °C for 60 minutes and resolved on pre-run 4.8% polyacrylamide native gels44. The band shifts were visualized by autoradiography.
B1H assay
Bacterial one-hydrid (B1H) system was used to investigate DNA-protein interaction in vivo in E. coli cells45. Briefly, plasmid constructs were created by cloning the bait DNA and target Crp into the pBXcmT and pTRG vectors, respectively, and verified by sequencing. The resultant plasmids were used to co-transform BacterioMatch II Validation Reporter Competent Cells on M9 salt agar plates containing 25 mg/ml chloramphenicol and 12.5 mg/ml tetracycline with or without 3-AT. A pair of pBXcmT-Pcyd/pTRG-Crp were used as positive control, and a 300 bp DNA fragment of the 16s rRNA gene promoter (pBXcmT-P16s) was used as negative control46,47. The plates were incubated for 24 h and then moved to room temperature for an additional 16 h (colonies indicating positive interaction usually appeared between 18 and 24 h). The positive interactions were confirmed by streaking colonies on plates containing both 3-AT and streptomycin (12.5 mg/ml).
Expression assay
Expression assay was performed using a single-copy integrative lacZ reporter system as described previously48. DNA fragments of interest were generated by PCR and cloned into the reporter vector pHGEI01, verified by sequencing, and the correct plasmid was then transferred into relevant S. oneidensis strains by conjugation. Once transferred into S. oneidensis strains, pHGEI01 containing promoter of interest integrates into the chromosome and the antibiotic marker is subsequently removed by an established approach38. Cells grown to the mid-log phase under experimental settings described in relevant figure legends were collected, four biological replicates in total, and levels of gene expression were determined by measuring β-Galactosidase activity essentially the same as before38,48. Other analyses. DNA and protein sequence similarity searches were performed using the BLAST program. Sequences of proteins of interest for alignments were obtained from Genbank. Alignments were performed using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo). Signal peptide was predicted using the SignalP program49. The relative intensity of specific protein signals was measured using ImageJ50. Experimental values are subjected to statistical analyses and presented as means ± SD (standard deviation). Student’s t-test was performed for pairwise comparisons of groups.
Other analyses
DNA and protein sequence similarity searches were performed using the BLAST program. Sequences of proteins of interest for alignments were obtained from Genbank. Alignments were performed using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo). Signal peptide was predicted using the SignalP program49. The relative intensity of specific protein signals was measured using ImageJ50. Experimental values are subjected to statistical analyses and presented as means ± SD (standard deviation). Student’s t-test was performed for pairwise comparisons of groups.
Results
Genes SO_3545 and SO_3896 encode two major porins in S. oneidensis
S. oneidensis is predicted to be equipped with a large number of OM β-barrel proteins29 (Table S1). As a first step to assess biological processes in which porins are involved, we determined the profile and abundance of S. oneidensis OM β-barrel proteins. OM fractions were prepared from mid-log phase cells, proteins in these fractions were resolved by SDS-PAGE, and visible bands were excised for protein determination by MS/MS analysis. The identified proteins included five predicted OM β-barrel proteins, SO_1215, SO_1821, SO_3099, SO_3545, and SO_3896 (Fig. 1). The identities of these proteins were confirmed with in-frame deletion mutants for their encoding genes (Fig. 1). SO_3099 is a homologue of E. coli FadL, which forms a ligand-gated channel for passive diffusion of long-chain fatty acids across the OM51. Given that this channel mediates hydrophobic molecules, it is unlikely to be involved in transport of hydrophilic molecules as porins. Apparently, two proteins migrated close to the 40 kDa marker were most abundant. One is SO_3545 (37.4 kDa after signal peptide removal), an OmpA-like porin, which migrated at ~39 kDa (Fig. 1). The discrepancy between migration and residue-based molecular weight is common to OmpA-like porins because of posttranslational modification (PTM)52,53. Sequence analysis revealed that SO_3545 contains a segment ‘LSLGVSYRFGQGE’, which resembles the highly conserved PTM motif established in OmpA porins of other bacteria (Fig. S1). Based on these similarities, we named SO_3545 as OmpA in S. oneidensis. The other is SO_3896, which was recently named as OmpS38 according to its molecular weight after signal peptide removal (37.6 kDa) deduced from the sequence30. The last two identified OM β-barrel proteins, SO_1215 and SO_1821, are produced significantly less than OmpA or OmpS38. Currently, little is known about these two proteins, either biochemically or physiologically.
Figure 1. Major porins in the outer membrane (OM) in S. oneidensis.
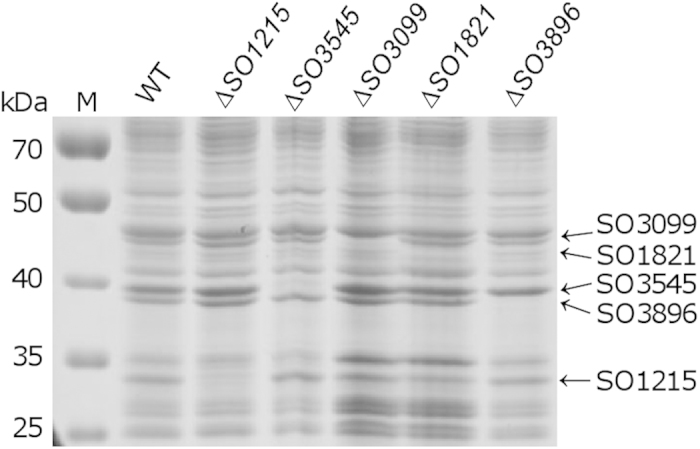
WT represents the wild-type strain. Cells were grown overnight in LB and subcultured 1:200 in fresh medium. OM proteins were extracted and separated on SDS-PAGE. Bands indicated are β-barrel proteins identified by MS/MS analysis and validated by comparing to the corresponding mutant. Experiments were repeated at least three times and representative results were shown.
Major porins may not be subjected to regulation by the EnvZ-OmpR system
There are several mechanisms adopted by bacterial cells to cope with changes in osmolarity of the environment, one of which is through the porin proteins1. We thus examined production of the major porins under various osmolarities. Mid-log phase cultures were adjusted to similar cell densities, the same volume of which (containing cells of the similar number) was subjected to extraction of OM proteins. As shown in Fig. 2A, no significant difference in levels of either OmpS38 or OmpA was visualized, under conditions of both low (0 to 0.5% NaCl) and high (5% NaCl and 5 to 20% sucrose) osmolarities, suggesting that neither OmpS38 nor OmpA responds to changes in osmolarity in a significant manner. To further confirm this, we evaluated the influence of the EnvZ-OmpR system on production of OmpS38 and OmpA. As shown in Fig. 2B, levels of OmpS38 and OmpA were comparable in the ∆envZ∆ompR strain in comparison with the wild-type under all tested conditions. These data manifest that either OmpS38 or OmpA may not be a porin under regulation of the EnvZ-OmpR system.
Figure 2. Neither OmpS38 nor OmpA responds to changes in osmolarity.
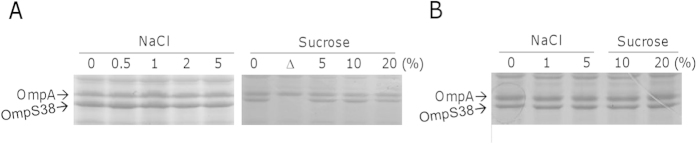
(A) NaCl-free LB media containing NaCl or sucrose at indicated concentrations were used. The level of NaCl in the standard LB is 1%. OM proteins from cells of the similar number were extracted and separated on SDS-PAGE, Similar data were obtained from OmpA. The ompS38 mutant (∆) was included as the control. (B) OM proteins extracted from the ∆envZ∆ompR strain grown under indicated conditions were applied to SDS-PAGE. Representative results were shown.
Major porins appear not important in diffusion of β-lactam antibiotics in S. oneidensis
As nonspecific channels, porins form the pathway through which small, hydrophilic antibiotics, such as β-lactams, diffuse across the OM of Gram-negative bacteria6,54. As a result, loss of major classic and OmpA-like porins generally reduces the susceptibility to certain β-lactam antibiotics55. To test whether OmpS38 and OmpA porins play a significant role in diffusion of β-lactam antibiotics, we assayed the sensitivities of porin mutants to three different β-lactam antibiotics, ampicillin, celfsulodin, and aztreonam, in comparison with the wild-type (Fig. 3). Our previous study revealed that susceptibilities of S. oneidensis to these three antibiotics differ significantly25,26,27. Strains lacking the ompS38 gene were indistinguishable from the wild-type in susceptibilities to all test antibiotics, indicating that OmpS38 is dispensable to diffusion of β-lactam antibiotics. Loss of OmpA resulted in a similar phenotype in general, except for slightly increased sensitivity to cefsulodin. Moreover, we constructed a strain (∆double) lacking both major porins and determined its susceptibility to these antibiotics. We found that the mutant displayed slightly increased sensitivity to ampicillin and cefsulodin (Fig. 3). One of possible explanations would be that loss of these two porins all together may impact the integrity of the OM, leading to faster diffusion of these antibiotics. To test this notion, we evaluated the susceptibility of these mutants to SDS, a detergent that primarily impairs the OM36. The results demonstrated that the lack of OmpA or both porins enhanced SDS sensitivity modestly (Fig. 3), implicating the role of OmpA in maintenance of the OM structure. In contrast, OmpS38 appeared to be negligible. Given that the loss of both major porins does not reduce susceptibilities to tested antibiotics, it is unlikely that these two major porins form the critical pathways through which β-lactams diffuse into the periplasm in S. oneidensis.
Figure 3. Effects of porin mutations on susceptibility to β-lactams and SDS.
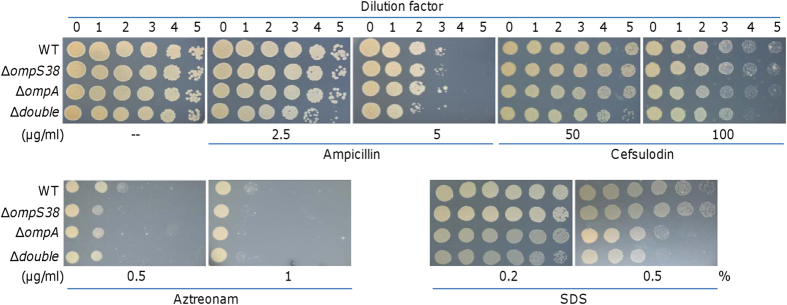
Strains were grown in LB to ~0.4 of OD600. Culture densities were normalized, 10-fold serial dilution were prepared for each, and 5 μl of each dilution was spotted onto LB without or with an chemical agent as indicated. ∆double represents a strain lacking both major porins. Results shown are representative of three independent experiments.
OmpS38 generally affects anaerobic respiration
OmpS38 has been previously implicated a role in respiration of Fe(III), nitrate, and fumarate8. To confirm this and to test whether OmpA also has a role in these processes, we compared abilities of the wild-type, ompS38, and ompA mutant strains to reduce various EAs, including oxygen, fumarate, TMAO, nitrate, ferric citrate, and insoluble FeO(OH). With all tested EAs, the ∆ompA strain behaved the same as the wild-type (Figs 4 and S2), eliminating the possibility that OmpA is involved in respiration of these EAs. In the case of the ∆ompS38 strain, in comparison with the wild-type no difference was observed under aerobic condition (Fig. S2), indicating that OmpS38 has no impact on respiration of oxygen. When fumarate, TMAO, nitrate, ferric citrate, or FeO(OH) was respired, however, the ∆ompS38 strain exhibited a consistent defect, a significant lag in respiration or growth supported by respiration (Figs 4A and S2). These data suggest that the loss of OmpS38 impedes respiration of non-oxygen EAs, at least at the early phase. Notably, the ompS38 mutation impaired fumarate respiration also by reducing respiration rate, leading to a ~1.8–fold difference in generation time, implicating a specific mechanism other than that underlying the growth lag. Investigation into this mechanism is currently underway and will be described as part of a separate report.
Figure 4. OmpS38 is involved in anaerobic respiration.
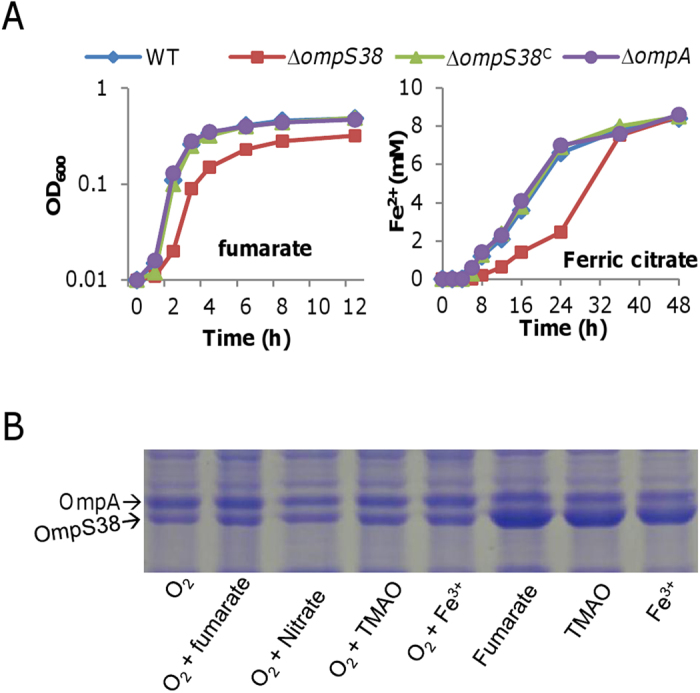
(A) Respiration of various EAs by strains as indicated. Furamate, oxygen, and TMAO (Fig. S2) were evaluated by measuring growth whereas ferric citrate, nitrate, and FeO(OH) (Fig. S2) were estimated by measuring the levels of products as growth supported by these EAs was too low to be reliably monitored. ∆ompS38C represents the mutant carrying a copy of the gene for complementation. Error bars (less than 10% of the average), representing S.D. from three independent experiments, were omitted for clarity. (B) Production of OmpS38 and OmpA under conditions indicated. Cells of mid-log phase grown on indicated EAs were subjected to SDS-PAGE. Presented are representative results of three independent experiments. Production in cultures supported by nitrate under anaerobic conditions was not assayed because growth was too poor.
To examine response of OmpS38 to these EAs, we used SDS-PAGE to assay its production. Compared to cells cultivated aerobically, those grown on fumarate, TMAO, and ferric citrate had significant enhanced production levels, which were indistinguishable from one another (Fig. 4B). Moreover, addition of these EAs to aerobic cultures did not increase OmpS38 production, indicating that oxygen, presumably simply as an EA, overwhelms the effect of these EAs. Overall, these data suggest that OmpS38 affects anaerobic respiration but may not specifically respond to non-oxygen EAs, at least in the context of expression.
Crp is involved in regulation of ompS38
Augmented OmpS38 production under anaerobic conditions implies an association with cellular anaerobic respiration but not with respiration of oxygen EAs. In S. oneidensis, Crp is the most important global regulator mediating adaptation of metabolic modes in response to the availability of electron acceptors35,38,43,56. Coincidently, a Crp-binding site was previously predicted to reside within the sequence upstream of the ompS38 gene, from −144 to −123 relative to the translation initiation code (Fig. 5A), implying that Crp may directly regulate ompS38 transcription35,57. To test this notion, we measured the activity of PompS38 in the crp− background using a markerless integrative lacZ reporter48. A fragment of 414 bp upstream of the coding sequence was amplified and placed in front of the full length E. coli lacZ gene within pHGEI01, and the resulting vector was introduced into the relevant strains. After chromosome integration and the antibiotic marker removal, activities of the promoter (PompS38) in a single copy were assayed (Fig. 5B). The PompS38 activity in the ∆crp strain was substantially reduced, approximately 20% relative to that in the wild-type, indicating that Crp is crucial for ompS38 expression. We then tested whether the ompS38 gene is also under control of Fnr and ArcA, two other global regulators established for mediating respiration in many bacteria58. However, the activity levels of PompS38 were hardly altered in either ∆arcA or ∆fnr mutant compared with the wild-type, suggesting that neither has a significant role in regulation of the ompS38 gene. These observations were supported by SDS-PAGE analysis of major porins (Fig. 5C). In the absence of either ArcA or Fnr, production levels of OmpS38 were statistically insignificant in comparison with the wild-type. In contrast, Crp was nearly essential for OmpS38 production as the porin was barely visible in gels stained with Coomassie Brilliant Blue. To confirm whether the Crp regulation is realized by direct interaction with the sequence upstream of the ompS38 gene, we used EMSA and B1H assays. The reaction mixture for EMSA included DNA fragments upstream of the ompS38 operon covering predicted Crp-binding sites, purified Crp proteins, and 10 μM cAMP, a condition proven to give the best result43. A similar-length fragment upstream of gyrB (encoding DNA gyrase subunit B) was included in the assay as negative control according to the method established previously44. A gel shift band was observed with the ompS38 upstream sequences when 0.5 μM of Crp was added to the reaction mixture and the intensity of the shifted band became stronger with 2 μM of Crp (Fig. 5D). In contrast, the fragments upstream of the gyrB and ompA genes were not affected by Crp. Consistently, in the B1H assay the strong interaction was detected between Crp with the ompS38 segment covering the predicted Crp-binding motif (Table 2). Collectively, these data converge on the idea that Crp is the regulator controlling expression of the ompS38 gene in a direct manner.
Figure 5. Expression of ompS38 is under the control of Crp.
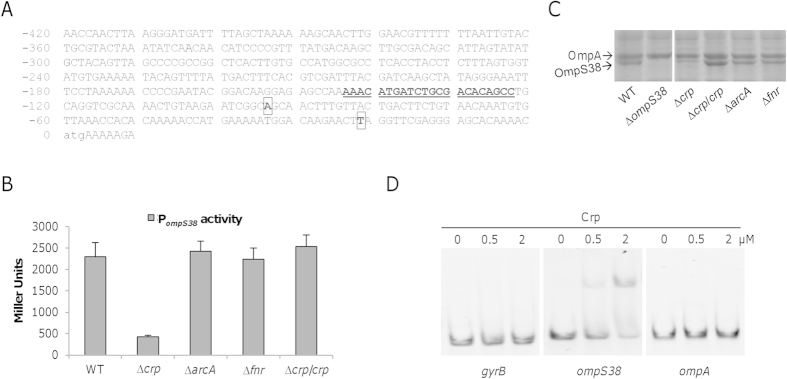
(A) Map of the S. oneidensis ompS38 promoter region. The Crp-binding site was in bold and underlined. The locations of transcription initiation sites are in box, which were determined by 5′-RACE described in the text (next section). (B) Activities of PompS38 in strains indicated. The promoter was placed in front of E. coli lacZ gene and integrated into the chromosome of the strains indicated. Cells of mid-log phase were assayed by measuring β-galactosidase activity. (C) Production of OmpS38 in indicated strains. The lanes shown here were from the same gel, which was given in Fig. S3A. (D) EMSA assay was performed in the presence of 0, 0.5, or 2 μM Crp, 10 μM cAMP, and 2 nM radiolabeled promoter DNA. 0.2 μg/μl poly(dI·dC) was used in all binding reactions to block nonspecific interactions. Promoter region of gyrB was used as negative control. In this figure, experiments were conducted independently at least three times and error bars represent S.D. or the representative was presented.
Table 2. Bacterial one-hybrid (B1H) assay of Crp with various promoters.
| Bait Vector pBXcmT | Target Vector pTRG | Colonies on nonselective platesa | Colonies on selective platesb | Confirmationc |
|---|---|---|---|---|
| /— | /— | 167 | 0 | — |
| /— | /Crp | 176 | 0 | — |
| /Pcyd | /Crp | 203 | 175 | 172 |
| /Pcyd | /— | 163 | 0 | — |
| /— | /Crp | 188 | 0 | — |
| /P16S | /Crp | 156 | 1 | 0 |
| /PompS38 | /Crp | 144 | 132 | 128 |
| /PompA | /Crp | 138 | 1 | 0 |
aM9 agar + 25 μg/ml chloramphenicol + 12.5 μg/ml tetracycline.
ba + 5 mM 3-AT.
cb + 12.5 μg/ml streptomycin.
Transcription of ompS38 is under control of two promoters
While loss of Crp results in substantially reduced production of OmpS38, the ompS38 gene is still evidently expressed as revealed by both the lacZ-reporter and SDS-PAGE. To investigate the source for this expression, we constructed a series of mutants carrying various truncations within the region upstream of the ompS38 coding sequence, including ∆414–258, ∆414–203, ∆414–101, and ∆414–55 (for reference, ∆414–258 represents the mutant lacking base-pairs from −414 to −259 relative to the ompS38 gene) (Fig. 6A). The abundance of OmpS38 produced in these mutants was analyzed with SDS-PAGE (Fig. 6B). As expected, ∆414–101 and ∆414–55, in which the Crp-binding motif remains, produced OmpS38 at levels similar to that from the ∆crp strain. While the protein was visible in the wild-type, ∆414–258, and ∆414–203, the abundance in the former two apparently exceeded that in ∆414–203, implying that the promoter P202 (a fragment covering 202 bp upstream of the ompS38 gene) is impaired with respect to activity. To confirm this, we placed these segments in front of the E. coli lacZ gene and measured their activities (Fig. 6A). As shown in Fig. 6C, P414 (from WT, 414 bp upstream of the ompS38 gene) and P259 (from mutant ∆414–258) behaved identically in the the wild-type and ∆crp strains, indicating that these two segments are sufficiently long to support full-scale expression. Consistent with SDS-PAGE data in Fig. 6B, the promoter P202 had significantly reduced activity, approximately 70% relative to P414 and P259 when Crp was present. In the absence of Crp, the activity of the ompS38 promoter within all segments tested was reduced to levels that were indistinguishable. Given that the Crp-binding motif remains, the segment probably loses another regulatory element, which is included in P414 and P259. In the case of P100 and P54, while activating effect of Crp mostly vanished, both segments drove the lacZ expression at levels lower than P414, P259, and P202 without Crp. The further reduction in activity is probably due to the complete loss of the Crp-dependent promoter. Nevertheless, the activity of approximately 300 Miller units was still obtained, which was not affected by Crp (Fig. 6C). These data together imply the presence of a weak promoter that is independent of Crp.
Figure 6. Transcription of ompS38 initiates from two sites.
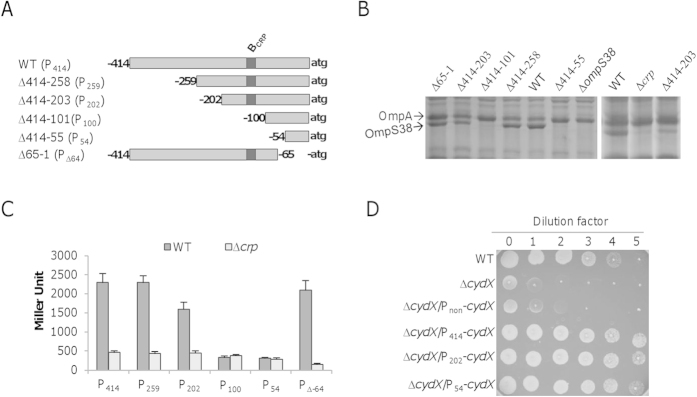
(A) Constructs of the S. oneidensis ompS38 promoter mutants. Constructs were named either as strains for OmpS38 production analysis shown in (B) or promoters for the lacZ reporter analysis shown in (C). For instance, ∆414-203 refers to a mutant lacking base-pairs from -414 to -203 relative to the ompS38 gene and P202 means the resulting segment (from -202 to -1) in ∆414-203 is used to drive the lacZ gene. P414 refers to the wild-type promoter PompS38 in Fig. 5, covering base-pairs from -414 to -1. (B) SDS-PAGE analysis of OM proteins in strains indicated. Gels shown here were run under the same experimental conditions. (C) Expression analysis of promoters indicated in WT and ∆crp strains. (D) Nitrite susceptibility assay of strains indicated. Cells were prepared as described in Fig. 3A. ∆cydX is hypersensitive to nitrite. Pnon, no promoter in front of cydX. Experiments were conducted independently at least three times and error bars represent S.D. or the representative was presented.
To provide evidence for the presence of the second ompS38 promoter, we took advantage of our previous finding that the cytochrome bd oxidase confers S. oneidensis nitrite resistance38. Given that the promoter for the bd oxidase operon is approximately 200 Miller units, the proposed Crp-independent promoter seems sufficiently strong. We used P54 to drive expression of the cydX gene, which encodes the third functionally essential subunit of the bd oxidase (in addition to CydA and CydB)59. As shown in Fig. 6D, the cydX gene under the control of P54 restored the nitrite resistance of the ∆cydX strain, a scenario also obtained with P414 and P202. Given that the promoterless control vector failed to do so, it is clear that there is a promoter within 54 bp upstream of the ompS38 gene. The presence of two promoters was then validated by 5′-RACE, as marked in Fig. 5A. The main start site (Crp-dependent) mapped to nucleotide −94 and represented 88% of the positive clones that were identified by 5′-RACE. A second site mapped to nucleotide −22. In support of these results, we constructed P∆64, which covers the segment from −65 to −414, and used it to drive ompS38 expression (Fig. 6A). Based on values of protein levels and β-galactosidase activity (Fig. 6B,C), it is apparent that this promoter within the segment is strong, comparable to that within P414 and P259. Altogether, these data conclude that transcription of the ompS38 gene initiates from two sites.
Fur is involved in regulation of ompS38
The difference in ompS38 expression levels between ∆414–203 (P202) and WT (P414) implies that the gene is subjected to additional regulation. In Vibrio cholerae, expression of major porin gene ompT is positively regulated through the direct binding by Fur, the global transcriptional regulator for iron-dependent expression of many genes60,61. Although a Fur-binding motif is not identified within the ompS38 upstream sequence by bioinformatics prediction57, we made attempts to investigate whether the ompS38 gene is transcriptionally influenced by Fur given that V. cholerae and S. oneidensis are most closely related in phylogeny, sharing extensive regions of similar gene order29. To this end, a ∆fur strain was constructed, which was validated by exhibiting a growth defect that was in excellent agreement with the results of a previous study and genetic complementation (data not shown)62. The influence of the Fur loss on production of OmpS38 was evident, approximately 55% relative to that in the wild-type, manifesting that the regulator functions as an activator (Fig. 7A). To examine whether the impact of Fur on expression of the ompS38 gene is affected by intracellular iron levels, we assayed OmpS38 levels. Unlike iron in excess (Fig. 4), iron in scarcity reduced production of OmpS38, to approximately 60% with iron-chelator dipyridyl at 50 μM (Fig. 7A). However, the negative effect of dipyridyl on OmpS38 production appeared additive to that resulting from the loss of Fur (Fig. 7B). In contrast, addition of exogenous iron did not interfere with the OmpS38 production in the absence of Fur. Consistent results were obtained from the lacZ reporter (Fig. 7C). Expression of ompS38 was not affected by the chelator in the ∆crp mutant, indicating that the loss of Crp overwhelms overall regulation. Changes in iron levels by adding dipyridyl but not exogenous iron had evident effect on the expression in the wild-type. At 50 μM, the chelator induced ∼50% reduction. A similar trend was observed in the ∆fur mutant. These data, collectively, suggest a possibility that the chelator and Fur function independently from each other.
Figure 7. Fur is involved in regulation of ompS38.
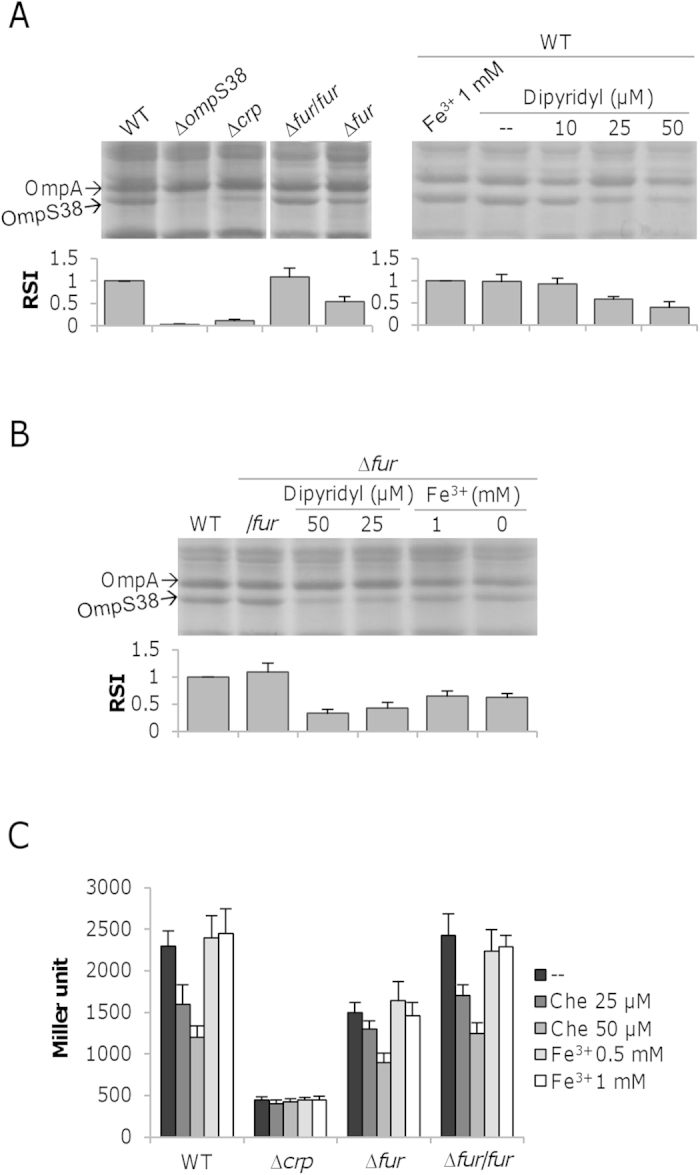
(A) SDS-PAGE analysis of OM proteins in strains indicated. The assay included the wild-type strain with iron at different levels, which were achieved by adding FeCl3 or iron-chelator dipyridyl. The lanes shown here were from the same gel, which was given in Fig. S3B. (B) SDS-PAGE analysis of OM proteins in the ∆fur strain under conditions indicated. In both (A,B), relative signal intensity (RSI) of OmpS38 in the indicated samples was estimated using software Image J. The averaged value for the wild-type in each gel was set to 1 as the standard, to which values of other samples were normalized. The values are the mean ± SD (error bars) (n = 4). (C) Expression analysis of PompS38 in strains under conditions indicated. Che represents chelator dipyridyl. Experiments were conducted independently at least three times and error bars represent S.D. or the representative was presented.
Discussion
The purpose of this study was to identify major porins and to explore how they influence the physiology in S. oneidensis. Our previous study had shown that the microorganism has an EnvZ-OmpR TCS, which is highly homologous to the E. coli counterpart31. Given that the system is conserved in regulation of osmolarity, we anticipated that the most abundant porins of S. oneidensis may share characteristics of E. coli OmpF and OmpC. We were surprised when this was not the case. Among five predicted β-barrel proteins revealed by combining MS/MS and mutational analyses, OmpS38 and OmpA are the most abundant but seemingly not responsive to either changes in osmolarity or the TCS. In E. coli, OmpA is estimated to present at ~100 000 copies per cell, making it a major OM protein crucial to cell structure and integrity63. Accordingly, it is not surprising that OmpA assumes the similar task in S. oneidensis. OmpS38 is certain to be a classic porin with 12–22 transmembrane segments as the most closely related proteins to this protein are all such porins, based on sequence similarities8. Additionally, the protein contains the conserved membrane core domain and variable loops typical of porins1. In addition to non-responsive to changes in osmolarity, the presented data showed that these two major porins are largely dispensable in allowing the entrance of β-lactam antibiotics, a function implicated in both classic and OmpA-like porins in some bacteria7,55. With respect to maintenance of the OM structure, OmpA is implicated to some extent whereas the significance of OmpS38 is negligible.
OM components are no doubt important in respiration of non-oxygen EAs, especially insoluble metal species. The OM c-type cytochromes complex MtrABC plays a central role in reduction of extracellular EAs acting not only as the reductase by direct contact but also as the electron station for cycling extracellular electron shuttles23,64. But how porins impact anaerobic respiration is unclear. Similar to OmpS38, a major porin OmpJ of ~49 kDa in Geobacter sulfurreducens was found to be essential for respiration of insoluble Fe (III) and Mn (IV) oxides9. Given that both OmpS38 and OmpJ lack any apparent electron transfer moieties, it is unlikely that they are involved in the final electron transfer to extracellular EAs. One possibility is that these proteins function as MtrB, which is a non-cytochrome component of the metal reduction complex. By forming a trans-OM pore of 3–4 nm in diameter65, MtrB allows electron transporter cytochrome c MtrA to span the OM such that electrons can be transferred from the periplasm to reductase MtrC, which is located on the surface of the OM (Ross et al. 2007). This mechanism for microbe-to-mineral electron transfer is termed as the ‘porin-cytochrome’ model66. In S. oneidensis, equally possible is that OmpS38 promotes indirect electron transfer mediated by electron shuttles such as riboflavin, another mechanism for extracellular electron transfer in this bacterium21,22. This notion gains support from a finding that E. coli cells producing the synthetic large pore porin protein are able to efficiently use riboflavin as the electron shuttle, resulting in significant improvement in extracellular electron transfer11.
The mechanism by which OmpS38 affects growth on and/or respiration of some soluble EAs, such as fumarate, nitrate, and TMAO, is not clear. Apparently, the proposal used to explain the defect in respiration of insoluble EAs does not work as these soluble EAs are reduced in the periplasm. It is possible that OmpS38 forms the major passage for extracellular small molecules across the OM to enter the periplasm. Although generally regarded to be non-specific, porins are in fact somewhat selective for small molecules, especially charged ones5,67. Thus, the OmpS38 channel may be particularly important for fumarate, leading to the substantial defect in growth on the EA in its absence. We are currently testing this possibility.
Although we have little knowledge of the mechanism by which OmpS38 is involved in anaerobic respiration, in the presented study we unraveled why expression of the porin is elevated under anaerobic conditions. In S. oneidensis, Crp, rather than Fnr or the Arc system, serves as the major protein mediating switch between aerobic and anaerobic respiration35,56. As activity of Crp relies on the cellular cAMP levels, which is higher under anaerobic conditions, genes under positive control of Crp are usually transcribed at augmented levels in the presence of oxygen43,68. This explains that increased expression of OmpS38 is consistently observed from cells grown on all non-oxygen EAs tested. Regulation by Crp on expression of major porins in a direct manner has been reported before in V. cholerae and Yersinia pestis69,70. However, in these enteric bacteria Crp functions as a prominent global regulator that controls the expression of an array of genes mainly in response to carbon and energy sources in the environment71. How these evolutionally diverse bacteria adopt the same strategy to regulate expression of major porins is of great interest.
OmpS38 is produced at reduced levels in the absence of Fur, resembling major porin OmpT of some V. cholerae strains61. Meanwhile, expression of these two porins is significantly reduced when iron is limited, suggesting a similar response to iron levels. However, differences are found. In contrast to that increased OmpT by addition of exogenous iron relies on Fur, the S. oneidensis regulator seems to exert positive regulation by a mechanism that is independent of iron levels. This notion is supported by a previous finding that a large number of S. oneidensis genes involved in energy metabolism are iron-responsive but Fur-independent62. Moreover, OmpS38, unlike OmpT, is probably not under direct control of Fur given the missing of a Fur-binding motif (Fur box) within its upstream region57,72.
The significance of OmpS38 expression controlled by two independent promoters is not clear. Apparently, transcription occurs from both promoters simultaneously, evidenced in the crp mutant strain in which the additive effect is observed. However, given that the Crp-independent promoter is substantially weaker than the Crp-dependent one, the contribution of the former to overall expression is conceivably minor. Nonetheless, because the Crp-independent promoter is seemingly constitutive, it is thus tempting to speculate that the low levels of the porin might be critical under yet unknown conditions. A goal for future experiments will be to investigate role of these promoters and to specify conditions under which their activity is required and/or altered.
Additional Information
How to cite this article: Gao, T. et al. Positive regulation of the Shewanella oneidensis OmpS38, a major porin facilitating anaerobic respiration, by Crp and Fur. Sci. Rep. 5, 14263; doi: 10.1038/srep14263 (2015).
Supplementary Material
Acknowledgments
This research was supported by National Natural Science Foundation of China (31270097, 41476105), Major State Basic Research Development Program (973 Program: 2010CB833803), and the Fundamental Research Funds for the Central Universities.
Footnotes
Author Contributions H.G. conceived the idea and designed the project. T.G., L.J. and J.Y. carried out the experiments. T.G., L.J. and H.G. analyzed data. T.G., L.J. and H.G. wrote the paper.
References
- Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeth K. & Thein M. Porins in prokaryotes and eukaryotes: Common themes and variations. Biochem. J. 431, 13–22 (2010). [DOI] [PubMed] [Google Scholar]
- Wimley W. C. The versatile β-barrel membrane protein. Curr. Opin. Struct. Biol. 13, 404–411 (2003). [DOI] [PubMed] [Google Scholar]
- Hong H., Patel D. R., Tamm L. K. & van den Berg B. The outer membrane protein OmpW forms an eight-stranded β-barrel with a hydrophobic channel. J. Biol. Chem. 281, 7568–7577 (2006). [DOI] [PubMed] [Google Scholar]
- Koebnik R., Locher K. P. & Van Gelder P. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37, 239–253 (2000). [DOI] [PubMed] [Google Scholar]
- Kojima S. & Nikaido H. Permeation rates of penicillins indicate that Escherichia coli porins function principally as nonspecific channels. Proc. Natl Acad. Sci. USA 110, E2629–E2634 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pages J.-M., James C. E. & Winterhalter M. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat. Rev. Micro. 6, 893–903 (2008). [DOI] [PubMed] [Google Scholar]
- Maier T. & Myers C. The outer membrane protein Omp35 affects the reduction of Fe(III), nitrate, and fumarate by Shewanella oneidensis MR-1. BMC Microbiol. 4, 23 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afkar E., Reguera G., Schiffer M. & Lovley D. A novel Geobacteraceae-specific outer membrane protein J (OmpJ) is essential for electron transport to Fe (III) and Mn (IV) oxides in Geobacter sulfurreducens. BMC Microbiol. 5, 41 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duperthuy M. et al. Use of OmpU porins for attachment and invasion of Crassostrea gigas immune cells by the oyster pathogen Vibrio splendidus. Proc. Natl Acad. Sci. USA 108, 2993–2998 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong Y.-C. et al. Enhancement of extracellular electron transfer and bioelectricity output by synthetic porin. Biotechnol. Bioeng. 110, 408–416 (2013). [DOI] [PubMed] [Google Scholar]
- Rumbo C. et al. The Acinetobacter baumannii Omp33-36 porin is a virulence factor that induces apoptosis and modulates autophagy in human cells. Infect. Immuni. 82, 4666–4680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slauch J. M. & Silhavy T. J. Genetic analysis of the switch that controls porin gene expression in Escherichia coli K-12. J. Mol. Biol. 210, 281–292 (1989). [DOI] [PubMed] [Google Scholar]
- Oshima T. et al. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol. Microbiol. 46, 281–291 (2002). [DOI] [PubMed] [Google Scholar]
- Wang L. C., Morgan L. K., Godakumbura P., Kenney L. J. & Anand G. S. The inner membrane histidine kinase EnvZ senses osmolality via helix-coil transitions in the cytoplasm. EMBO J. 31, 2648–2659 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Cruz M. A. & Calva E. The complexities of porin genetic regulation. J. Mol. Microbiol. Biotechnol. 18, 24–36 (2010). [DOI] [PubMed] [Google Scholar]
- Vogel J., & Papenfort K. Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol. 9, 605–611 (2006). [DOI] [PubMed] [Google Scholar]
- Fredrickson J. K. et al. Towards environmental systems biology of Shewanella. Nat. Rev. Micro. 6, 592–603 (2008). [DOI] [PubMed] [Google Scholar]
- Logan B. E. Exoelectrogenic bacteria that power microbial fuel cells. Nat. Rev. Micro. 7, 375–381 (2009). [DOI] [PubMed] [Google Scholar]
- Aracic S., Semenec L. & Franks A. E. Investigating microbial activities of electrode-associated microorganisms in real-time. Front. Microbiol. 5, 663 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsili E. et al. Shewanella secretes flavins that mediate extracellular electron transfer. Proc. Natl Acad. Sci. USA 105, 3968–3973 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloski N. J. & Gralnick J. A. Flavin electron shuttles dominate extracellular electron transfer by Shewanella oneidensis. mBio 4, e00553–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirbadian S. et al. Shewanella oneidensis MR-1 nanowires are outer membrane and periplasmic extensions of the extracellular electron transport components. Proc. Natl Acad. Sci. USA 111, 12883–12888 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Héritier C. & Nordmann P. Chromosome-encoded Ambler class D β-lactamase of Shewanella oneidensis as a progenitor of carbapenem-hydrolyzing oxacillinase. Antimicro. Agents Chemother. 48, 348–351 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J. et al. Expression of blaA underlies unexpected ampicillin-induced cell lysis of Shewanella oneidensis. PLoS ONE 8, e60460 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J. et al. Distinct roles of major peptidoglycan recycling enzymes in β-lactamase production in Shewanella oneidensis. Antimicro. Agents Chemother. 58, 6536–6543 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Sun Y., Mao Y., Jin M. & Gao H. PBP1a/LpoA but not PBP1b/LpoB are involved in regulation of the major β-lactamase gene blaA in Shewanella oneidensis. Antimicro. Agents Chemother. 10.1128/AAC.04669-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda J. M. & Abbott S. L. The genus Shewanella: from the briny depths below to human pathogen. Crit. Rev. Microbiol. 40, 293–312 (2014). [DOI] [PubMed] [Google Scholar]
- Heidelberg J. F. et al. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotech. 20, 1118–1123 (2002). [DOI] [PubMed] [Google Scholar]
- Yin J. et al. Regulation of nitrite resistance of the cytochrome cbb3 oxidase by cytochrome c ScyA in Shewanella oneidensis. MicrobiologyOpen 4, 84–99 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Wei B., Shi M. & Gao H. Functional assessment of EnvZ/OmpR two-component system in Shewanella oneidensis. PLoS ONE 6, e23701 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Wan F., Mao Y. & Gao H. Unraveling the mechanism for the viability deficiency of Shewanella oneidensis oxyR null mutant. J. Bacteriol. 10.1128/JB00154-15. (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M. et al. Unique organizational and functional features of the cytochrome c maturation system in Shewanella oneidensis. PLoS ONE 8, e75610 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Wang J., Tang P., Chen H. & Gao H. Genetic and molecular characterization of flagellar assembly in Shewanella oneidensis. PLoS ONE 6, e21479 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H. et al. Physiological roles of ArcA, Crp, and EtrA and their interactive control on aerobic and anaerobic respiration in Shewanella oneidensis. PLoS ONE 5, e15295 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan F. et al. Impaired cell envelope resulting from arcA mutation largely accounts for enhanced sensitivity to hydrogen peroxide in Shewanella oneidensis. Sci. Rep. 5, 10228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L. et al. Posttranslational modification of flagellin FlaB in Shewanella oneidensis. J. Bacteriol. 195, 2550–2561 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H. et al. Crp-dependent cytochrome bd oxidase confers nitrite resistance to Shewanella oneidensis. Environ. Microbiol. 15, 2198–2212 (2013). [DOI] [PubMed] [Google Scholar]
- Viollier E., Inglett P. W., Hunter K., Roychoudhury A. N. & van Cappellen P. The ferrozine method revisited: Fe(II)/Fe(III) determination in natural waters. Appl. Geochem. 15, 785–790 (2000). [Google Scholar]
- Miranda K. M., Espey M. G. & Wink D. A. A rapid, simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide 5, 62–71 (2001). [DOI] [PubMed] [Google Scholar]
- Gao H. et al. Global transcriptome analysis of the heat shock response of Shewanella oneidensis. J. Bacteriol. 186, 7796–7803 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H. et al. Generation and validation of a Shewanella oneidensis MR-1 clone set for protein expression and phage display. PLoS ONE 3, e2983 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G. et al. Combined effect of loss of the caa3 oxidase and Crp regulation drives Shewanella to thrive in redox-stratified environments. ISME J. 7, 1752–1763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Wang X., Yang Z., Palzkill T. & Zhou J. Probing regulon of ArcA in Shewanella oneidensis MR-1 by integrated genomic analyses. BMC Genomics 9, 42 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M. et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 19, 1301–1308 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. et al. Protection from oxidative stress relies mainly on derepression of OxyR-dependent KatB and Dps in Shewanella oneidensis. J. Bacteriol. 196, 445–458. (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Luo Q., Jiang Y., Wu G. & Gao H. Managing oxidative stresses in Shewanella oneidensis: intertwined roles of the OxyR and OhrR regulons. Environ. Microbiol. 16, 1821–1834 (2014). [DOI] [PubMed] [Google Scholar]
- Fu H., Jin M., Ju L., Mao Y. & Gao H. Evidence for function overlapping of CymA and the cytochrome bc1 complex in the Shewanella oneidensis nitrate and nitrite respiration. Environ. Microbiol. 16, 3181–3195 (2014). [DOI] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G. & Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Meth. 8, 785–786 (2011). [DOI] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S. & Eliceiri K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Meth. 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepore B. W. et al. Ligand-gated diffusion across the bacterial outer membrane. Proc. Natl Acad. Sci. USA 108, 10121–10126 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian M., Fuerst M. M., Shabalin Y. & Reusch R. N. Sorting signal of Escherichia coli OmpA is modified by oligo-(R)-3-hydroxybutyrate. Biochim. Biophys. Acta 1768, 2660–2666 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharian E. & Reusch R. N. Haemophilus influenzae outer membrane protein P5 is associated with inorganic polyphosphate and polyhydroxybutyrate. Biophys. J. 92, 588–593 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández L. & Hancock R. E. W. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 25, 661–681 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara E. & Nikaido H. OmpA is the principal nonspecific slow porin of Acinetobacter baumannii. J. Bacteriol. 194, 4089–4096 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffarini D. A., Schultz R. & Beliaev A. Involvement of cyclic AMP (cAMP) and cAMP receptor protein in anaerobic respiration of Shewanella oneidensis. J. Bacteriol. 185, 3668–3671 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novichkov P. S. et al. RegPrecise 3.0—A resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genomics 14, 745 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J. & Paget M. S. Bacterial redox sensors. Nat Rev Micro 2, 954–966 (2004). [DOI] [PubMed] [Google Scholar]
- Chen H., Luo Q., Yin J., Gao T. & Gao H. Evidence for the requirement of CydX in function but not assembly of the cytochrome bd oxidase in Shewanella oneidensis. Biochim. Biophys. Acta 1850, 318–328 (2015). [DOI] [PubMed] [Google Scholar]
- Andrews S. C., Robinson A. K. & Rodríguez-Quiñones F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237 (2003). [DOI] [PubMed] [Google Scholar]
- Craig S. A., Carpenter C. D., Mey A. R., Wyckoff E. E. & Payne S. M. Positive regulation of the Vibrio cholerae porin OmpT by iron and Fur. J. Bacteriol. 193, 6505–6511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. et al. Characterization of the Shewanella oneidensis Fur gene: roles in iron and acid tolerance response. BMC Genomics 9, S11 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusch R. N. Insights into the structure and assembly of Escherichia coli outer membrane protein A. FEBS J. 279, 894–909 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto A. et al. Cell-secreted flavins bound to membrane cytochromes dictate electron transfer reactions to surfaces with diverse charge and pH. Sci. Rep. 4, 5628 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartshorne R. S. et al. Characterization of an electron conduit between bacteria and the extracellular environment. Proc. Natl Acad. Sci. USA 106, 22169–22174 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson D. J. et al. The ‘porin–cytochrome’ model for microbe-to-mineral electron transfer. Mol. Microbiol. 85, 201–212 (2012). [DOI] [PubMed] [Google Scholar]
- García-Giménez E., Alcaraz A. & Aguilella V. M. Divalent metal ion transport across large biological ion channels and their effect on conductance and selectivity. Biochem. Res. Inter. art. 245786, 10.1155/2012/245786 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charania M. A. et al. Involvement of a membrane-bound class III adenylate cyclase in regulation of anaerobic respiration in Shewanella oneidensis MR-1. J. Bacteriol. 191, 4298–4306 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. C., Merrell D. S., Camilli A. & Kaper J. B. ToxR interferes with CRP-dependent transcriptional activation of ompT in Vibrio cholerae. Mol. Microbiol. 43, 1577–1589 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H. et al. Regulatory effects of cAMP receptor protein (CRP) on porin genes and its own gene in Yersinia pestis. BMC Microbiol. 11, 40 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorke B., & Stulke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Micro. 6, 613–624 (2008). [DOI] [PubMed] [Google Scholar]
- Seo S. W. et al. Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat. Commun. 5, 4910 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


