Abstract
In order to investigate the association between the iNOS gene polymorphisms and susceptibility to cancer, a search of English papers was done using Pubmed, the Cochrane Library, Embase, ISI Web of Science, Google (scholar) database, and all Chinese reports were conducted using CBMDisc, Chongqing VIP database, and CNKI database. A total of eight studies were included in this meta-analysis including 1,920 cases and 2,373 controls. The results indicated that the polymorphisms in iNOS gene (C150T(Ser608 Leu) polymorphism and polymorphic (CCTTT)n repeats) had no association with cancer risk for all genetic models. This meta-analysis suggested that the polymorphisms in the iNOS gene were not associated with cancer risk.
It has been widely accepted that cancer was one of the important causes of dead all over the world, characterized by angiogenesis and inflamation1. To date, the etiology of cancer remains unknown and disease-modifying medical treatments are limited. Nevertheless, since the involvement of cytokines in cancer was hypothesized2, there were a lot of candidate genes approach in a case-control study of single nucleotide polymorphisms (SNPs) including inducible nitric oxide synthase (iNOS)3,4,5,6,7,8,9,10.
According to previous records, iNOS has played an important role in angiogenesis, stimulation of proliferation, and inhibition of apoptosis11. Previous research indicated that the iNOS gene might be involved in the development of cancer through disrupting carcinogenesis12. The chromosome allocation of iNOS is 17q11.2, which has a genomic size of 48 kb and encodes a protein of 131 kDa13. There were some known iNOS gene polymorphisms such as C150T (Ser608 Leu) polymorphisms and polymorphic (CCTTT)n repeats.
Thus far, previous studies concerning association between iNOS gene polymorphism and risk of cancer are limited and rather conflicting3,4,5,6,7,8,9,10. Because lack the evidence to provide a reliable conclusion in a single study, we conducted a meta-analysis on these eligible studies, to evaluate the strength relationship between the iNOS gene polymorphism and risk for cancer, which would have a much greater possibility of reaching reasonably strong conclusions.
Results
Study inclusion and characteristics
The process of the search was shown in Fig. 1. After a literature search, eight studies3,4,5,6,7,8,9,10(six studies3,4,6,7,8,9 with 1512 cases and 1942 controls for C150T (Ser608 Leu) polymorphism and two studies5,10 with 408 cases and 431 controls for polymorphic (CCTTT)n repeats) were included in this meta-analysis. The identified studies and their main characteristics were shown in Table 1 and Table 2. Genotype distribution of any polymorphism did not differ from HWE within control groups.
Figure 1.
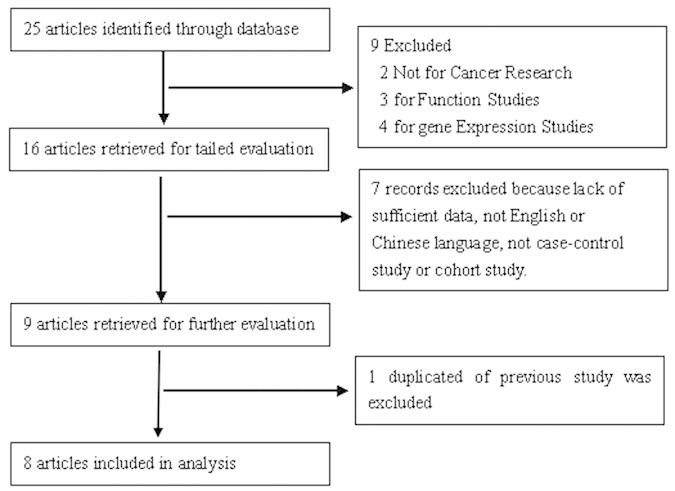
Flow chart demonstrating those studies that were processed for inclusion in the meta-analysis.
Table 1. Characteristics of the Included Studies for Meta-analysis.
| First Author | PublicationYear | Location | Ethnicity | Histology | GenotypingMethod | InosPolymorphism | Cases(n) | Controls(n) | Cconflicts |
|---|---|---|---|---|---|---|---|---|---|
| Rafiei A3 | 2012 | Iran | Asian | Gastric cancer | PCR | C150T(Ser608 Leu) | 159 | 170 | Significant association |
| Ferguson HR4 | 2008 | Ireland | Caucasian | Esophageal adenocarcinoma | TaqMan | C150T(Ser608 Leu) | 209 | 248 | No significant association |
| Shen CH5 | 2007 | Taiwan | Asian | Bladder carcinoma | PCR | (CCTTT)n | 250 | 250 | Significant association |
| Lee TS7 | 2007 | Korea | Asian | Cervical cancer | PCR | C150T(Ser608 Leu) | 176 | 172 | No significant association |
| Li C9 | 2007 | USA | Caucasian | Cutaneous melanoma | PCR | C150T(Ser608 Leu) | 602 | 603 | No significant association |
| Goto Y8 | 2006 | Japan | Asian | Gastric cancer | PCR-RFLP | C150T | 201 | 454 | Significant association |
| Tatemichi M10 | 2005 | Japan | Asian | Gastric cancer | PCR | (CCTTT)n | 158 | 181 | Significant association |
| Shen J6 | 2004 | China | Asian | Gastric cancer | PCR | C150T(Ser608 Leu) | 165 | 295 | Significant association |
PCR: polymerase chain reaction
Table 2. Distributions of the inducible nitric oxide synthase Genotype and Allele among Cases and Controls.
| Distribution of C150T(Ser608 Leu)genotypes Case/comtrol(n) |
Frequency of C150T(Ser608Leu)allelesCase/comtrol(n) |
Distribution of (CCTTT)nrepeats *genotypesCase/comtrol(n) |
Frequency of (CCTTT)n repeats*alleles Case/comtrol(n) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| first author | CC | CT | TT | HWE forcontrol | C | T | first author | SS | SL | LL | HWE forcontrol | S | L |
| Ferguson HR4 | 148/167 | 55/72 | 6/9 | 0.72 | 351/406 | 67/90 | Shen CH5 | 45/56 | 113/110 | 87/79 | 0.14 | 203/222 | 287/268 |
| Lee TS7 | 135/115 | NA | NA | - | NA | NA | Tatemichi M10 | 60/82 | 75/78 | 23/21 | 0.71 | 195/242 | 121/120 |
| Li C9 | 404/393 | 184/187 | 14/23 | 0.89 | 992/973 | 212/233 | - | - | - | - | - | - | - |
| Shen J6 | 108/189 | 33/49 | 2/8 | 0.06 | 249/427 | 37/65 | - | - | - | - | - | - | - |
| Rafiei A3 | 89/92 | 59/72 | 11/6 | 0.07 | 237/256 | 81/84 | - | - | - | - | - | - | - |
| Goto Y8 | 175/403 | 25/48 | 1/3 | 0.24 | 375/854 | 27/54 | - | - | - | - | - | - | - |
NA/-: Not applicable. HWE: Hardy-Weinberg equilibrium. *repeat numbers divided into two groups: S (9-11repeats) and L (12-18repeats).
Quantitative data synthesis
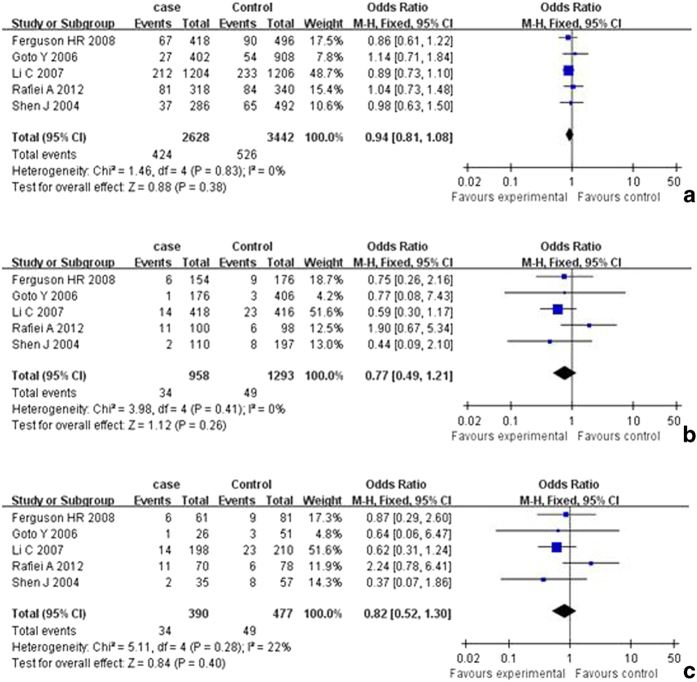
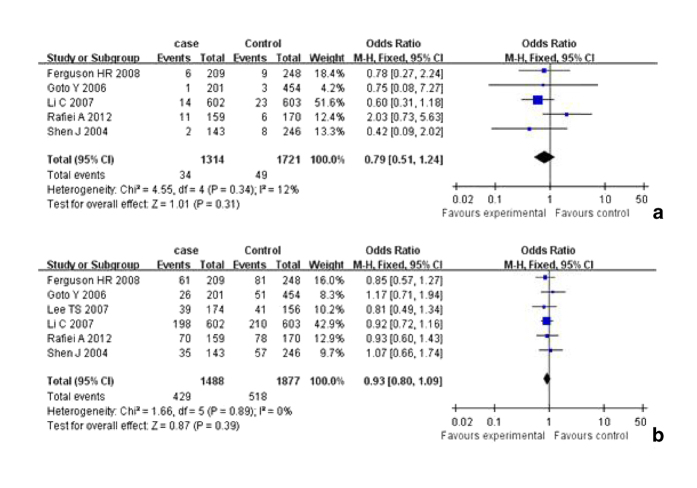
The results of analysis for different polymorphisms by Revman software was shown in Fig. 2 and Fig. 3. As shown in Table 3, the results indicated that there was no association between polymorphisms in the iNOS gene including C150T (Ser608 Leu) polymorphisms and polymorphic (CCTTT)n repeats and risk of cancer. In subgroup analysis for C150T (Ser608 Leu) polymorphism by ethnicity (OR = 1.04, 95% CI = 0.82-1.32 for T vs. C; OR = 1.10, 95% CI = 0.52-2.33 for TT vs. CC; OR = 1.13, 95% CI = 0.53-2.41 for TT vs. CT; OR = 1.14, 95% CI = 0.54-2.38 for recessive model; OR = 0.98; 95% CI = 0.77-1.25 for dominant model) among Asians. There was no significant publication bias according to funnel plot (Fig. 4) and Egger’s test (p = 0.65).
Figure 2.
a. Forest plot of the association between cancer and the C150T(Ser608 Leu) mutation (T vs C). b. Forest plot of the association between cancer and the C150T(Ser608 Leu) mutation (TT vs CC). c. Forest plot of the association between cancer and the C150T(Ser608 Leu) mutation (TT vs CT).
Figure 3.
a. Forest plot of the association between cancer and the C150T(Ser608 Leu) mutation (TT vs CT+CC). b. Forest plot of the association between cancer and the C150T(Ser608 Leu) mutation (CT+TT vs CC).
Table 3. Summary ORs and 95% CI of the C150T(Ser608 Leu) Polymorphism and the polymorphic (CCTTT)n repeats in the inducible nitric oxide synthase Gene and Cancer Risk.
| Statistical models | Genotype/Allele | Numberof Study | OR | 95%CI | I2% | P | Z | P forZ test |
|---|---|---|---|---|---|---|---|---|
| C150T(Ser608 Leu) | ||||||||
| Allele Model | T vs. C | 5 | 0.94 | 0.81-1.08 | 0 | 0.83 | 0.88 | 0.38 |
| Codominant model | TT vs. CC | 5 | 0.77 | 0.49-1.21 | 0 | 0.41 | 1.12 | 0.26 |
| TT vs. CT | 5 | 0.82 | 0.52-1.30 | 22 | 0.28 | 0.84 | 0.4 | |
| Recessive model | TT vs. CT+CC | 5 | 0.79 | 0.51-1.24 | 12 | 0.34 | 1.01 | 0.31 |
| Dominant model | TT+CT vs. CC | 6 | 0.93 | 0.80-1.09 | 0 | 0.89 | 0.87 | 0.39 |
| Subgroup | ||||||||
| C150T(Ser608 Leu) in Asian | ||||||||
| Allele Model | T vs. C | 3 | 1.04 | 0.82-1.32 | 0 | 0.90 | 0.35 | 0.72 |
| Codominant model | TT vs. CC | 3 | 1.10 | 0.52-2.33 | 0 | 0.29 | 0.24 | 0.81 |
| TT vs. CT | 3 | 1.13 | 0.53-2.41 | 46 | 0.16 | 0.31 | 0.75 | |
| Recessive model | TT vs. CT+CC | 3 | 1.14 | 0.54-2.38 | 31 | 0.23 | 0.34 | 0.74 |
| Dominant model | TT+CT vs. CC | 4 | 0.98 | 0.77-1.25 | 0 | 0.74 | 0.14 | 0.89 |
| (CCTTT)n repeats* | ||||||||
| Allele Model | L vs. S | 2 | 1.20 | 0.99-1.46 | 0 | 0.75 | 1.83 | 0.07 |
| Codominant model | LL vs. SS | 2 | 1.41 | 0.95-2.11 | 0 | 0.84 | 1.69 | 0.09 |
| LL vs. SL | 2 | 1.09 | 0.77-1.54 | 0 | 0.88 | 0.49 | 0.63 | |
| Recessive model | LL vs. SL+SS | 2 | 1.19 | 0.86-1.65 | 0 | 0.76 | 1.07 | 0.29 |
| Dominant model | LL+SL vs. SS | 2 | 1.33 | 0.98-1.82 | 0 | 0.93 | 1.83 | 0.07 |
OR: odds ratio; CI: confidence interval. *repeat numbers divided into two groups: S (9-11 repeats) and L (12-18 repeats).
Figure 4.
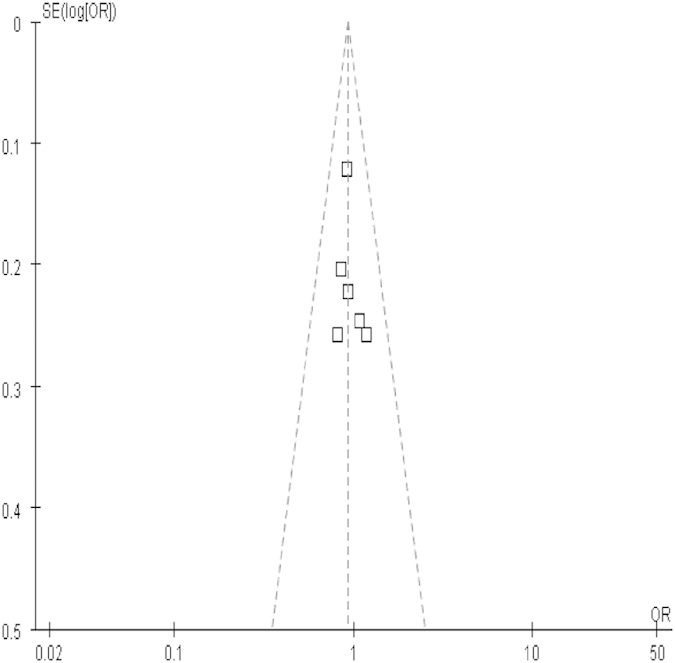
Funnel plot of studies conducted on the association between C150T(Ser608 Leu) mutation and cancer risk.
Sensitivity Analysis
In order to examine the influence of the individual data set to the pooled ORs, every single study was deleted each time. Sensitivity analysis indicated our results were statistically robust (data not shown).
Discussion
As a production of nitric oxide (NO), iNOS is produced during inflammation by macrophages14. Expression of iNOS in response to cytokines is one of the important sections for inflammatory reaction and relates to angiogenesis, suggesting its potential role in the process of carcinogens15. The polymorphism (C150T) in exon 16 of iNOS gene results in an amino acid substitute, Ser608 Leu. The iNOS Ser608 Leu allele (C > T polymorphism) leads an amino acid alteration in a regulatory domain of the enzyme16. Furthermore, a polymorphic pentanucleotide (CCTTT)n repeat probably 2.5 kilobase substream the transcription initiation site has been clarified to affect iNOS expression17. In our meta-analysis, the associations between these two polymorphisms in iNOS gene and risk of cancer were studied.
Except detection the expression of iNOS in vivo or in vitro with tumor18,19, there were some other researches in cancer patients that susceptibility to carcinogenesis may be correlated with the existence of particular alleles at the iNOS locus. However, the results are inconsistent and inconclusive due to limited sample size and different study methods. To the best of our knowledge, this is the first meta-analysis to evaluate iNOS gene polymorphism in development and growth of cancer. In order to acquire a more reliable and comprehensive evidence on both variants, we conducted this meta-analysis to evaluate the association between the polymorphism in iNOS gene and risk of cancer on the basis of data from eight studies. The results indicated that there was no significant association for iNOS gene polymorphism and risk of cancer. In addition, another iNOS gene (iNOS974) was studied in the relationship with cancer. However, there was no significant difference between iNOS974 polymorphism and cancer risk9.
Although we have not found a significant association between iNOS gene polymorphisms and cancer risk, some studies revealed that the risk of cancer is increasing among smoking or drinking individuals with polymorphism in iNOS gene5,6. In addition, studies also reported that iNOS gene polymorphism was associated with the risk of H pylori-related gastric cancer3,8. We could provide a hypothesis that there was an association between iNOS gene and risk of cancer combined with risk factors including lifestyle and H pylori infection. As the reason for heterogeneity (including different study methods and outcomes) among studies, we could not use meta-analysis to analyze the relationship between iNOS genes combined with those risk factors and cancer.
This meta-analysis has pooled the available data from the eligible studies, which has significantly increased the statistical power. Nevertheless, the results of the present meta-analysis should also be explained within the context of its limitations. First, cancer is a multi-factorial illness from complex interactions between environmental exposure and genetic factors. In this meta-analysis, we had insufficient data to conduct an evaluation of such interactions for the role of iNOS polymorphisms and factors in cancer development. Second, the number of current studies is relative limited. Thus, investigations involving large number of different races are necessary for a more reliable assessment on their associations. Third, our meta-analysis is based on unadjusted estimate because lacking of sufficient data. Forth, although we have made our best efforts to avoid all potential publications, it is likely that some are missed or displayed erroneously. Furthermore, we did not include studies published in language other than English or Chinese.
In conclusion, this meta-analysis revealed that the polymorphisms in iNOS gene could not be regarded as a strong genetic risk factor for cancer but it might be association with cancer combined with additional risk factors. In addition, this result should be interpreted cautiously. In order to better understand the potential etiology for cancer in human, large well-designed studies in the susceptibility of cancer evidence are needed to perform in future. It also will be necessary to combine genetic factors and other environmental risk factors.
Methods
Selection of eligible studies
Studies on the associations between iNOS gene polymorphisms and cancer were scrutinized by two reviewers (L.L. and J.H.J.) independently. We searched Pubmed, Embase, the Cochrane Library, ISI Web of Science, Google (scholar) database, Chinese Biological Medicine Disc (CBMDisc), China National Knowledge Infrastructure (CNKI), and Chongqing VIP database (Last search was updated on December 10, 2014) using the terms “inducible nitric oxide synthase or iNOS”, “cancer or tumor or carcinoma” and “polymorphism, variant or mutation”. Reference lists were checked and additional literatures were contacted by researchers. Authors of publications were contacted when results were unclear or when sufficient data were not reported. The search was performed without restriction on language, but we only included articles written in English or Chinese.
Selection Criteria
Studies were included if they fulfilled all of the following entry criteria: (1) it must be a case-control or cohort study design; (2) there were sufficient data for iNOS gene mutations with risk of cancer; (3) the genotype distribution in the controls of all studies should be in agreement with Hardy-Weinberg equilibrium (HWE); and (4) in the case of multiple publications from the same study group, the most complete and recent results were used.
Exclusion criteria
The exclusion criteria were defined as: (1) abstracts, reviews and studies on animal; (2) useless data reported, genotype number or frequency was not included; and (3) genotype distribution in the control population not consistent with HWE.
Data extraction
After studies selection, two investigators independently extracted data from each study with a standard form and entered into a database. When discrepancies were appeared, all investigators were recruited to evaluate the data. The following information was collected: First author, publication year, location, ethnicity, characteristics, sample sizes of patients and controls, genotype numbers.
The review and analysis were guided to conduct by the PRISMA statement for preferred reporting of systematic reviews and meta-analysis20.
Statistical analysis
Odds ratios (ORs) with 95% confidence intervals (CIs) for genotypes and alleles were used to assess the strength of association between iNOS gene polymorphisms and risk of cancer. The ORs were conducted for the allele contrast, codominant model, recessive model, and dominant model, respectively. Heterogeneity was examined with I2 statistic interpreted as the proportion of total variation contributed by between-study variation. If there was significant heterogeneity, the random effects model would be used to analyze the pooled ORs21,22. Otherwise, the pooled ORs were analyzed by the fixed effects model23. The potential publication bias was assessed with funnel plot and Egger’s test. An asymmetric plot suggests a possible publication bias and the P value of Egger’s test being considered representative of significant publication bias if it was less than 0.0524. All statistical tests were performed by RevMan software (version 5.0, Review Manager, Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration, 2010) and Comprehensive Meta-Analysis software version 2.0 (Biostat, Englewood Cliffs, I.N.J., USA). P value less than 0.05 for any test was considered to be statistically significant.
Additional Information
How to cite this article: Jiao, J. et al. Lack of association of the iNOS gene polymorphism with risk of cancer: a systematic review and Meta-Analysis. Sci. Rep. doi: 10.1038/srep09889 (2015).
Acknowledgments
Thanks to our English teacher Dr. Sharon Forsyth who is the Director of Biomedical Editing International in Australia to copyedite this article. Thanks to Xiaomei Wu, Ph.D, Professor of the Department of Clinical Epidemiology and Evidence Medicine in the First Affiliated Hospital of China Medical University.
Footnotes
Author Contributions J.H.J. and L.L. have contributed to the design of the study, analysis and interpretation of data and drafting a part of manuscript. J.H.J. and D.S.H. also took part in analyzing data, and drafting a part of manuscript. L.L. and J.H.J. investigated and collected data. L.L., J.H.J. and J.Y.W. carried out statistical analysis. L.L., J.H.J. and J.Y.W. prepared all figures and tables. All authors reviewed the manuscript.
References
- Hammam O. et al. A Possible Role for TNF-α in Coordinating Inflammation and Angiogenesis in Chronic Liver Disease and Hepatocellular Carcinoma. Gastrointest Cancer Res. 6, 107–114 (2013). [PMC free article] [PubMed] [Google Scholar]
- Li J. et al. Identification of high-quality cancer prognostic markers and metastasis network modules. Nat Commun. 1, 1–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiei A. et al. Inducible nitric oxide synthetase genotype and Helicobacter pylori infection affect gastric cancer risk. World J Gastroenterol. 18, 4917–4924 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson H. R. et al. Cyclooxygenase-2 and inducible nitric oxide synthase gene polymorphisms and risk of reflux esophagitis, Barrett’s esophagus, and esophageal adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 17, 727–731 (2008). [DOI] [PubMed] [Google Scholar]
- Shen C. H. et al. Inducible nitric oxide synthase promoter polymorphism, cigarette smoking, and urothelial carcinoma risk. Urology 69, 1001–1006 (2007). [DOI] [PubMed] [Google Scholar]
- Shen J. et al. A novel genetic polymorphism of inducible nitric oxide synthase is associated with an increased risk of gastric cancer. World J Gastroenterol. 10, 3278–3283 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. S. et al. Lack of association of the cyclooxygenase-2 and inducible nitric oxide synthase gene polymorphism with risk of cervical cancer in Korean population. Ann N Y Acad Sci. 1095, 134–142 (2007). [DOI] [PubMed] [Google Scholar]
- Goto Y. et al. Inducible nitric oxide synthase polymorphism is associated with the increased risk of differentiated gastric cancer in a Japanese population. World J Gastroenterol. 12, 6361–6365 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. et al. Polymorphisms of the neuronal and inducible nitric oxide synthase genes and the risk of cutaneous melanoma: a case-control study. Cancer 109, 1570–1578 (2007). [DOI] [PubMed] [Google Scholar]
- Tatemichi M. et al. Increased risk of intestinal type of gastric adenocarcinoma in Japanese women associated with long forms of CCTTT pentanucleotide repeat in the inducible nitric oxide synthase promoter. Cancer Lett. 217, 197–202 (2005). [DOI] [PubMed] [Google Scholar]
- Wang D., Mann J. R. & DuBois R. N. The role of prostaglandins and other eicosanoids in the gastrointestinal tract. Gastroenterology, 128, 1445–61 (2005). [DOI] [PubMed] [Google Scholar]
- Lala P. K. & Chakraborty C. Role of nitricoxide in carcinogenesis and tumour progression. Lancet Oncol. 2, 149–156 (2001). [DOI] [PubMed] [Google Scholar]
- Marsden P. A. et al. Localization of the human gene for inducible nitric oxide synthase (NOS2) to chromosome 17q11.2-q12. Genomics 19, 183–185 (1994). [DOI] [PubMed] [Google Scholar]
- Felley C. P. et al. Oxidative stress in gastric mucosa of asymptomatic humans infected with Helicobacter pylori: effect of bacterial eradication. Helicobacter 7, 342–348 (2002). [DOI] [PubMed] [Google Scholar]
- Vallance P. & Collier J. Biology and clinical relevance of nitri coxide. Br Med J. 309, 453–457 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannesen J. et al. Linkage of the human inducible nitric oxide synthase gene to type 1 diabetes. J Clin Endocrinol Metab. 86, 2792–2796 (2001). [DOI] [PubMed] [Google Scholar]
- Xu W. et al. Evolution of a homopurine homopyrimidine pentanucleotide repeat sequence upstream of the human inducible nitric oxide synthase gene. Gene. 204, 165–170 (1997). [DOI] [PubMed] [Google Scholar]
- Brandão M. M., Soares E., Salles T. S. & Saad S. T. Expression of inducible nitric oxide synthase is increased in acute myeloid leukaemia. Acta Haematol. 106, 95–99 (2001). [DOI] [PubMed] [Google Scholar]
- Takahashi M., Mutoh M., Kawamori T., Sugimura T. & Wakabayashi K. Altered expression of beta-catenin, inducible nitric oxide synthase and cyclooxygenase-2 in azoxymethane-induced rat colon carcinogenesis. Carcinogenesis 21, 1319–1327 (2000). [PubMed] [Google Scholar]
- Moher D. et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 6, e1000097 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DerSimonian R. & Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials 28, 105–114 (2007). [DOI] [PubMed] [Google Scholar]
- DerSimonian R. & Laird N. Meta-analysis in clinical trials. Control Clin Trials 7, 177–188 (1986). [DOI] [PubMed] [Google Scholar]
- Mantel N. & Haenszel, W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 22, 719–748 (1959). [PubMed] [Google Scholar]
- Egger M. et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–634 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]