Abstract
Cardiac arrhythmias are often associated with mutations in ion channels or other proteins. To enable drug development for distinct arrhythmias, model systems are required that allow implementing patient-specific mutations. We assessed a muscular pump in Caenorhabditis elegans. The pharynx utilizes homologues of most of the ion channels, pumps and transporters defining human cardiac physiology. To yield precise rhythmicity, we optically paced the pharynx using channelrhodopsin-2. We assessed pharynx pumping by extracellular recordings (electropharyngeograms—EPGs), and by a novel video-microscopy based method we developed, which allows analyzing multiple animals simultaneously. Mutations in the L-type VGCC (voltage-gated Ca2+-channel) EGL-19 caused prolonged pump duration, as found for analogous mutations in the Cav1.2 channel, associated with long QT syndrome. egl-19 mutations affected ability to pump at high frequency and induced arrhythmicity. The pharyngeal neurons did not influence these effects. We tested whether drugs could ameliorate arrhythmia in the optogenetically paced pharynx. The dihydropyridine analog Nemadipine A prolonged pump duration in wild type, and reduced or prolonged pump duration of distinct egl-19 alleles, thus indicating allele-specific effects. In sum, our model may allow screening of drug candidates affecting specific VGCCs mutations, and permit to better understand the effects of distinct mutations on a macroscopic level.
The heart beat results from a highly regulated series of physiological events. Cardiac depolarization triggers voltage gated Na+ -channels and VGCCs that shape the rise and plateau phases (T-type VGCCs and L-type VGCCs, respectively) of the cardiac action potential (AP)1,2. The AP is followed by repolarization, involving several types of K + -channels, and a resting phase3. The period between ventricular depolarization and repolarization defines the QT interval in the electrocardiogram4. The rise in cytosolic Ca2+ triggers ryanodine receptor 2 (RyR2), a Ca2+-activated Ca2+ release channel in the sarcoplasmic reticulum (SR), thus evoking contraction. Following repolarization, Ca2+ is extruded from the cytosol, by SR membrane Ca2+-pumps and the Na+/Ca2+-exchanger (NCX) in the plasma membrane, jointly leading to muscular relaxation5.
Cardiac arrhythmias constitute prevalent forms of heart disease. Arrhythmias can affect heart rate, i.e. faster or slower than normal (tachycardia or bradycardia, with >100 or <60 beats per minute, respectively), or they disturb the regular rhythm and sequence of physiological events constituting a heartbeat, for example in aged hearts6, or following damage (e.g. artery disease, heart attack)7. Last, arrhythmias are caused by mutations in genes relevant for generation, maintenance and termination of the cardiac AP, as well as intracellular Ca2+ handling8. For example, Long QT (LQT) syndromes involve a number of genes affecting the duration of the QT interval (prolonging it), like K+ channels, and also the L-type VGCC, Cav1.2 (CACNA1C). Mutations in Cav1.2 lead to Timothy syndrome (LQT8), and usually delay inactivation of L-type channels, thus extending the AP plateau phase9,10,11.
Drugs that accelerate inactivation of L-type VGCCs, like 1,4-dihydropyridines (DHPs), are effective in LQT8 patients. However, DHPs broadly affect L-type channels, thus causing side effects. As more specific mutations in Cav1.2 are being identified in LQT8 patients (and for other genes affecting other arrhythmias; see e.g. a database at http://triad.fsm.it/cardmoc/), a demand for new effective drugs, even acting specifically on distinct mutations is generated12,13. Therefore, genetically amenable systems allowing for straightforward drug screening, in a mutation-specific manner, would be beneficial. Patient-specific cardiomyocytes derived from induced pluripotent stem cells can be used to assess aspects of arrhythmia on a cellular basis, but are too expensive for screening purposes and cannot recapitulate cardiac arrhythmias in the context of the whole heart. A LQT model based on mutations in the KCNH2 channel was previously generated in zebrafish14; yet, introducing mutations and transgenes is slow. The nematode Caenorhabditis elegans is a genetically accessible experimental system with a 2.5 day life cycle, enabling straightforward (and cheap) mass cultivation, i.e. properties beneficial for transgenesis and drug screening. The C. elegans feeding organ, the pharynx, is a rhythmically active muscular pump15, with many properties similar to the vertebrate heart (but also dissimilarities)16. It consists of 20 muscle cells (and accessory cells), connected by gap junctions as in the heart, forming an elongated structure with an axial lumen and anterior and terminal bulbs (Fig. 1a, lower panel). The mostly autonomous pharyngeal nervous system is required for (fast) pumping and modulation of pharynx activity in presence or absence of bacterial food17,18,19. The pharynx pumps with up to 4 Hz in the presence of bacteria, which are sucked in and moved through procorpus and anterior bulb, to be collected in the anterior isthmus. Isthmus peristalsis then transports bacteria to the terminal bulb, where they are mechanically lysed20. Like in the heart, pharynx contractions continue in the absence of coordinate neuronal input, but require acetylcholine. Ion channels facilitating pharynx action are known and the pharyngeal AP (well resembling the human cardiac AP, as it exhibits a plateau) was thoroughly characterized. Fast pharynx pumping requires EAT-2 nicotinic acetylcholine receptors (nAChRs)21, replacing voltage gated sodium channels. T-type and L-type VGCCs (encoded by cca-1 and egl-19, respectively) shape the rise- and plateau phases of the pharyngeal AP22. Repolarization is modulated by glutamate-gated chloride channels, and executed by voltage-gated K+-channels, encoded by exp-2 (ref. 23) which, though homology is low, is functionally similar to the mammalian HERG channel, but also UNC-103 K+-channels. Pharynx muscle also expresses the UNC-68 ryanodine receptor and the SR Ca2+ storage protein CSQ-1, similar to human CASQ2 (refs. 24, 25, 26). Mutations in these genes affect pharynx pumping and its physiological and electrical properties. They may thus serve to establish a model system for homologous mutations found in humans. Some aspects of pharynx cell biology, however, point to convergent development of the two organs, like origin from ectodermal (pharynx) vs. mesodermal tissue (heart)16.
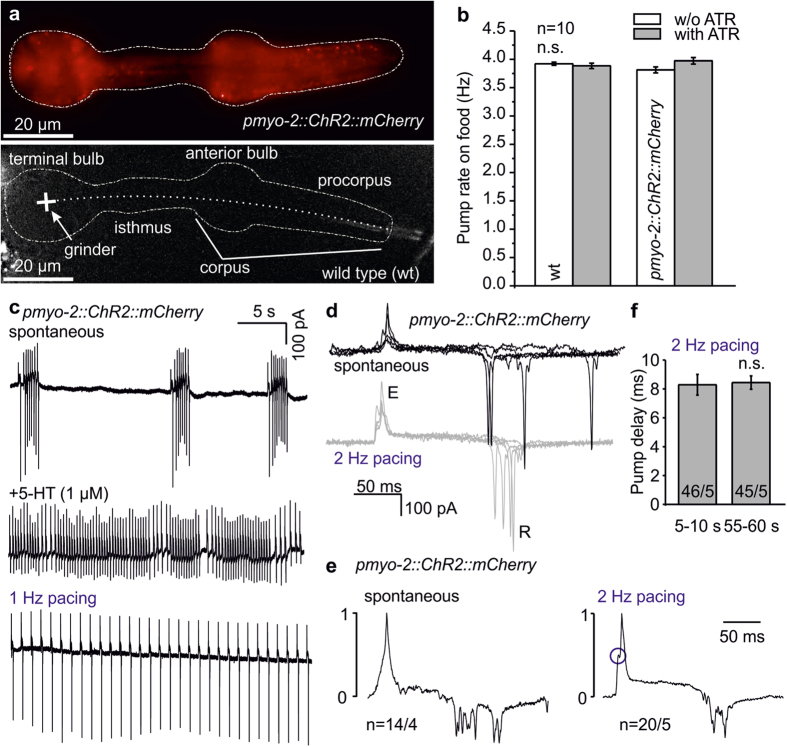
Figure 1. ChR2 expressed in pharyngeal muscle cells (PMCs) enables photostimulation and light-induced pumping with high reproducibility.
(a) Expression of ChR2(H134R)::mCherry in PMCs, compared to a non-transgenic control. Dashed lines indicate pharynx position, dotted line indicates pharynx lumen, structural features of the pharynx are named. Scale bars: 20 μm. (b) The pharynx pump rate on food (visually counted) is unaffected by ChR2 expressed in PMCs (compared to wild type—wt), regardless of cultivation in absence or presence of 10 μM ATR (n = 10 animals each). (c) EPG recordings of activity in pharynxes of animals expressing ChR2::mCherry in PMCs. Compared are spontaneous activity (upper trace), activity stimulated by addition of serotonin (5-HT, 1 μM; middle trace), or in response to 1 Hz optical pacing (lower panel). (d) Comparison of overlaid original EPG recordings of spontaneous (black) and 2 Hz (10 ms, 470 nm) stimulated (grey) EPGs from four pharynxes expressing ChR2::mCherry. E (excitation/ contraction of pharynx) and R (relaxation/repolarization) peaks are indicated, traces have been aligned to the E spike. (e) Averaged, E-peak normalized EPG traces of spontaneous (14 traces) or 2 Hz paced pumps (20 traces) from different pharynxes (n = 4–5). A peak preceding the E-spike is observed in averaged 2 Hz paced pump traces, likely elicited by ChR2-photocurrents in PMCs, indicated by a blue circle. (f) Pump delay (time between start of stimulation pulse and occurrence of E-peak) is consistent during long recording periods, and was averaged from the indicated number of pumps/animals, for seconds 5–10 or 55–60 of one minute paced pumping.
Pharynx pumping can be recorded by EPGs, analogous to electrocardiograms. The head of the animal is sucked into a pipette for extracellular recording. Either whole animals, or cut head preparations are used, the latter enabling more accurate measurements of pump rate and duration, and distinguishing muscle activity of different parts of the pharynx, as well as activity of pharyngeal neurons27,28. Pharynx pumping is influenced by the presence of food, but also by neuromodulators like serotonin29,30. Drugs can be applied either to intact animals, or to cut heads, where drug access is more direct. Spontaneous pharynx pumping is not as regular as required to assess rhythmicity and the consequences of arrhythmogenic mutations. Thus, experimental means of pacing the pharynx to a precisely regular beat are necessary to generate a C. elegans test system for analyzing arrhythmogenic mutations, as well as anti-arrhythmogenic drugs. For high throughput, non-invasive methods for pacing would be beneficial, which may be achieved by optogenetics in the transparent animal, using the light-gated ion channel channelrhodopsin (ChR2)31,32,33. Here, we establish the pharynx as an optogenetically paced system. By ChR2-mediated depolarization, we achieved pharynx pumping up to 6 Hz. Mutations in the L-type VGCC EGL-19 affected pump duration and ability of the pharynx to achieve high pump rates, similar to a LQT syndrome. Video microscopy and kymographic analyses enabled high throughput, while deducing key parameters of the AP, and also revealed arrhythmic episodes during prolonged pacing. Importantly, the DHP analog nemadipine A (from now on, nema-A) permitted reverting the prolonged pump duration of a specific allele of egl-19, while it had opposing effects on wild type (wt) and other egl-19 alleles. Optogenetic pharynx pacing thus facilitates functional analysis of VGCC alleles in the context of a whole organ, and enables straightforward screening for novel anti-arrhythmogenic drugs.
Results
ChR2 enables optogenetic pacing of the C. elegans pharynx
We expressed ChR2(H134R)::mCherry (a gain-of-function mutant31; from now on, ‘ChR2’ refers to this variant) in pharyngeal muscle cells (PMCs), using the pmyo-2 promoter. Expression was expected in intracellular and plasma membranes. Diffuse mCherry-fluorescence could be observed in the whole pharynx, whereas no fluorescence was detected in wt animals (Fig. 1a). Spontaneous pump rate of the pharynx, in presence of bacteria, in PMC-ChR2 animals or wt, cultivated with or without the ChR2 chromophore all-trans retinal (ATR) was unaltered, indicating that PMCs were not affected by ChR2 expression (Fig. 1b).
We tested whether pharynx pumping could be paced optically. To assess electrical events associated with pharynx pumping, we used EPG recordings. Spontaneous pharynx activity is unsteady (Fig. 1c, upper panel): Periods of fast pumping alternate with periods of low or no pump activity. Addition of serotonin stimulates pumping30, leading to ongoing activity, yet the pump rate is too variable to allow detecting ‘arrhythmic’ events (Fig. 1c, middle panel). In contrast, when we applied pacing light pulses (1 Hz, 470 nm, 10 ms, 1.5 mW/mm2), ChR2-transgenic pharynxes of animals showed a steady 1 Hz rhythm (Fig. 1c, lower panel). We analyzed the EPGs in detail. Compared to spontaneous pumping, the pump duration, i.e. period between E- (excitation, contraction of terminal bulb) and R (relaxation/repolarization of corpus27,28) peaks was much more reproducible between animals and between different pumps in pharynxes optically paced at 2 Hz (Fig. 1d), and in averaged, E-peak normalized traces, R-peaks were much less distributed in paced pharynxes (Fig. 1e). Paced pharynxes exhibited an extra peak in the averaged EPG preceding the E-peak, representing ChR2 photocurrents. The delay between the light pulse and the E-peak was very reproducible with low jitter for 2 Hz paced pharynxes, regardless of whether this was determined early (5–10 s) or late (55–60 s) during 1 min, 2 Hz paced pump trains (8.29 ± 0.72 and 8.44 ± 0.46 ms, respectively), showing high reliability (Fig. 1f).
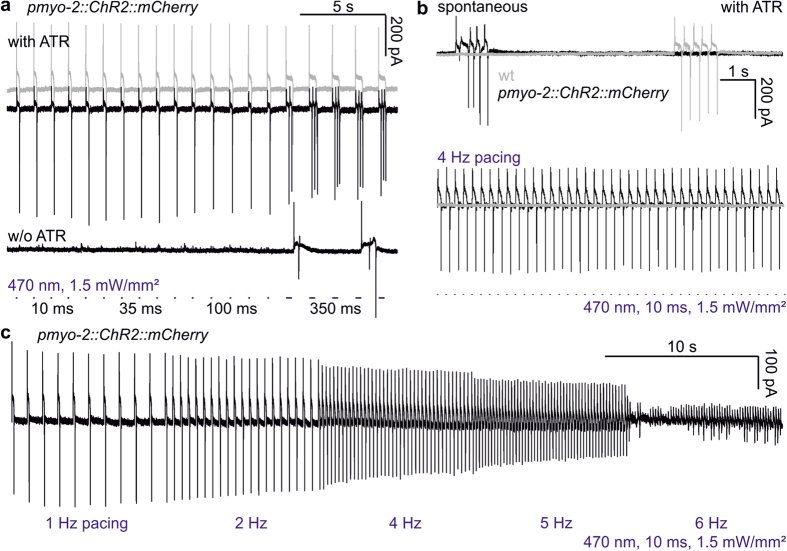
Next, we assessed if and how duration of light pulses would affect pump duration or the number of evoked pumps. At 10, 35 or 100 ms, no difference was observed (n = 6; Fig. 2a; Supplementary Fig. S1). However, 350 ms light pulses caused multiple, or long-lasting pumps. Animals cultivated without ATR showed no pumps in response to short light pulses, though 350 ms light pulses sometimes evoked pumping, possibly an indirect consequence of photophobic signaling by LITE-1 and/or GUR-3 UV-/blue light receptors34,35. We thus used light pulses of 10 or 35 ms in subsequent experiments. 4 Hz pumping could be readily evoked in ChR2-transgenic animals, but not in non-transgenic animals (Fig. 2b). In a ‘stress test’, pacing was increased every 10 s in 1 Hz increments up to 6 Hz (Fig. 2c; Supplementary Video 1). Most PMC-ChR2 pharynxes could achieve 5 Hz maximal frequency. Many pharynxes would also follow 6 Hz reliably, though they sometimes alternated between 6 and 3 Hz pumping. Rarely, pharynxes could be paced up to 7 Hz (10 ms pulses, 1.5 mW/mm2). To enable pacing for long periods (up to 65 s), we settled with 1.5–2 mW/mm2 light intensity.
Figure 2. ChR2-mediated stimulation of the pharynx induces highly regular pumping at frequencies up to 7 Hz.
(a) Pumping in ChR2::mCherry expressing pharynxes, recorded as EPGs, stimulated by light pulses of different length (10, 35, 100, 350 ms), causing accordingly increased pump duration (grey trace), or multiple pumps (top black trace) during 350 ms stimuli. Pharynxes of animals raised without ATR exhibit non-specific pump events (bottom black trace). (b) Pumping can be stimulated at 4 Hz, (470 nm, 10 ms, 1.5 mW/mm2; lower black EPG trace), but not in non-transgenic animals (lower grey trace). Upper traces: Non-transgenic (grey) or transgenic pharynxes in the presence of ATR (black) exhibit normal bouts of spontaneous pumping. (c) Pumping (single EPG trace) stimulated by increasing frequencies of light pulses as in b, as a ‘stress test’ protocol (pacing frequency increased stepwise up to 6 Hz, as indicated below the trace). This pharynx could not lock in to 6 Hz pacing.
The pharyngeal nervous system does not influence pump duration in the paced pharynx
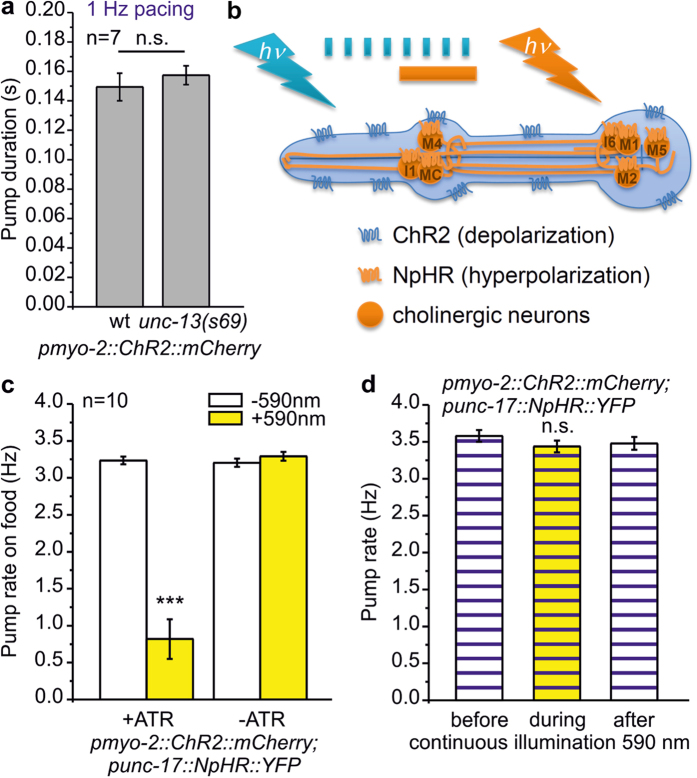
The pharynx is innervated by 20 neurons17,18,19. To establish the pharynx as an arrhythmia model, we needed to assess whether neuronal signaling may influence paced pharyngeal pumping. Recent work showed that optogenetic stimulation of pharynx neurons induces pumping36. Pharyngeal neurons may also negatively regulate pump rate or duration, possibly counteracting ChR2-mediated depolarization. Pump duration in unc-13(s69) mutants, which have abolished chemical synaptic transmission37, was no different during 1 Hz pacing (157 ± 6, compared to 149 ± 9 ms in wt; n = 7; Fig. 3a). To look for acute effects of cholinergic neurons on paced pumping in PMC-ChR2 animals, we expressed the hyperpolarizing, light-driven chloride pump halorhodopsin (NpHR)38 in cholinergic cells, some of which innervate the pharynx (MC, M1, M2, M4, M5, I1 and I6 neurons; Fig. 3b). We first assessed the effect of neuronal silencing on spontaneous pumping. Photoinhibition of cholinergic neurons (590 nm, 0.5 mW/mm2), which led to body relaxation due to silencing of cholinergic neuromuscular junctions38 (Supplementary Fig. S2), significantly reduced the pump rate on food (3.24 ± 0.05 vs. 0.82 ± 0.27 Hz; n = 10; Fig. 3c). However, upon optogenetic pacing (3.7 Hz, 35 ms, 470 nm), 1 min photo-inhibition of cholinergic neurons had no significant effect on the pump rate (Fig. 3d). Thus, neuronal malfunction, which could be effected by mutations in genes we aim to test in pharynx muscle, but which may also affect neuronal function, did not influence optogenetic pacing of pharynx muscle.
Figure 3. The pharyngeal nervous system does not influence EPG parameters during optogenetically paced pumping.
(a) Pump duration during 1 Hz pacing compared in wild type (wt) and unc-13(s69) mutant (n = 7 pharynxes). (b) Schematic depicting transgenic pharynxes used in experiments in (c,d). The pharynx expresses ChR2 in PMCs and cholinergic pharyngeal neurons express halorhodopsin (NpHR). While ChR2 is activated by 470 nm light pulses (blue tick marks) to induce pumping, cholinergic neurons are hyperpolarized by continuous illumination (1 min) with 590 nm light (yellow bar) (c) Spontaneous pump rate in presence of bacteria, in pharynxes as described in B, from animals raised in the absence or presence of ATR, as indicated. Yellow-light induced hyperpolarization of cholinergic neurons affects spontaneous pump rate (t-test with Bonferroni correction, ***P < 0.001). (d) Mean pump rate, visually counted for 30 s during 4 Hz optical pacing (35 ms, 470 nm) is not impaired by hyperpolarization of cholinergic neurons using yellow light (middle). Compared is frequency before (left) or after yellow light (right).
Mutations in the EGL-19 L-type voltage gated Ca2+channel affect pump duration
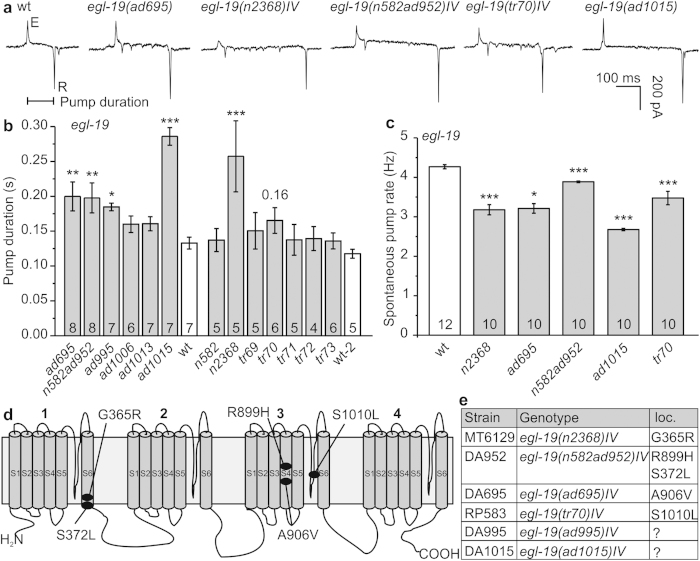
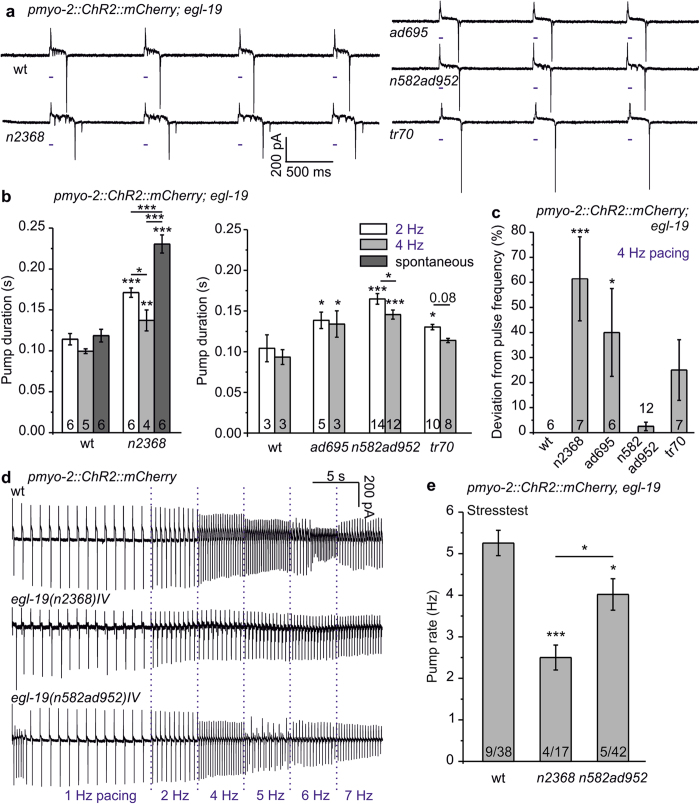
We next explored whether mutations in a gene required for function and rhythmicity of the pharynx may be analyzed by optogenetic pacing. L-type VGCCs shape the plateau phase of cardiac, as well as pharyngeal APs. EGL-19 is homologous to human Cav1.2. Numerous alleles of egl-19 have been isolated, and some of them characterized and classified as reduction- or gain-of-function (r.o.f., g.o.f.) mutants, following different assays (in some cases by electrophysiology), based on EGL-19’s roles in development, pharynx, locomotion and egg-laying muscles22,39,40,41,42,43,44. We determined pump durations for a range of these mutants: Alleles ad695, n582ad952, ad995, ad1015, and n2368 showed increased pump duration (Fig. 4a,b), which was accompanied by a significantly reduced pump rate for these alleles, as well as for tr70 (Fig. 4c). The most pronounced increases of pump duration (~2.1-fold) were found for ad1015 (counterintuitively, a r.o.f. mutant; another r.o.f. allele, n582, did not show altered pump duration) and n2368 (g.o.f.; slowed inactivation43), the latter of which carries the mutation G365R in the S6 transmembrane (TM) segment of the first module (Fig. 4d,e; Supplementary Fig. S6, S7). This mutation affects the equivalent residue as an established LQT8 mutation in Cav1.2, G402S, thus both mutations replace glycine with charged or polar amino acids11,45. In other g.o.f. alleles, the effects were less pronounced, i.e. ~1.5-fold for ad695 and n582ad952, harboring mutations A906V and the double mutation S372L; R899H39, respectively. Allele tr70 carries the mutation S1010L42. All of these residues are within TM segments across modules I or III (R899 and A906 are in the voltage sensor helix S4 of the third module; S1010 is in the pore loop of the third module), or close to the cytosolic membrane face (S372; Fig. 4d,e; Supplementary Figs. S6 and S7, structural model based on the NavAb voltage gated Na+ channel46). How these mutations may affect EGL-19 desensitization requires detailed analysis; however ad695 and n2368 g.o.f. alleles showed reduced desensitization in electrophysiology43.
Figure 4.
Mutations in the Cav1.2 homolog EGL-19 prolong spontaneous pump duration and reduce pump rate. (a) Original EPG recordings of wild type (wt) and egl-19 mutants (alleles are indicated) with prolonged pump duration (interval E-R spike is indicated). (b) Group data, pump durations (mean ± s.e.m.) during spontaneous pumping, of several egl-19 alleles; statistically significant differences to wt are indicated (***P < 0.001; **P < 0.01; *P < 0.05; one way ANOVA with Fisher post-hoc test). (c) Group data, spontaneous pump rate on food of selected egl-19 mutants, compared to wt (t-test with Bonferroni correction: ***P < 0.001; **P < 0.01; *P < 0.05). (d) Schematic of the EGL-19 α1 subunit of the L-type VGCC with the amino acid changes in the alleles listed in (e).
We combined egl-19 alleles with known molecular lesion with PMC-ChR2 for pacing. Stable pharynx pumping could be induced in all of these strains (Fig. 5a), but the mean pump duration was increased, which was statistically significant for all alleles at 2 Hz pacing (Fig. 5b). Interestingly, mean pump duration in tr70 was not significantly prolonged at 4 Hz pacing, while n2368, ad695, and n582ad952 had increased pump duration also at this pace frequency. Pacing affected pump duration particularly for egl-19(n2368): spontaneous pumps lasted ~1.7-fold longer than during 4 Hz pacing (p ≤ 0.001; Figs 4b and 5b); thus, enforced, regular depolarization may affect EGL-19(G365R) such that it desensitizes faster.
Figure 5. EGL-19 L-type VGCC mutants affect pump parameters and pacing ability of the optically paced pharynx.
(a) Comparison of EPG recordings of 1 Hz optically paced egl-19 mutants and wild type (wt). Light pulses are indicated by blue tick marks. (b) Group data of pump durations (mean ± s.e.m.; n = 3–10 pharynxes, as indicated in each bar) in pharynxes of egl-19 alleles, paced at 2 Hz or 4 Hz, as indicated, and compared to wild type (wt) pump durations. Left panel shows recordings of allele n2368 with 10 ms pacing and exemplary spontaneous pump duration, right panel contains other alleles with 35 ms pacing. (c) Deviation of observed pharynx pumping from the pacing frequency, for the indicated fraction of the stimulation period, given in %, for wt and the indicated egl-19 alleles (n = 6–12 pharynxes, as indicated). (d) Maximum pump frequency determined from pacing stress test (optical stimulation: 1 Hz 15s, 2, 4, 5, 6, 7 Hz each 5s) of the indicated wt or egl-19 mutant pharynxes. (e) The mean maximal pump rate achieved in the stress test protocol is shown for wt, as well as the indicated egl-19 mutants (n = 4–9 experimental days and n = 17–42 pharynxes). Statistically significant differences (***P < 0.001; **P < 0.01; *P < 0.05) were determined by one way ANOVA with Fisher post-hoc test in (b,c,e).
While pacing at 4 Hz, compared to wt, deviations from pacing frequency occurred 61 ± 17% of the time in n2368, 40 ± 18% (ad695), and 25 ± 12%; (tr70; the latter was not significant; Fig. 5c). Deviation from 4 Hz pacing did not correlate with increased pump duration in all cases, i.e. despite increased pump duration for n582ad952 at 4 Hz, it did not deviate from pace frequency (Fig. 5c). To better understand these different behaviors, we assessed the performance of n2368 and n582ad952 in the ‘stress test’. Above distinct frequencies, AP duration and repolarization time should exceed the interstimulus interval, which could lead to cessation of pumping, e.g. through depolarization block, expressing a form of ‘arrhythmia’. Alternatively the pharynx may adapt to the pace rate and reduce the pump duration, or it could skip stimuli and pump at reduced frequency. 15 stimuli at 1 Hz induced steady pumping of the pharynx, then the pacing frequency was increased every 5 s up to 7 Hz (Fig. 5d). Across 4–9 experiments on 17–42 animals, on average, wt reached a maximum pump rate of 5.26 ± 0.3 Hz, whereas n2368 (2.5 ± 0.3 Hz) and n582ad952 (4.0 ± 0.3 Hz) mutants showed a reduced ability to pump at higher rates (Fig. 5e). More specifically, 5 Hz was reached by 76.3% of wt, 33.3% of n582ad952 and only 17.6% of n2368 animals. 6 Hz pacing was achieved by 44.7% and 11.9% (wt and n582ad952, respectively). 7 Hz was achieved by 13.2% of wt, but none of the mutants. Instead, these pharynxes pumped at reduced rate (i.e. only every 2nd or 3rd stimulus evoked a pump; Fig. 5d). Thus, optogenetic pacing facilitates analyzing parameters of rhythmic pharyngeal activity under steady, regular conditions, particularly for mutations affecting pump duration. Several egl-19 mutations evoked a prolonged ‘Q-T’ (E-R in the pharynx) interval, and at least one allele (n2368), resembling the human LQT8 mutation G402S, evoked pharyngeal ‘arrhythmia’ while pacing.
Achieving high throughput investigation of pumping by video microscopy and kymograph analysis
To further explore using the pharynx as a simple, genetically amenable model for (homologous) mutations affecting human cardiac physiology and arrhythmia, we wanted to analyze drug effects. Conceivably, even drugs specifically addressing distinct molecular lesions may thus be identified, but high throughput would be required, which cannot be easily provided by EPG recordings. Though higher throughput electrophysiological methods, involving microfluidic devices, were described47, these are experimentally demanding, problematic with mutant strains growing to small sizes, and not available in many labs. We thus turned to video microscopy, of many animals in parallel, for simple, reliable and versatile analysis of pharynx pumping, enabling access for drugs, and still providing all relevant information about pump rate and duration like EPG recordings.
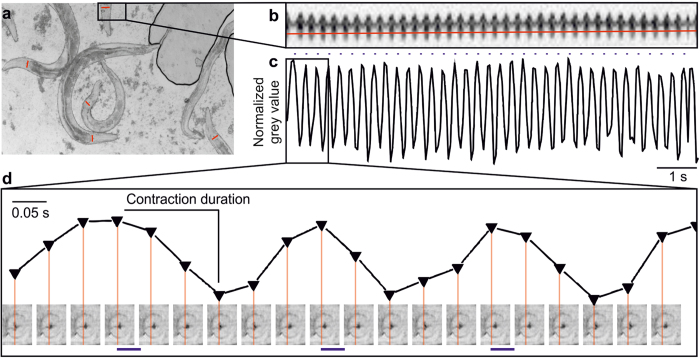
We immobilized intact animals on high-percentage agarose pads with polystyrene beads48, and photostimulated them at 3.7 Hz (35 ms, 470 nm; Fig. 6a). Videos were obtained with a high-speed, high-resolution sCMOS camera with large chip size, to film 5–10 animals simultaneously. Pharynxes could easily be visualized at sufficient resolution to quantify pumping, using kymographic analysis. A line drawn across the terminal bulb (Fig. 6a) was used to analyze changes in grey values in each video frame (at 20 fps). Grinder movement could be extracted for periods of 30 s or more, while pumping was photostimulated. Grey values changed synchronously with pumping (Fig. 6b,c) and were quantitatively analyzed using a custom-written KNIME49 script. Contraction duration was determined from plotting normalized grey values of kymographic line scans, which could be assigned to single frame images of grinder movement. Onset of grinder displacement corresponds to the start (i.e. the large E-peak in the EPG), and maximal displacement matches the end of the contraction duration period (Fig. 6d; corresponding to the small r-peak in the EPG, which is often not well resolved; thus, generally the E-R duration is extracted from EPGs, see below).
Figure 6. Optical recording of pharynx pumping enabled by kymographic analysis in many animals in parallel.
(a) Micrograph showing several intact animals, immobilized on polystyrene beads. Red lines drawn across the pharynx terminal bulb indicate lines for which kymographic analysis is performed (grey values along the line obtained from each video frame). (b) Kymograph showing grey values of the line depicted in a (a still image of the respective video), upon optogenetic stimulation at 3.7 Hz frequency (35 ms, 470 nm light), for about 30 s. A line scan (red line) across the kymograph is used for detection of pump events, in a plot of grey values as shown in (c,d) Pump parameters are analyzed with a custom written KNIME script, using maximum and minimum grey values as an indication of axial movement of the pharynx grinder, during a pump cycle. Correlation of grey values with the actual images from the corresponding video frames is shown in the lower half. Red lines represent the line for detection of grinder movement (x-axis—time, y-axis—normalized grey value).
Arrhythmic pumping behavior of egl-19 L-type VGCC alleles under pacing conditions
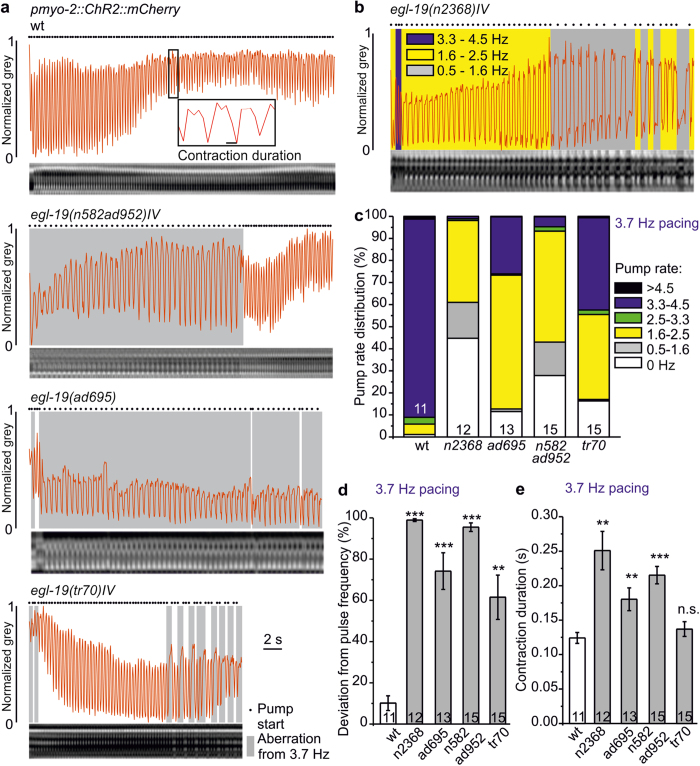
We compared pumping parameters obtained from kymographs using 3.7 Hz pacing for wt and egl-19 alleles n582ad952, ad695, tr70 and n2368 (Fig. 7a,b), to data obtained from EPGs. egl-19 mutants were often unable to lock in to 3.7 Hz, thus pumping at reduced frequency for variable periods before returning to pace frequency. We statistically analyzed these periods of different pump rates, and the overall deviation from pace frequency (Fig. 7c,d). Resulting pump rate distributions (Fig. 7c) showed that wt pumped at pace frequency ~90% of the time, while egl-19 mutants showed characteristic distributions of pump frequencies (>4.5 Hz, 4.5–3.3 Hz, 3.3–2.5 Hz, 2.5–1.6 Hz, 1.6–0.5 Hz, and no pumping, 0 Hz), that differed for each of the alleles. This was most obvious for n2368 (G365R, most similar to the LQT8 mutation G402S45), which achieved ~4 Hz pump rate for only ~1% of the time, while it pumped below 2.5 Hz for 98%, and below 1.6 Hz for ~61% of the time. n582ad952, ad695 and tr70 animals pumped around 4 Hz only 4.6, 25.8 and 41.8% of the time, respectively. Consequently, egl-19 defective animals pumped at ~2 Hz more often than wt: 37.1% (n2368), 60.6% (ad695), 50.3% (n582ad952) and 38.5% (tr70), compared to 4.7% (wt) Overall, all tested egl-19 mutants deviated significantly from the 3.7 Hz pacing frequency (Fig. 7d). Contraction duration (kymographs, intact wt animals: 124 ± 8 ms; Fig. 7e; EPG recordings, 4 Hz, cut head preparations: 93 ± 9 ms to 100 ± 3 ms, Fig. 5b), was increased in egl-19 mutants also in kymographs (~2-fold in n2368, 1.5-fold in ad695 and 1.7-fold in n582ad952; Fig. 7e). When averaged for periods of similar pump rates (e.g. for ~2 and ~4 Hz; Supplementary Fig. S3), contraction durations of egl-19 mutants were strongly increased only for 2 Hz pumping, consistent with EPG-derived pump durations. Importantly, altered pump durations and frequency distributions were not due to neuronal effects of egl-19 mutations, as neuronal photoinhibition in n2368 mutants neither altered contraction duration nor induced further deviation from pacing frequency (Supplementary Fig. S4).
Figure 7. egl-19 mutants exhibit highly arrhythmic pumping despite steady optical pacing, as shown by kymographic analyses.
(a) Kymographic analyses of egl-19 mutant animals compared to wild type (wt), as indicated, during 3.7 Hz pacing (35 ms light pulses, over 30 s). Kymographs are shown below in each panel; the derived normalized grey values are shown as red traces. Periods of observed pumping deviating from 3.7 Hz are shaded grey. Pump starts are shown by black dots. (b) Pump rate distribution at 3.7 Hz pacing in a egl-19(n2368)IV animal represented by shaded areas (blue: 3.3–4.5 Hz, yellow: 1.6–2.5 Hz, grey: 0.5–1.6 Hz. (c) Group data, distributions of pump rates (black: >4.5 Hz, blue: 3.3–4.5 Hz, green: 2.5–3.3 Hz, yellow: 1.6–2.5 Hz, grey: 0.5–1.6 Hz, white: 0 Hz) in the indicated egl-19 mutants, compared to wild type (wt), obtained from animals paced at 3.7 Hz (n = 11–15 animals). (d) Mean deviation (±s.e.m.) of observed pharynx pumping from the pacing frequency, for the indicated fraction of stimulation period, given in % for wild type and the indicated egl-19 alleles (n = 11–15 animals). (e) Contraction duration (mean ± s.e.m.) of the animals analyzed in d. Statistically significant differences in (d,e): t-test with Bonferroni correction (***P < 0.001; **P < 0.01; *P < 0.05).
Nema-A, a dihydropyridine VGCC antagonist, alters pump duration egl-19 allele-specifically
Last, we tested optogenetic pharynx pacing for analyzing drugs affecting relevant proteins. Nema-A, a DHP analog (Fig. 8a, inset), is a specific antagonist of egl-19 in C. elegans, but also acts on vertebrate L-type VGCCs41,42. Nema-A induces various defects in wt growth, morphology and egg-laying, and egl-19 g.o.f. alleles rescue nema-A induced phenotypes (and vice versa, nema-A rescues some egl-19 g.o.f. defects)41,42. Effects on pharynx pumping were not described yet.
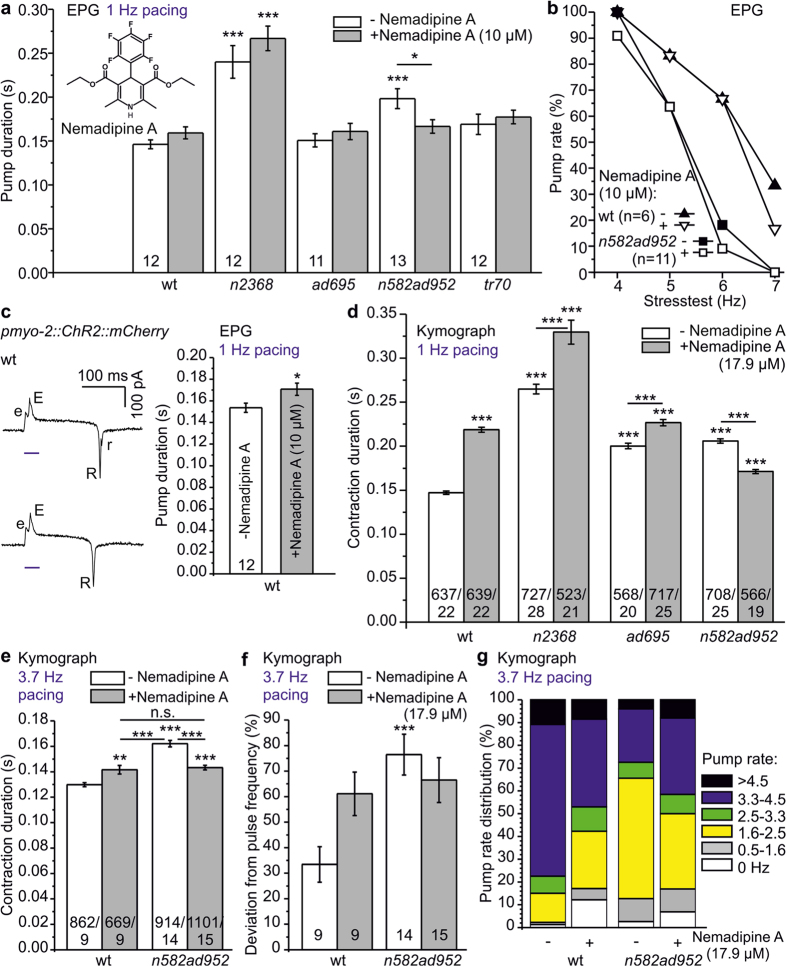
Figure 8. Nema-A ameliorates egl-19(n582ad952) mutant defects in paced pumping in dissected pharynxes and intact animals.
(a) Pump duration deduced from EPG recordings of dissected, optically paced pharynxes (1 Hz), before (white), or following 2 min incubation with nema-A (grey bars, 10 μM; molecular structure shown in the inset), in egl-19 mutants and wild type (wt) as indicated (n = 11–13 pharynxes). (b) Pacing stress test of wt (triangles) and egl-19(n582ad952) (squares), following incubation with (10 μM, open symbols) or without nema-A (closed symbols). Shown is the fraction of pharynxes achieving pumping at the indicated pace frequency. (c) Not every EPG shows a distinct r-peak. Exemplary analysis of pump duration, as time between E- and r-peak, points out a significant effect of nema-A on wt. (d) Contraction duration for the indicated wt or egl-19 mutant animals, during 1 Hz optical pacing, after 30 min incubation in the absence or presence of 17.9 μM nema-A (n = 19–28 animals, 523–727 contractions). (e) Contraction duration, (f) mean deviation from pace frequency, and (g) pump rate distributions for the indicated wt or egl-19 mutant animals, during 3.7 Hz optical pacing, after 30 min incubation in the absence or presence of 17.9 μM nema-A, dissolved in M9 buffer, were analyzed by kymographic analysis for higher throughput and detailed pump rate assessment (n = 9–15 animals, 669–1101 contractions). Pump rate distribution in (g) is color coded: black: >4.5 Hz, blue: 3.3–4.5 Hz, green: 2.5–3.3 Hz, yellow: 1.6–2.5 Hz, grey: 0.5–1.6 Hz, white: 0 Hz. Statistically significant differences: one way ANOVA with Fisher post-hoc test in (a), and t-test with Bonferroni correction in (c–f) (***P < 0.001; **P < 0.01; *P < 0.05).
Buffer, with or without 0.1% DMSO (used to keep nema-A dissolved), had no effects on pump duration (Supplementary Fig. S5). In EPG analyses at 1 Hz pacing (Fig. 8a), and in a stress test (Fig. 8b), application of nema-A (10 μM, 2 min) to exposed pharynxes in the cut-head preparation, surprisingly, had no significant effect on pump duration of wt or egl-19 alleles n2368, ad695 and tr70. However, as the ‘Auto EPG’ analysis software we used cannot reliably detect small r-peaks in EPGs (i.e. terminal bulb relaxation; Fig. 8c), EPG pump duration is calculated as E-R interval. This is different from kymographic analysis, where onset of terminal bulb contraction and relaxation are measured, corresponding to the E-r interval, which is longer than E-R intervals. To exemplify this for wt, we manually analyzed EPGs with clearly detectable r-peaks, unraveling significantly increased pump duration after nema-A exposure (Fig. 8c). This agreed well with kymographic analyses, where nema-A (17.9 μM for 30 min, as intact animals were used) evoked significantly increased contraction duration by ~33%, ~12% and ~20% in wt, ad695 and n2368, at 1 Hz (Fig. 8d,e), and ~7% for wt at 4 Hz pacing (Fig. 8e). Interestingly, n582ad952 pump duration was significantly reduced after nema-A treatment in EPG and kymograph analyses at 1 Hz (~16% and ~17%, respectively; Fig. 8a,d) and ~12% at 3.7 Hz (kymograph; Fig. 8e). The ameliorating effect of nema-A on egl-19(n582ad952), a g.o.f. allele combined with a r.o.f. mutation (n582 had no pumping phenotype on its own; Fig. 4b), occurred in an apparently allele-specific manner: Despite similarly prolonged pump durations in n2368 and ad695 g.o.f. animals without nema-A (Figs 4b and 5b), the antagonist further prolonged pumps, just as in wt. Nema-A (10 μM) did not significantly enhance the pump rate of egl-19(n582ad952) in the EPG stress test (Fig. 8b), but deviation from pulse frequency, as well as pump rate distribution were reverted to wt (Fig. 8f,g; kymographs, 17.9 μM).
Discussion
We established the C. elegans pharynx as a model for analysis of arrhythmogenic mutations in the EGL-19 L-type VGCC, some of which closely related to human mutations effecting Timothy syndrome. Optogenetic pacing enabled steady pumping up to 7 Hz for short periods, and at 2 or 4 Hz for times >1 min. High reproducibility of stimulation, with ~8 ms between light pulse and terminal bulb contraction (E-peak), as well as low jitter emphasize the quality of the model. For high throughput, we established a video analysis assay of pumping, using kymographic quantification. EPG as well as kymographic analyses demonstrated effects of nema-A, a DHP analog affecting EGL-19 function, in restoring normal pump parameters of one g.o.f. allele.
The optogenetically paced pharynx was not appreciably affected by neuronal input. Three pharyngeal neurons affect normal feeding15: M4 is required for isthmus peristalsis17, while M3 influences timing of terminal bulb relaxation50, and MC may modulate intrinsic pacing of pumping51. M4 and MC are cholinergic, and their hyperpolarization (NpHR-mediated), while significantly reducing spontaneous pump rate, had no effect on pump frequency or duration upon PMC-ChR2-pacing. As EGL-19 is expressed also in M4 (ref. 39), we assessed whether egl-19 mutations may affect paced pumping neuronally. However, while egl-19(n2368) pharynxes showed highly aberrant pumping, cholinergic neuron hyperpolarization did not exacerbate their pump phenotypes (Supplementary Fig. S4). Moreover, unc-13(s69) animals with abolished chemical synaptic transmission37 had unaltered pump duration during pacing.
To probe EGL-19 mutants in modeling LQT8-like arrhythmia in the pharynx, we analyzed several alleles. ad695, n582ad952, ad995, ad1015, n2368 and tr70 showed increased pump duration and reduced spontaneous pump rate on food. Optogenetic pacing of these alleles uncovered pumping phenotypes resembling LQT8 syndrome: Prolonged pump duration, equivalent to prolonged APs, was likely caused by decreased desensitization of the channel, as established for n2368 and ad695 alleles43. However, also a r.o.f. allele, ad1015 prolonged pump duration, for unknown reasons, but possibly pointing to specific roles of EGL-19 in pharynx vs. other muscles.
All of these alleles reduced pump rate, despite the mutated residues being located in different regions of the channel (Supplementary Figs. S6, S7). Gain-of-function alleles ad695, n582ad952 and n2368 were previously described to be myotonic and to cause terminal bulb relaxation deficiency; the extent of the effects was n2368 > ad695 ≫ n582ad95239, in line with patch clamp measurements for n2368 and ad69543. In the context of paced pumping, this translated into arrhythmia. n2368 showed highest pump duration in spontaneous and paced pumping, and also had the highest deviation from pacing frequency. The n2368 mutation G365R locates near the cytosolic end of S6 of module I, affecting the equivalent residue of a confirmed human LQT8 mutation (G402S45). Given its position in the protein, in structural alignments to the known NavAb structure46,52 (a surrogate for VGCC structures; Supplementary Figs. S6, S7), this mutation could affect channel desensitization by destabilizing the channel gate. Despite similarly increased pump duration, n582ad952 did not show aberrant pumping in EPGs, while it did so in kymographic analyses. Possibly, regular, forced pumping induces beneficial effects for this double allele; alternatively, the E-R interval duration is not as well-suited for uncovering arrhythmias as the E-r interval (which became obvious in our nema-A assays). S372L (in n582ad952, together with R899H in the S4 helix of the voltage sensor of module III) is very close to G365R. The mutated residue (A906V) in ad695, also located in the S4 helix of the voltage sensor of module III, may affect channel gating or repolarization-dependent closure. The S1010L mutation in tr70, prolonging pump duration and previously described as mildly myotonic and nema-A resistant (however, with respect to phenotypes other than pharynx pumping)42, is located in the pore loop of the third module. Residues in the analogous region of voltage gated K + channels affect C-type inactivation53; however, this type of inactivation has been not reported for VGCCs. Interestingly, also the r.o.f. mutation ad1015 led to prolonged pump duration, pointing to differences in EGL-19 function in pharynx versus other muscle.
egl-19 mutations affected the ability of the pharynx to lock-in to fast pacing. In stress tests, using cut head preparations, these mutants could not follow 5–6 Hz pacing, and in intact animals, they often switched from the induced pace rate to lower pump rates, following only every 2nd or 3rd stimulus. Similar effects are observed in LQT8 patients upon increased heart rate in the form of T-wave alternans. Pharynx anatomy is probably too simple to show such effects, yet missed pharynx pumps and T-wave alternans likely have the same origin, i.e. both LQT8 hearts and the pharynx model have prolonged Q-T and E-r interval, respectively. Using the pharynx analysis methods we established, egl-19 mutants can be recognized as abnormal in paced pumping, and relevant egl-19 mutations may thus serve as approximated LQT8 models: If novel anti-arrhythmogenic drugs ameliorate aberrant pharynx pumping in such mutants, they may likely act also in LQT8 patients. As we showed, Nema-A reverted prolonged contraction and arrhythmogenic behavior of the n582ad952 allele in paced pharynxes. In other g.o.f. alleles (ad695, n2368), nema-A, surprisingly, prolonged pump duration. As nema-A enhances desensitization, it should have effected a r.o.f. phenotype—yet, since the r.o.f. allele ad1015 also prolonged pump duration, this may be a specific consequence of EGL-19 r.o.f. in (paced) pharynx pumping. Our findings indicate allele-specific drug effects of nema-A, at least in our assay. Thus, patient-specific mutations may be introduced into the pharyngeal EGL-19 channel, and patient-specific drugs could be identified from drug libraries devised for Cav1.2.
In sum, we established an optogenetic pacing system in the pharynx of the genetically amenable animal model C. elegans, enabling to (introduce and) study mutations in genes required for normal pharynx pumping and rhythmicity, including orthologous human cardiac disease associated arrhythmogenic mutations. Our approach enables drug-testing and, using kymograph analysis, even to screen drug libraries. It should be transferable to other arrhythmia types with genetic causes, e.g. catecholaminergic polymorphic ventricular tachycardia (CPVT)10.
Methods
C. elegans strains
C. elegans strains were cultivated at 20 °C either on nematode growth medium (NGM)54 or for electrophysiological experiments on high growth medium (HGM), fed with E. coli strain OP-50-1. We used the following strains: N2 (wild type), DA695: egl-19(ad695), DA952: egl-19(n582ad952), DA995: egl-19(ad995), DA1006: egl-19(ad1006), DA1013: egl-19(ad1013), DA1015: egl-19(ad1015), MT1212: egl-19(n582), MT6129: egl-19(n2368), RP582: egl-19(tr69), RP583: egl-19(tr70), RP584: egl-19(tr71), RP585: egl-19(tr72), RP586: egl-19(tr73). unc-13(s69) was kindly provided by E. Jorgensen.
We generated these transgenic strains: ZX1652: egl-19(n2368); zxIs20[pmyo-2::ChR2(H134R)::mCherry; pges-1::nls::GFP], ZX1661: egl-19(n582ad952); zxIs20[pmyo-2::ChR2(H134R)::mCherry; pges-1::nls::GFP], ZX1662: N2; zxIs20[pmyo-2::ChR2(H134R)::mCherry; pges-1::nls::GFP], ZX1808: egl-19(ad695); zxIs20[pmyo-2::ChR2(H134R)::mCherry; pges-1::nls::GFP], ZX1809: egl-19(tr70); zxIs20[pmyo-2::ChR2(H134R)::mCherry; pges-1::nls::GFP], ZX1810: unc-13(s69); zxIs20[pmyo-2::ChR2(H134R)::mCherry; pges-1::nls::GFP], ZX1811: zxEx796[punc-17::NpHR::YFP;pelt-2::mCherry]; zxIs20[pmyo-2::ChR2(H134R)::mCherry; pges-1::nls::GFP], ZX1812: egl-19(n2368); zxEx797[punc-17::NpHR::YFP;pelt-2::mCherry]; zxIs20[pmyo-2::ChR2(H134R)::mCherry; pges-1::nls::GFP]
Plasmids
Plasmid pmyo-2::ChR2(H134R)::mCherry was generated by PCR amplification of the myo-2 promotor (primers oEF3: 5′-ACATGTCAGGTCGAGGCATTTG-3′ and oEF4: 5′-GGATCCCCCGAGGGTTAAAATGA-3′) from pPD132.102 (Addgene). The amplicon was digested with BamHI and PciI and ligated into plasmid pdat-1::ChR2::mCherry55. Punc-17::NpHR::YFP was described previously38.
Microinjection and generation of transgenic animals
Animals expressing ChR2 in PMCs were generated by microinjection into the germline (3 ng/μL pmyo-2::ChR2::mCherry, 20 ng/μL ges-1::nls::gfp, and 100 ng/μL pUC-19 plasmid). For chromosomal integration, 100 transgenic L4 larvae were irradiated with 66.6 mJ UV light. After starving, F2 generation was washed off with M9 buffer, 1000 transgenic animals were singled and integration events were identified by screening the F3 generation56. The integrated transgene zxIs20 was outcrossed 4x, and then crossed into the candidate egl-19 mutant strains.
Genotyping of egl-19 mutants
Genotyping of egl-19(n2386) occurred by phenotype, and was confirmed by sequencing of PCR products (primers oEF100: 5′-TCGAGCCATGATTCCTTTGC-3′, oEF101: 5′-TCTAGCTGCCCATTTACTCG-3′). egl-19(ad695) was genotyped after PCR (oEF102: 5′-AGCTGCTGAAGATCCTCTAC-3′ and oEF103: 5′-TGGCCATCCTTCGAAAGTTG-3′), the PCR product was digested with HpyCH4V and analyzed by 2% polyacrylamide gel electrophoresis. egl-19(n582ad952) was genotyped by MseI digestion of the oEF100/oEF101 amplicon. Genotyping of egl-19(tr70) was by differential annealing temperature binding of primers carrying mutated or wt bases at the 3‘-end. Primers for the mutation were oEF104: 5′-GCGATGATTTCACTTTTCGTAGTTCT-3′; oEF106: 5′-TGCTTGCACGCCAAAGATAC-3′, and oEF105: 5′-GCGATGATTTCACTTTTCGTAGTTTC-3′; oEF106: 5′-TGCTTGCACGCCAAAGATAC-3′ for wt. Last, all alleles were verified by sequencing.
Fluorescence microscopy
Expression of pmyo-2::ChR2::mCherry was analyzed on a Zeiss Axio Observer, with an 40x/0.25 Zeiss ∞/- APlan oil objective and mCherry filter set (Ex 580/23 nm, BS 605, Em 625/15 nm). Animals were transferred on 2% agarose pads in M9 buffer (K2PO4, 20 mM; Na2HPO4, 40 mM; NaCl, 80 mM ; MgSO4, 1 mM) and immobilized with 1 μL freshly prepared 50 mM NaN3 solution (Sigma-Aldrich, USA, St. Louis) in M9 from a 1 M stock in water.
Determination of spontaneous pump rate on food
One day before experiments, L4 larvae were placed on NGM dishes (55 mm, 8 mL NGM) seeded with 320 μL OP50 culture. For measurements requiring ATR (Sigma-Aldrich) 1:1000 of a 100 mM stock in ethanol was added to OP50. Pumping of animals on food was visually counted for 1 min. Measurements of animals co-expressing pmyo-2::ChR-2::mCherry and punc-17::NpHR::YFP (strains ZX1811 and ZX1812) were performed on a fluorescence stereo microscope (MZ16F; Leica, Germany) with continuous illumination (590 nm, 1 mW/mm2). Mean values, s.e.m. and further statistics (t-test and Bonferroni correction) were calculated with Excel (Microsoft, USA).
EPG-Recording and Optical Pacing
About 16–24 hours prior to experiments, young adult hermaphrodites were placed on fresh HGM plates with or without ATR. ATR (0.65 μl of a 100 mM stock in ethanol) was added to 650 μl of OP50 culture and spread onto 94 mm culture dishes (vented, Greiner Bio-One) containing 25 mL of HGM. For cut head preparations, animals were transferred into a recording chamber containing a Sylgard-coated coverslip (Ø25 mm) and filled with 1.5 mL of EmD50 buffer (NaCl, 140 mM; KCl, 3 mM; CaCl2, 3 mM; MgCl2, 1 mM; Hepes, 10 mM; D-Mannitol, 50 mM; pH 7.3 adjusted with NaOH). Dissection was performed under a stereomicroscope (Stemi 2000-C, Zeiss). The head was cut away from the body with a scalpel (Braun Aesculap, Germany) directly posterior to the terminal bulb. Upon dissection, the body wall muscles contract and expose the posterior pharynx57.
Electrophysiology was performed on an Olympus BX51W microscope. After positioning the recording chamber, the tip of the worm head was sucked under 100-fold magnification into an EPG-suction electrode, connected via a silver-chloride coated silver wire to a EPC-10 amplifier (Heka, Germany). The EPG-suction electrode was pulled from Borosilicate glass 1B100-4 (World Precision Instruments) by using a horizontal puller (Sutter P97) to ~20 μm inner diameter tip. As a bath electrode an Ag/AgCl pellet electrode (World Precision Instruments) was used27,28. For optical pacing with a 470 nm LED (KSL-70, Rapp Optoelectronics, Germany) the pharynx was positioned below a 60x water-immersion objective (LUMIplan FI/IR, 0.9 NA); an EGFP-ET filter set (AHF Analysentechnik AG, Germany) was used. EPG recording and triggering of light pulses was synchronized by PatchMaster software (Heka).
We recorded spontaneous pumping (at least for 1 minute) or stimulated the pharynx with 470 nm light pulses. 1.5–2 mW/mm2 intensity and pulse duration of 10 or 35 ms were most suitable. In every paced experiment, the first five (1 Hz stimulation) or ten pumps (>1 Hz stimulation) were excluded from analysis, to obtain stable pumping. For drug tests, 10 s of 1 Hz stimulation were recorded before and two minutes after drug application. Nema-A (Sigma-Aldrich) stock was prepared in DMSO before dilution in EmD50. 500 μl nema-A (30 μM in EmD50) was applied to the recording chamber containing 1 mL EmD50, resulting in a concentration of 10 μM. Final DMSO concentration was ~0.1%.
We used Review software (Bruxton Corporation, USA) to translate PatchMaster files to ABF files. Pump rate and duration were analyzed by AutoEPG58 (kindly provided by C. James, Southampton, UK). Excel was used for calculation of means and s.e.m., 1-way ANOVA with Fisher post-hoc test was performed using OriginPro (OriginLab, USA).
High-throughput kymograph recording and optical pacing
Transgenic L4 larvae cultivated with ATR were placed on fresh NGM dishes, seeded with OP-50-1 and ATR, one day prior to the assay. 1 μL of polystyrene microspheres (Polysciences 00876–15, 2.5% w/v suspension) were added on pads composed of 10% agarose59. About 10 animals were transferred into the beads and gently overlaid with a coverslip48.
Measurements were performed on a Zeiss Axio Observer, equipped with a 100 W HBO lamp, EGFP Filter (Ex. 472/30, beam splitter 570, Em. 675/50) and an 10x/0.25 Zeiss ∞/- APlan objective. Animals were stimulated with blue light pulses (35 ms, 1.5 mW/mm2) with a computer-controlled shutter (Sutter Instruments) every 250 ms (~4 Hz) or second (~1 Hz) over a 30 s period. Recording was via an ORCA Flash 4.0 sCMOS camera (Hamamatsu, Japan; 20 fps, 10 ms exposure time, 2 × 2 binning, 1024 × 1024 Pixel) and μManager v1.4 software. For drug tests, animals were incubated 30 min in 10 μL 17.9 μM nema-A solution (Sigma-Aldrich) with 0.3% DMSO in M9 (Stock solution 47.7 mM in DMSO). To determine influence of cholinergic neurons on pacing, measurements were performed during continuous illumination (0.5 mW/mm2) with a 590 nm LED (KSL-054, Rapp Optoelectronics) by 3.7 Hz stimulation, 35 ms blue light pulses, over 30 seconds.
Multi-kymographs of grinder movements from each animal were prepared using ImageJ. The region of interest was chosen next to the grinder, such that pump movements showed as bright or dark parts in the kymograph. After setting a line scan through the kymograph, pump movements were extracted as periodic grey value changes. Traces were normalized and if necessary inverted to achieve a consistent representation of pump movements, by a custom KNIME script49. Start points and end of minima were defined in each trace using manual correction and a machine learning process based on fixed rules. Calculation was restricted to the first 100 light pulses for measurements with stimulation frequencies of ~4 Hz. Pumping frequency was calculated from the beginning of consecutive peaks. Time dependent aberration from 4 Hz was determined if pump rate was >3.3 Hz and <4.5 Hz (±1 frame). Pumping frequencies were classified in groups: 0 Hz, 0.5–1.6 Hz, 1.6–2.5 Hz, 2.5–3.3 Hz, 3.3–4.5 Hz and >4.5 Hz. Contraction duration was calculated from fractions of time between beginning of peak and the end of the subsequent minima in Excel (Microsoft). Furthermore, data was correlated to particular ranges of pumping frequencies to assess correlation between observed pumping frequency and contraction duration. Statistically significant differences were determined by t-test and Bonferroni correction.
Structural model of EGL-19
The x-ray structure of the homo-tetrameric voltage gated sodium channel NavAb from Arcobacter butzleri (PDB file 3RVZ) was used as 3D model for positioning of mutated residues in the egl-19 alleles tested in this work. Each module of EGL-19 (a monomer) was aligned with NavAb (Supplementary Fig. S7, showing only the first 3 modules), and the likely positions of the residues determined and highlighted in the NavAb structure, rendered in different orientations, using PyMOL (www.pymol.org; Supplementary Fig. S6). Some views were given as 30 Å slabs for better visualization.
Additional Information
How to cite this article: Schüler, C. et al. Arrhythmogenic effects of mutated L-type Ca2+-channels on an optogenetically paced muscular pump in Caenorhabditis elegans. Sci. Rep. 5, 14427; 10.1038/srep14427 (2015).
Supplementary Material
Acknowledgments
We thank Mona Hoeret and Heike Fettermann for expert technical assistance, and Sebastian Wabnig for help in setting up kymograph measurements. We thank C. James (University of Southampton, UK) for the Auto EPG software, and E. Jorgensen for strains. Some strains were obtained from the Caenorhabditis Genetics Center (CGC). This research was funded by the EU FP7/2007–2013 under grant agreement No HEALTH-F2-2009-241526 (EUTrigTreat), and by the Deutsche Forschungsgemeinschaft (DFG), GO1011/3-1 and GO1011/3-2, and Cluster of Excellence Frankfurt—Macromolecular Complexes (EXC115), to A.G.
Footnotes
The authors declare no competing financial interests.
Author Contributions Designed research: C.S., E.F., L.S. and A.G. Performed experiments: C.S., E.F. and L.S. Performed data analysis: C.S., E.F., L.S. and W.S.C. Wrote the paper: C.S., E.F. and A.G., with help of the other authors.
References
- Shaw R. M. & Colecraft H. M. L-type calcium channel targeting and local signalling in cardiac myocytes. Cardiovascular research 98, 177–186 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesirca P., Torrente A. G. & Mangoni M. E. T-type channels in the sino-atrial and atrioventricular pacemaker mechanism. Pflugers Arch 466, 791–799 (2014). [DOI] [PubMed] [Google Scholar]
- Schmitt N., Grunnet M. & Olesen S. P. Cardiac potassium channel subtypes: new roles in repolarization and arrhythmia. Physiol Rev 94, 609–653 (2014). [DOI] [PubMed] [Google Scholar]
- Postema P. G. & Wilde A. A. The measurement of the QT interval. Current cardiology reviews 10, 287–294 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despa S. & Bers D. M. Na(+) transport in the normal and failing heart—remember the balance. Journal of molecular and cellular cardiology 61, 2–10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nattel S., Shiroshita-Takeshita A., Brundel B. J. & Rivard L. Mechanisms of atrial fibrillation: lessons from animal models. Progress in cardiovascular diseases 48, 9–28 (2005). [DOI] [PubMed] [Google Scholar]
- Roberts-Thomson K. C., Lau D. H. & Sanders P. The diagnosis and management of ventricular arrhythmias. Nature reviews Cardiology 8, 311–321 (2011). [DOI] [PubMed] [Google Scholar]
- Marsman R. F., Tan H. L. & Bezzina C. R. Genetics of sudden cardiac death caused by ventricular arrhythmias. Nature reviews Cardiology 11, 96–111 (2014). [DOI] [PubMed] [Google Scholar]
- Dixon R. E., Cheng E. P., Mercado J. L. & Santana L. F. L-type Ca2+channel function during Timothy syndrome. Trends in cardiovascular medicine 22, 72–76 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venetucci L., Denegri M., Napolitano C. & Priori S. G. Inherited calcium channelopathies in the pathophysiology of arrhythmias. Nature reviews Cardiology 9, 561–575 (2012). [DOI] [PubMed] [Google Scholar]
- Splawski I. et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell 119, 19–31 (2004). [DOI] [PubMed] [Google Scholar]
- Nattel S. & Carlsson L. Innovative approaches to anti-arrhythmic drug therapy. Nat Rev Drug Discov 5, 1034–1049 (2006). [DOI] [PubMed] [Google Scholar]
- Kumar K. & Zimetbaum P. J. Antiarrhythmic drugs 2013: state of the art. Current cardiology reports 15, 410 (2013). [DOI] [PubMed] [Google Scholar]
- Arnaout R. et al. Zebrafish model for human long QT syndrome. Proc Natl Acad Sci USA 104, 11316–11321 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L. & You Y. J. C. elegans feeding. WormBook; 10.1895/wormbook.1.150.1 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mango S. E. The C. elegans pharynx: a model for organogenesis. WormBook; 10.1895/wormbook.1.129.1 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L. & Horvitz H. R. A cell that dies during wild-type C. elegans development can function as a neuron in a ced-3 mutant. Cell 51, 1071–1078 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L. & Horvitz H. R. Pharyngeal pumping continues after laser killing of the pharyngeal nervous system of C. elegans. Neuron 3, 473–485 (1989). [DOI] [PubMed] [Google Scholar]
- Franks C. J., Holden-Dye L., Bull K., Luedtke S. & Walker R. J. Anatomy, physiology and pharmacology of Caenorhabditis elegans pharynx: a model to define gene function in a simple neural system. Invert Neurosci 6, 105–122 (2006). [DOI] [PubMed] [Google Scholar]
- Fang-Yen C., Avery L. & Samuel A. D. Two size-selective mechanisms specifically trap bacteria-sized food particles in Caenorhabditis elegans. Proc Natl Acad Sci USA 106, 20093–20096 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay J. P., Raizen D. M., Gottschalk A., Schafer W. R. & Avery L. eat-2 and eat-18 are required for nicotinic neurotransmission in the Caenorhabditis elegans pharynx. Genetics 166, 161–169 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtonda B. & Avery L. CCA-1, EGL-19 and EXP-2 currents shape action potentials in the Caenorhabditis elegans pharynx. J Exp Biol 208, 2177–2190 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. W., Fleischhauer R., Dent J. A., Joho R. H. & Avery L. A mutation in the C. elegans EXP-2 potassium channel that alters feeding behavior. Science 286, 2501–2504 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T., Sakube Y., Ahnn J., Kim do H. & Kagawa H. Molecular Dissection, Tissue Localization and Ca(2+) Binding of the Ryanodine Receptor of Caenorhabditis elegans. J Mol Biol 324, 123–135 (2002). [DOI] [PubMed] [Google Scholar]
- Maryon E. B., Saari B. & Anderson P. Muscle-specific functions of ryanodine receptor channels in Caenorhabditis elegans. J Cell Sci 111, 2885–2895 (1998). [DOI] [PubMed] [Google Scholar]
- Cho J. H., Eom S. H. & Ahnn J. Analysis of calsequestrin gene expression using green fluorescent protein in Caenorhabditis elegans. Mol Cells 9, 230–234 (1999). [PubMed] [Google Scholar]
- Raizen D. M. & Avery L. Electrical activity and behavior in the pharynx of Caenorhabditis elegans. Neuron 12, 483–495 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook A., Franks C. J. & Holden-Dye L. Electrophysiological recordings from the pharynx. WormBook; 10.1895/wormbook.1.110.1 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers C. M., Franks C. J., Walker R. J., Burke J. F. & Holden-Dye L. Regulation of the pharynx of Caenorhabditis elegans by 5-HT, octopamine, and FMRFamide-like neuropeptides. J Neurobiol 49, 235–244 (2001). [DOI] [PubMed] [Google Scholar]
- Horvitz H. R., Chalfie M., Trent C., Sulston J. E. & Evans P. D. Serotonin and octopamine in the nematode Caenorhabditis elegans. Science 216, 1012–1014 (1982). [DOI] [PubMed] [Google Scholar]
- Nagel G., Brauner M., Liewald J. F., Adeishvili N., Bamberg E. & Gottschalk A. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr Biol 15, 2279–2284 (2005). [DOI] [PubMed] [Google Scholar]
- Nagel G. et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci USA 100, 13940–13945 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruegmann T. et al. Optogenetic control of heart muscle in vitro and in vivo. Nat Methods 7, 897–900 (2010). [DOI] [PubMed] [Google Scholar]
- Bhatla N. & Horvitz H. R. Light and Hydrogen Peroxide Inhibit C. elegans Feeding through Gustatory Receptor Orthologs and Pharyngeal Neurons. Neuron 85, 804–818 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S. L. et al. A novel molecular solution for ultraviolet light detection in Caenorhabditis elegans. PLoS Biol 6, 0060198 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojanowski N. F., Padovan-Merhar O., Raizen D. M. & Fang-Yen C. Neural and genetic degeneracy underlies Caenorhabditis elegans feeding behavior. J Neurophysiol 112, 951–961 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond J. E., Davis W. S. & Jorgensen E. M. UNC-13 is required for synaptic vesicle fusion in C. elegans. Nat Neurosci 2, 959–964 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F. et al. Multimodal fast optical interrogation of neural circuitry. Nature 446, 633–639 (2007). [DOI] [PubMed] [Google Scholar]
- Lee R. Y., Lobel L., Hengartner M., Horvitz H. R. & Avery L. Mutations in the alpha1 subunit of an L-type voltage-activated Ca2+channel cause myotonia in Caenorhabditis elegans. Embo J 16, 6066–6076 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jospin M., Jacquemond V., Mariol M. C., Segalat L. & Allard B. The L-type voltage-dependent Ca2+channel EGL-19 controls body wall muscle function in Caenorhabditis elegans. J Cell Biol 159, 337–348 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok T. C. et al. A genetic screen for dihydropyridine (DHP)-resistant worms reveals new residues required for DHP-blockage of mammalian calcium channels. PLoS Genet 4, 1000067 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok T. C. et al. A small-molecule screen in C. elegans yields a new calcium channel antagonist. Nature 441, 91–95 (2006). [DOI] [PubMed] [Google Scholar]
- Laine V., Segor J. R., Zhan H., Bessereau J. L. & Jospin M. Hyperactivation of L-type voltage-gated Ca2+channels in Caenorhabditis elegans striated muscle can result from point mutations in the IS6 or the IIIS4 segment of the alpha1 subunit. J Exp Biol 217, 3805–3814 (2014). [DOI] [PubMed] [Google Scholar]
- Liu P., Ge Q., Chen B., Salkoff L., Kotlikoff M. I. & Wang Z. W. Genetic dissection of ion currents underlying all-or-none action potentials in C. elegans body-wall muscle cells. J Physiol 589, 101–117 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splawski I. et al. Severe arrhythmia disorder caused by cardiac L-type calcium channel mutations. Proc Natl Acad Sci USA 102, 8089–8096 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J., Scheuer T., Zheng N. & Catterall W. A. The crystal structure of a voltage-gated sodium channel. Nature 475, 353–358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockery S. R. et al. A microfluidic device for whole-animal drug screening using electrophysiological measures in the nematode C. elegans. Lab Chip 12, 2211–2220 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E., Sun L., Gabel C. V. & Fang-Yen C. Long-term imaging of Caenorhabditis elegans using nanoparticle-mediated immobilization. PLoS One 8, 3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold M. et al. KNIME: The Konstanz Information Miner. In: Data Analysis, Machine Learning and Applications (eds Preisach C., Burkhardt H., Schmidt-Thieme L., Decker R. ). (Springer, 2008). [Google Scholar]
- Avery L. Motor neuron M3 controls pharyngeal muscle relaxation timing in Caenorhabditis elegans. J Exp Biol 175, 283–297 (1993). [DOI] [PubMed] [Google Scholar]
- Raizen D. M., Lee R. Y. & Avery L. Interacting genes required for pharyngeal excitation by motor neuron MC in Caenorhabditis elegans. Genetics 141, 1365–1382 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J., Gamal El-Din T. M., Scheuer T., Zheng N. & Catterall W. A. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature 486, 135–139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero-Morales J. F. et al. Molecular driving forces determining potassium channel slow inactivation. Nat Struct Mol Biol 14, 1062–1069 (2007). [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal-Gadea A. et al. Caenorhabditis elegans selects distinct crawling and swimming gaits via dopamine and serotonin. Proc Natl Acad Sci USA 108, 17504–17509 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T. C. Transformation and microinjection. WormBook; 10.1895/wormbook.1.108.1 (2006). [DOI] [Google Scholar]
- Avery L., Raizen D. & Lockery S. Electrophysiological methods. Methods in cell biology 48, 251–269 (1995). [PMC free article] [PubMed] [Google Scholar]
- Dillon J. et al. AutoEPG: software for the analysis of electrical activity in the microcircuit underpinning feeding behaviour of Caenorhabditis elegans. PLoS One 4, 0008482 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham S. Methods in Cell Biology. WormBook; 10.1895/wormbook.1.49.1 (2006). [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.