Abstract
A non-autonomous DNA transposon in rice, nDart1, is actively transposed in the presence of an autonomous element, aDart1, under natural conditions. The nDart1-promoted gene tagging line was developed using the endogenous nDart1/aDart1 system to generate various rice mutants effectively. While the dominant mutants were occasionally isolated from the tagging line, it was unclear what causes dominant mutations. A semidominant mutant, Bushy dwarf tiller1 (Bdt1), which has the valuable agronomic traits of multiple tillering and dwarfism, was obtained from the tagging line. Bdt1 mutant carried a newly inserted nDart1 at 38-bp upstream of transcription initiation site of a non-protein-coding gene, miR156d. This insertion caused an upstream shift of the miR156d transcription initiation site and, consequently, increased the functional transcripts producing mature microRNAs. These results indicate that the total amount of miR156d is controlled not only by transcript quantity but also by transcript quality. Furthermore, transgenic lines introduced an miR156d fragment that flanked the nDart1 sequence at the 5′ region, suggesting that insertion of nDart1 in the gene promoter region enhances gene expression as a cis-element. This study demonstrates the ability of nDart1 to produce gain-of-function mutants as well as further insights into the function of transposable elements in genome evolution.
Although transposable elements consisting of highly repetitive DNA sequences were once thought to be junk DNA, they play an important role in genome reorganization and evolution1. In the 1940s, Barbara McClintock discovered the mobile element in maize, and various genome projects have revealed that transposable elements occupy large fractions of eukaryotic genomes. For example, approximately 10%, 35%, and 84% of Arabidopsis, rice, and Maize genomes, respectively, are transposon-derived sequences2,3,4. Active transposons are considered to be harmful to the host genomes because transposition induces chromosomal breakage and rearrangements and genetic changes. Only a limited number of transposons that escape from genetic or epigenetic regulation by host genomes can move5. Active transposons are often observed in plants as mutable phenotypes6,7,8.
Rice is an excellent model for the study on transposons because the whole genome sequences have been decoded with high accuracy and its genome is relatively transposon abundant3,8. An endogenous non-autonomous DNA transposon nDart1-0 was identified in a rice virescent mutant (Oryza sativa japonica L., cv. Taichung 65 background), pale-yellow leaf-variegated (pyl-v)9. The nDart1 elements are actively transposed over most of the genome in specific lines that carry an active autonomous element, aDart1–27, that encodes a transposase gene10. The nDart1 elements, a member of hAT superfamily, excise from inserted chromosome sites and transpose to another site. nDart1-0 and its closely related non-autonomous elements, nDart1–1 to nDart1–12, exhibit different transposition frequencies in spite of their high sequence similarity11. Previously, we revealed that the most active element, nDart1-0, has a low cytosine methylation status, and the active aDart1–27 possesses the hypomethylated promoter region12. Active transposons have been widely used as a tagging tool to elucidate gene functions. We developed the nDart1/aDart1 tagging line using the endogenous DNA transposon nDart1 elements. The tagging system is a powerful tool because nDart1 elements are actively transposed under natural growth conditions and tend to be integrated into genic regions, particularly into promoter-proximal genic regions, 0.5 kb in front of the putative initiation codons13. To effectively identify nDart1 insertion sites of the genome, the AFLP-based transposon display (TD) procedure, nDart-TD, and the iPCR-based procedure, nDart1-0-iPCR, were also developed14,15.
MicroRNAs (miRNAs) are important regulators of development in various organisms, including yeasts, fungi, animals, and plants16,17,18. In plants, transcripts transcribed from miRNA genes are processed into primary miRNAs (pri-miRNAs) with a 5′ cap and a poly-A tail. The pri-miRNAs form a stem-loop structure and then produce pre-miRNA by cleavage. The pre-miRNAs are processed into single-stranded small non-coding RNA molecules (~24 nucleotides). By interacting with the targeted genes, miRNAs induce post-transcriptional gene silencing. miRNAs were discovered by experimental approaches or bioinformatics predictions. To date, 592 miRNAs in 333 families have been annotated in rice (miRBase; http://www.mirbase.org/, Release 16). miR156 is well known as a quantitative repressor of vegetative phase change. miR156 targets SQUAMOSA promoter-binding-like (SPL) genes, and regulates the phase transition from the juvenile-to-adult phase19,20. The miR156 family is conserved in plants, and miR156-overexpressing transgenic rice, maize, Arabidopsis, and other plants demonstrated characteristic morphological changes: an increased number of branches, defects of the reproductive organs, a decrease in the number of seeds, and a reduction of tuber yields21,22,23,24. Overexpression of miR156 reduces the plastochron length by suppressing SPL genes that delay juvenile-to-adult phase transition.
Several reports have described loss-of-function mutants isolated from the nDart1-promoted tagging line. Herein, we report a gain-of-function mutant designated Bushy dwarf tiller1 (Bdt1) caused by insertion of nDart1–3 38 bp upstream of a non-coding RNA gene, miR156d. The nDart1 insertion increased the functional miR156d transcription level and conferred the multiple tillering and dwarf phenotype. The mechanism of the gain-of-function mutant is not well known so far. This study provides further insight into the function of active transposons and also clues regarding the mechanism of gain-of-function mutations and genome evolutions.
Results
Phenotype of semidominant Bdt1 mutant
A semidwarf mutant with an increasing number of tillers, designated as Bdt1, was isolated from the nDart1-promoted tagging line. A founder mutation in 07nD45-4 observed as normal or short panicles in individual tillers. The next generation plants that were derived from 13 panicles in independent tillers in 07nD45-4 were grown. Further genetic analysis showed that Bdt1 is a semidominant mutation for bushy and dwarf phenotypes in a single locus (Table 1). The homozygote mutant exhibited a more severe phenotype than that of the heterozygous mutant (Fig. 1a). The Bdt1 plants showed a bushy phenotype by developing secondary branches from irregular positions of culms. Although the internode length gradually increases from the basal to the upper part of the culm in rice (Fig. 1b), Bdt1 plants had nearly equal lengths of internodes (Fig. 1c). As compared with the wild-type plant, Bdt1 plants developed smaller leaves and more compact panicles. The homozygous Bdt1 plant bore fewer spikelets, and some panicles were morphologically aberrant and sterile. Overgrown bracts and leafy organs were observed above the panicle nodes (Fig. 1d–f). Some of the panicles appeared not to be exerted (Fig. 1f). Generally, a spikelet develops an awn from the lemma, but a pair of awns developed in the abnormal Bdt1 spikelet (Fig. 1g). A spikelet without a lemma and palea was also observed (Fig. 1h). The morphological differences between wild-type and Bdt1 plants are schematically represented in Fig. 1i.
Table 1. Segregation of dwarf and semi-dwarf phenotype in Bdt1 mutant progeny.
| Panicle-row lines | Panicle type in a founder | Segregation in next generation | ||
|---|---|---|---|---|
| Normal | Intermediate | Mutant | ||
| 3 | Short | 2 | 5 | 3 |
| 4 | Short | 1 | 5 | 2 |
| 5 | Short | 1 | 2 | 2 |
| 6 | Short | 1 | 3 | 4 |
| 7 | Short | 4 | 3 | 2 |
| 8 | Short | 2 | 2 | 3 |
| 9 | Short | 2 | 5 | 3 |
| 10 | Short | 2 | 2 | 1 |
| 11 | Short | 2 | 4 | 2 |
| 12 | Short | 1 | 2 | 1 |
| Total* | 18 | 33 | 23 | |
| 1 | Normal | 10 | 0 | 0 |
| 2 | Normal | 10 | 0 | 0 |
| 13 | Normal | 10 | 0 | 0 |
| Total | 30 | 0 | 0 | |
*Chi-square value = 1.54, df = 2, 0.25 < P < 0.50
Figure 1. Semidominant phenotypes of Bdt1 plants.
(a) Three-month old wild-type (BDT1/BDT1), Bdt1 heterozygous (BDT1/Bdt1), and Bdt1 homozygous (Bdt1/Bdt1) plants. Bar = 10 cm. Primary culms of the WT (b) and Bdt1/Bdt1 plants (c). Bar = 10 cm. (d–h) Abnormal panicles of Bdt1/Bdt1 plants. Bar = 5 cm in D–F, bar = 1 cm in G and H. White and red arrowheads indicate overgrown bracts and leaf-like structures, respectively. (i) Morphological phenotypes of WT and Bdt1 plants. Each broken line and each circle represents an internode and a panicle, respectively.
Identification of the Bdt1 allele
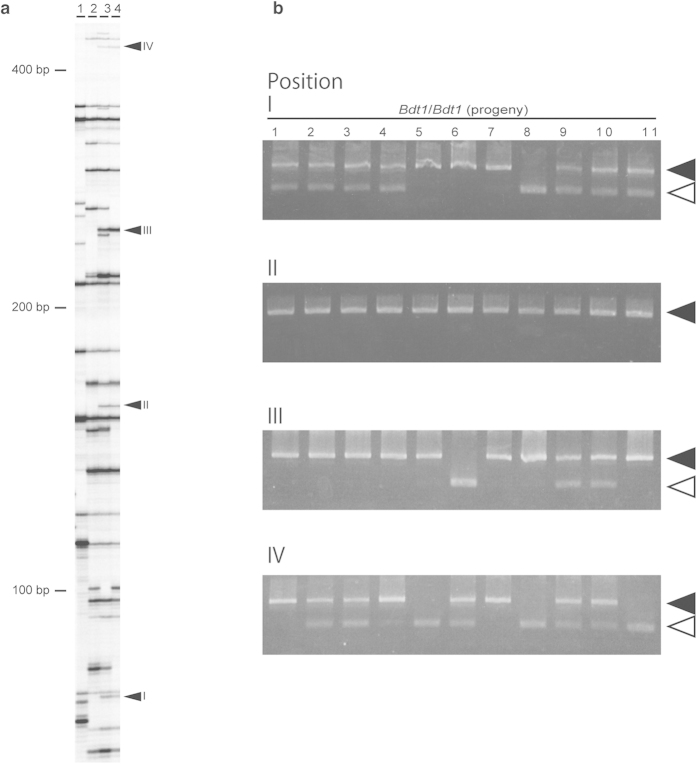
Because we predicted that a new insertion of the nDart1 element caused the Bdt1 mutation, nDart-TD was performed to identify unique insertion sites of nDart1 elements in Bdt1 plants. Using nDart-TD, 4 newly inserted nDart1 elements were detected as common bands between Bdt1/Bdt1 and BDT1/Bdt1 mutants (Fig. 2a). To eliminate non-Bdt1-related insertions, the genotype of the 4 genes was identified among the Bdt1 progeny. The results showed that the Bdt1 phenotype associated with the insertion was band II (Fig. 2b). This new insertion was nDart1–3, one of the active nDart1 elements. We identified the new insertion 38 bp upstream of the transcription initiation site (TIS) of the Os02g0180800 gene (AK073452, full-length cDNA in Nipponbare). This gene consists of five exons, and the first exon contains a 22-nt sequence corresponding to mature miR156d (Fig. 3a). Subsequent PCR analysis with primers flanking the nDart1–3 revealed that the nDart1–3 insertion in the Bdt1/Bdt1 plants was in the homozygous condition, and the amplified fragments from the BDT1/Bdt1 plant were identical in size to those from wild-type and Bdt1/Bdt1 plants (Fig. 3b). The mutant phenotype cosegregated with the nDart1–3 insertion. These results suggest that nDart1–3 insertion upstream of miR156d conferred the Bdt1 mutant phenotype.
Figure 2. Identification of the Bdt1 allele.
(a) TD analysis with TaqI-digested DNA are lane 1, Nipponbare; lane 2, BDT1/BDT1; lane 3, BDT1/Bdt1; lane 4, Bdt1/Bdt1. 1 The arrowheads represent polymorphic bands (positions 1–4) among WT and Bdt1 plants. (b) Genotyping of progeny plants obtained from Bdt1/Bdt1 by PCR analysis. The filled and open arrowheads indicate PCR-amplified bands using primer sets that detect nDart1–3 insertion at positions I–IV. Insertion of nDart1 was detected in all of the progeny only at position II among four candidates.
Figure 3. Characterization of the Bdt1 allele.
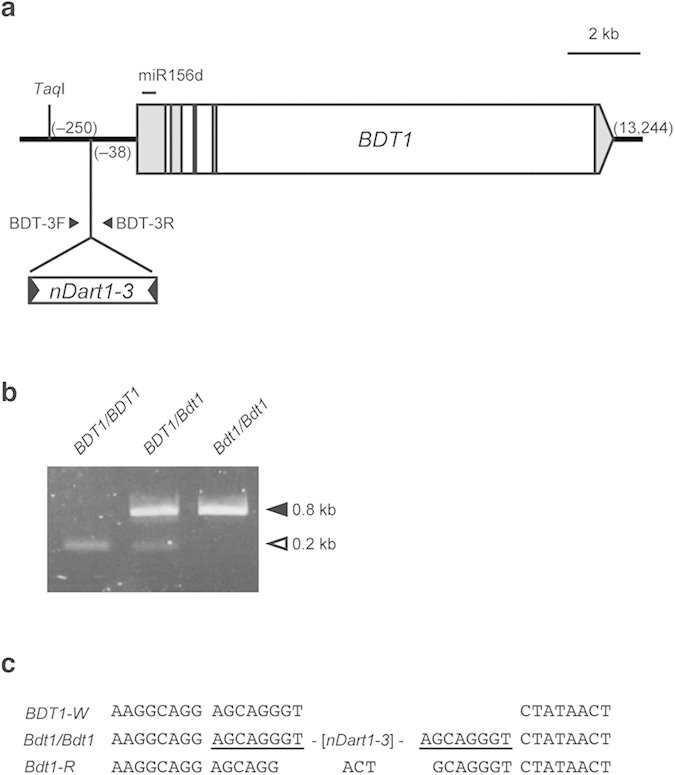
(a) Structure of the BDT1 gene carrying the nDart1–3 insertion. Shaded and white boxes represent BDT1 exons and introns, respectively. The horizontal line above the box represents the position where mature miR156d is transcribed. The numbers in parentheses represent the nucleotide positions from the transcription initiation sites of full-length cDNA AK073452. The small horizontal arrowheads indicate positions of the primers used for PCR amplification to characterize the insertions of nDart1–3 (Fig. 2b). (b) Genotyping of WT and Bdt1 plants by PCR analysis. The filled and open arrowheads indicate PCR-amplified bands with and without nDart1 insertion, respectively. (c) Sequence of the footprints generated by nDart1–3 excisions. The WT, homozygous Bdt1, and germinal revertant alleles are indicated by BDT1-W, Bdt1/Bdt1, and Bdt1-R, respectively. Target site duplication generated by nDart1–3 insertion at the Bdt1 locus is underlined.
Excision events of DNA transposons often leave sequence alternation called footprint at the flanking sites. Both perfect excision of the transposon sequence and a footprint that does not impair gene function(s) can generate revertants (i.e., nucleotide additions or deletions in multiples of three nucleotides generate wild-type revertants). nDart1-promoted mutants often produce germinal revertants in their progeny. To assess whether the Bdt1 mutation was indeed caused by the insertion of nDart1–3, we isolated a germinal revertant with a wild-type phenotype and examined the excision of nDart1–3 from the Bdt1 allele. The germinal revertant with the Bdt1-R allele was found to carry an 8-bp insertion and nucleotide substitutions as the footprint (Fig. 3c). Based on these results, we can conclude that the BDT1 gene corresponds to a non-coding RNA gene, Os02g0180800.
Transcript analysis of Bdt1
nDart1–3 insertion upstream of miR156d caused a semidominant phenotype. To investigate the effects of nDart1–3 insertion on the expression of the miR156d gene, real-time quantitative PCR (qPCR) analysis was performed. The expression levels of microRNA genes vary according to the developmental stage and the tissue type. We evaluated the expression levels in 3rd leaves, flag leaves, and lowermost nodes of Bdt1/Bdt1 plants using a primer set to specifically amplify the 5′ region of BDT1 where miR156d corresponding sequence. The results showed that the miR156d expression level in 3rd leaves of Bdt1/Bdt1 seedlings was more than 3-fold higher than that of wild-type seedlings (Fig. 4a). miR156d was also highly expressed in the flag leaves and nodes of mutants (Fig. 4a). nDart1–3 insertion increased the miR156d transcripts containing pre-miR156d through juvenile and adult stages in Bdt1 plants.
Figure 4. Transcription analysis of functional miR156d.
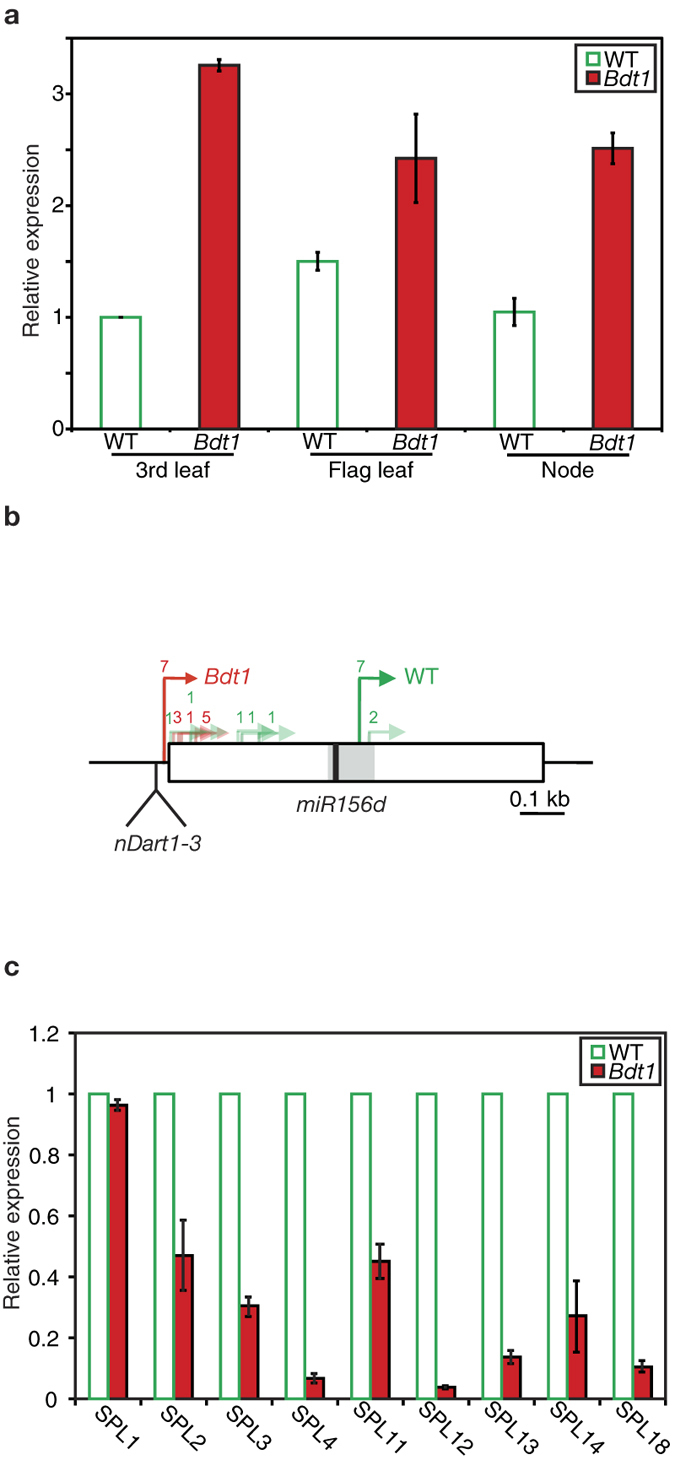
(a) Tissue-specific expression patterns of the miR156d gene in WT and Bdt1 plants. (b) Major transcription initiation sites of miR156d in WT and Bdt1 plants. Red and green arrows indicate transcription initiation sites of miR156d in WT and Bdt1 plants, respectively. Numbers above the arrows represent the numbers of clones that correspond to the transcription initiation site. The left end of the white box indicates the reported 5′ terminal of the full-length cDNA (AK073452) in Nipponbare (http://rapdb.dna.affrc.go.jp/). The gray and black boxes show the corresponding positions of the pre-miR156d and miR156d sequences. (c) Relative expression of SPL1 gene and miR156-targeted SPL genes in 3rd leaves of WT and Bdt1 plants. Error bars indicate SD.
In order to clarify the regulation mechanism of miR156d expression in Bdt1 mutants, transcription initiation sites (TIS) of the miR156d gene in wild-type and Bdt1/Bdt1 plants were identified by 5′-RACE analysis. In leaf blades of wild-type seedlings, some of the transcription started from a few different sites upstream of the pre-miR156d region, but the major TIS was downstream of the mature miR156d sequence. In contrast, the TISs in leaf blades of Bdt1/Bdt1 seedlings were only detected upstream of the pre-miR156d region. All Bdt1 transcripts contained the pre-miR156d region. The detected TISs are summarized in Fig. 4b. These results suggest that nDart1–3 insertion altered the TIS of miR156d, and the upstream shift in the transcripts increased functional miR156d expression.
Expression of miR156d-targeted SPL genes
In rice, 11 of 19 SPL genes contain miR156 target sites23. To confirm an increase of mature miR156d levels in Bdt1, the expression levels of miR156d-targeted SPL genes were examined in the 3rd leaves of wild-type and Bdt1 seedlings. qPCR analysis showed that a non-miR156d-targeted gene, SPL1, was expressed at the same level in wild-type and Bdt1 seedlings (Fig. 4c). In contrast, expression levels of miR156d-targeted SPLs (SPL2, SPL3, SPL4, SPL11, SPL12, SPL13, SPL14, and SPL18) were significantly decreased in Bdt1 seedlings (Fig. 4c). The expression levels of SPL4 and SPL12 were reduced more than 90%. These results suggest that BDT1 transcripts induced by nDart1–3 insertion generate functional microRNA and effectively suppress expression of SPL genes.
Effects of nDart1 insertion as a cis-element
Expression analysis of Bdt1 plants revealed that nDart1–3 insertion activated its downstream gene expression. To evaluate the effects of nDart1–3 on its downstream gene, we constructed three types of vectors, generated rice transgenic lines, and compared the phenotypes. Vectors pWT-BDT1 and pBdt1-BDT1 carried 1.6-kb BDT1 cDNA sequences (AK073452) that flanked the upstream region of wild-type and Bdt1 plants, respectively (Fig. 5a). Both constructs were postulated to contain the BDT1 promoter region. In pBdt1-BDT1, the 607-bp nDart1–3 sequence presents within the promoter region (Fig. 5a). Transgenic lines with CaMV35S promoter-driven BDT1 were generated to evaluate Bdt1 expression levels (Fig. 5a). By Agrobacterium-mediated transformation with 10 pCambia1305.1 (Control), 8 pWT-BDT1 lines, 12 pBdt1-BDT1 lines, and 10 p35S-BDT1 lines were obtained (Supplemental Table S1). Typical examples of T0 plants are shown in Fig. 5b. Transgenic plants transformed with pWT-BDT1 were moderately short. pBdt1-BDT1 and p35S-BDT1 plants exhibited morphological changes observed in plants that overexpressed miR156. The pBdt1-BDT1 lines had aberrant panicles and significantly reduced fertility. p35S-BDT1 transgenic lines showed more significant changes in phenotype. p35S-BDT1 plants generated no T1 generation due to the lack of inflorescences and a sterile spikelet on a branch. These observations indicate that the nDart1–3 sequence indeed increased the amount of mature miR156d in Bdt1, without disruption of miR156d gene. The miR156 function is quantitative. Based on the phenotype, the effect of nDart1–3 on miR156d expression was modest as compared to that of the CaMV35S promoter.
Figure 5. Effects of nDart1–3 insertion on miR156d expression.
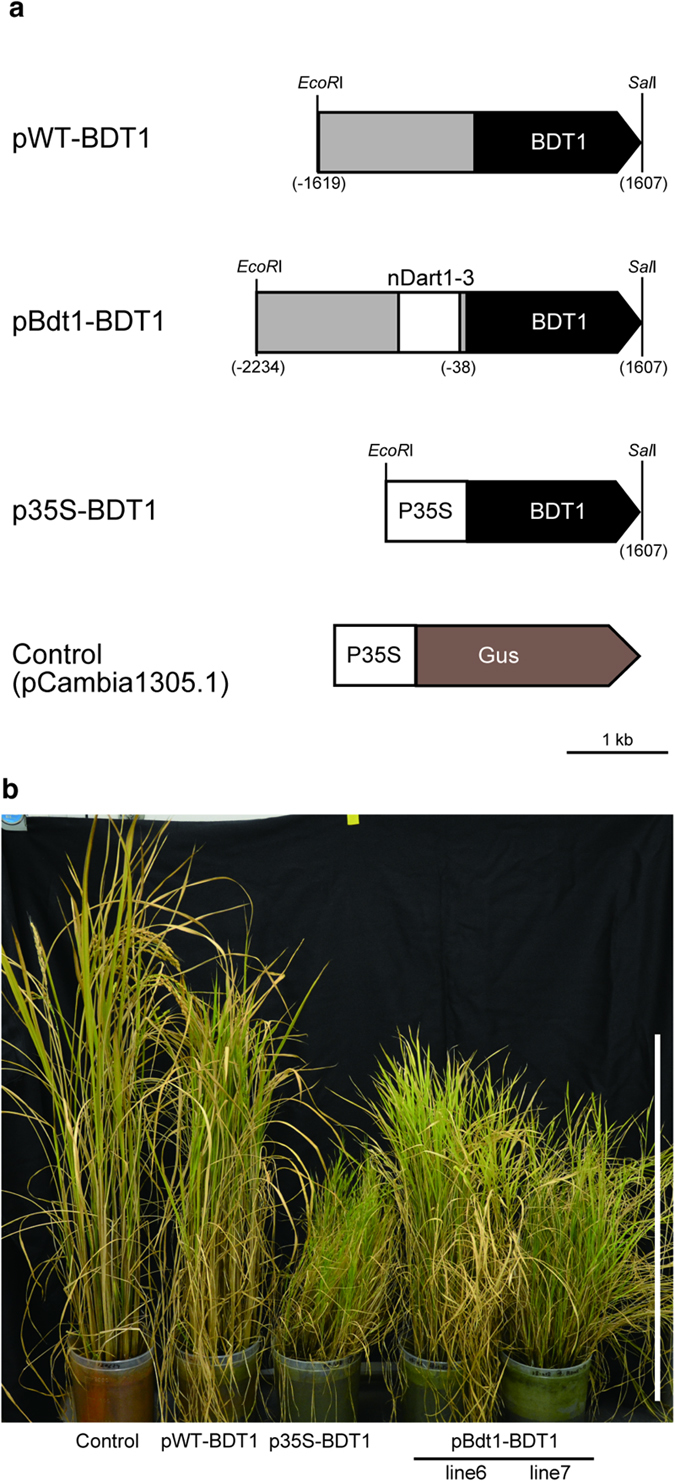
(a) Constructs of plasmid vectors used to generate transgenic lines. (b) Three-month old transgenic T0 lines. Scale bar = 50 cm.
Discussion
A semidominant mutant Bdt1 that displayed a semidwarf phenotype, enhanced tillering, and compact (and aberrant) panicles was isolated from the nDart1-promoted tagging line (Fig. 1). nDart-TD analysis revealed that the mutant allele Bdt1 was caused by the insertion of nDart1–3 into the promoter region of the microRNA gene, miR156d (Fig. 3a). Afterward, the mutant allele was confirmed by the fact that the germinal revertant exhibited the wild-type phenotype. The expression level of the miR156d transcripts in leaf blades of Bdt1 seedlings was approximately threefold higher than that of the wild type (Fig. 4a). 5′-RACE analysis showed that miR156d transcripts without mature miR156d sequences were not a small amount in wild-type leaves. On the other hand, all miR156d transcripts contained a pre-miR156d region, and the major TIS was relatively upstream in Bdt1 leaves (Fig. 4b). These results suggest that miR156d transcripts were originally a mixture that included and excluded the pre-miR156d region. The upstream shift of the TIS by nDart1–3 insertion increased the functional transcripts that produced mature miR156d. The quantity of functional miR156d transcripts is controlled not only by the transcription activity but also by the TIS of miR156d. The miR156d would have a complex regulation mechanism for the generation of mature miR156d. The major TIS may vary based on the plant organ and the developmental stage in plants.
In rice, the miR156 family consists of 12 members (a–l), and miR156 genes (a–j) produce the identical 20-nt mature miR15625,26. These microRNA genes show different spatial and temporal patterns of expression and different interaction patterns with SPLs in spite of the total match of miRNA sequences26. To confirm the increase of mature miR156d in Bdt1, the expression levels of miR156d-targeted SPLs were examined. RT-qPCR analysis showed that the expression of the targeted SPLs was obviously decreased. These results suggest that the increased miR156d transcripts in Bdt1 seedlings were efficiently processed into mature microRNA and indeed suppressed the targeted genes. The reduced ratio in a series of SPLs was not equal, but these patterns were similar to the results in miR156d-overexpressing plants26. The suppression level of miR156d varies according to the organ and the developmental stage. miR156d would not primarily contribute to the regulation of SPL2 and SPL11.
Subsequently, to evaluate the effects of nDart1–3 insertion on downstream gene expression, three types of transgenic plants were constructed. As part of a complementation assay, a BDT1 fragment that flanked the wild-type promoter region and a BDT1 fragment that flanked the Bdt1 upstream region were introduced into Nipponbare (Fig. 5a). The transgenic lines indicate that the nDart1 sequence in the promoter upregulated miR156d expression. The pBdt1-BDT1 lines had aberrant panicles and significantly reduced fertility. The CaMV35S-driven transgenic lines exhibited a more severe phenotype that resulted in complete sterility. The effect of nDart1 insertion was lower than that of a constitutive promoter, CaMV35S. The modest semidwarf phenotype in pWT-BDT1 lines can be interpreted as the co-expression of endogenous and exogenous BDT1 gene(s). Our previous study, using GUS reporter genes, showed that nDart1 itself does not have promoter activity10. Therefore, nDart1 elements require an endogenous promoter to upregulate flanking gene expressions. The increase of miR156d in Bdt1 mutation would mainly result from TIS alternation by recognition of RNA polymerase II. Generation patterns of miR156d in endogenous promoter-dependent Bdt1 plants must be different from those of constitutive promoter-driven miR156d-overexpressing plants in developmental stages and tissue types.
Several reports described gain-of-function mutants caused by transposons. The teosinte branched1 (tb1) gene in maize was identified as the major contributor to domestication of modern corn from the grass-like ancestor teosinte (Zea mays ssp. parviglumis)27. Recent studies have revealed that insertions of LTR-retrotransposon, Hopscotch 58–69 kb upstream of tb1 are associated with a quantitative increase in the expression of the maize tb1 allele28,29. Further genetic analysis demonstrated that the expression level in maize with transposon insertions upstream of the tb1 regulatory region was higher than that of teosinte. Cg1 is a maize miR156-overexpressing mutant caused by the insertion of a retrotransposon, STONER, 42 bp upstream of the TIS of tandem miR156b/c genes21. Bdt1 and Cg1 appeared to be the results of similar events in the genes and transposon insertion sites. However, nDart1–3 is a non-autonomous element that does not encode transposase, and the TIS of miR156d was downstream of nDart1–3. On the other hand, STONER is a long autonomous element, and Cg1 transcription is initiated within the STONER element. The activation mechanism of the downstream gene by nDart1 insertion must be different from that of Cg1. In the case of a dominant rice mutant apo1-D, nDart1-0 was inserted 3.5 kb upstream of the APO1 region and caused an increase of APO1 expression30. They suggested that the nDart1-0 insertion conferred the loss of negative regulation of APO1 expression. Because the nDart1 insertion sites were not adjacent to the APO1 TIS, the nDart1 must disrupt the silencer region of APO1.
The nDart1-promoted tagging line is highly expected to produce both loss-of-function and gain-of-function mutants to characterize various unknown genes because uncharacterized gene functions by loss-of-function mutation may be clarified by studying gain-of-function mutants. For example, pyl and snow-white leaf1 are recessive loss-of-function mutants caused by nDart1 insertion into the 5′UTR. These insertions caused a downshift of the TIS, and their transcripts lacked the proper initiation codons9,15. In Bdt1, nDart1 caused an upstream shift of the TIS and increased gene expression, including the pre-miR156d region. However, the mechanism of gain-of-function mutation is not well understood. Further investigation of nDart1 insertion sites and their impact on gene expression are necessary to reveal the alternation mechanism of TIS. It is well known that transposons are important sources of evolution. Some transposons can enhance neighboring gene expressions30,31,32. On the other hand, nDart1 elements did not have such promoter/enhancer activity10, but this study demonstrated that transposons without such activity contribute to genome evolution and genetic variation not only by gene disruption but also by activating gene expressions. We believe that this valuable feature of nDart1 will promote fine-tuning the useful properties for breeding.
Materials and Methods
Plant materials and growth conditions
Plants were grown in a growth chamber (12 h of light and 12 h of darkness) at 30 °C or in pots under natural conditions as previously described9.
Identification of the Bdt1 allele by nDart-TD
The nDart-TD was carried out according to the procedure previously reported with some modifications14. The genomic DNA was cleaved with the TaqI restriction enzyme and ligated to the adapter in a 25-μl reaction mixture. The initial amplification was performed in a 20-μl reaction mixture that contained adapter-ligated DNA and a set of adapter and Dart5′-1-1st primers. Subsequently, a set of primers composed of the adapter primer and the Dart5′-1,2,3-2nd primer labeled with 6-carboxyfluorescein (4.8 pmol each) was subjected to nested amplification. The 0.5 μl of amplified fragments mixed with a loading cocktail containing 9 μl of Hi-Di Formamide was heated at 95 °C for 3 min and then chilled on ice. The samples injected into an ABI 3130xl genetic analyzer (Applied Biosystems, Foster City, CA, USA) were analyzed by using GeneMarker (SoftGenetics, LLC, State College, PA, USA) genotyping software to detect TD patterns. To read the nucleotide sequences of the candidate nDart1 integration sites where wild-type and Bdt1 plants show polymorphism, the rest of the amplification products were concentrated by ethanol precipitation and electrophoresed on a 5% denaturing polyacrylamide gel. The bands were visualized using FMBIO II Multi-View (Hitachi Software Engineering, Tokyo, Japan). Unique bands in the Bdt1 mutants were extracted from the gel and were amplified with a set of adapter and Dart5′-1,2,3-2nd primers. The reaction products were subjected to sequencing analysis. The primer and adapter sequences used for TD analysis are listed in Supplemental Table S2.
General nucleic acid procedures
General nucleic acid procedures, including the preparation of genomic DNA and RNA, PCR and RT-PCR amplifications, and 5′-RACE analysis, were performed as previously described9,15.
RT-qPCR
Primers used to examine mRNA expressions are listed in Supplemental Table S2. Real-time RT-qPCR for expressions of SPL genes was performed as described in Xie et al. 2012. The expression levels of miR156d and SPL genes were normalized to the reference genes U6 (RNA) and AK071592, respectively, and their relative expression levels were calculated using the 2−ΔΔCT method31. Each set of primers was tested with cDNA from wild-type and Bdt1/Bdt1 plants with three technical32,33 replicates per sample using the THUNDERBIRD SYBER qPCR Mix (Toyobo, Osaka, Japan).
Constructs and transformation
For p35S-BDT1, the AK073452 fragment carrying PstI-SalI sites was obtained by amplification of full-length cDNA of AK073452 (the Rice Genome Resource Center RGRC, http://www.rgrc.dna.affrc.go.jp/index.html), using primers AK073452-PstI and AK073452-SalI, and the fragment was fused to the CaMV35S-promoter sequence carrying EcoRI-PstI sites amplified with primers 35S-EcoRI and 35S-XhoI. For pWT-BDT1 and pBdt1-BDT1, the AK073452 fragment carrying BamHI-SalI sites was fused to 1.6- and 2.2-kb fragments carrying EcoRI-BamHI sites amplified from the AK073452 upstream regions of wild-type and Bdt1 plants with primers BDT1-F (−) 1616-EcoRI and BDT1-R177, respectively. These three fragments were introduced into multiple cloning sites of the pCAMBIA1305.1 vector. As a negative control, the pCAMBIA1305.1 vector was used.
Agrobacterium-mediated rice transformation was performed using Agrobacterium tumefaciens strain EHA105, as described previously10.
Additional Information
How to cite this article: Hayashi-Tsugane, M. et al. A gain-of-function Bushy dwarf tiller 1 mutation in rice microRNA gene miR156d caused by insertion of the DNA transposon nDart1. Sci. Rep. 5, 14357; doi: 10.1038/srep14357 (2015).
Supplementary Material
Acknowledgments
Some experiments were performed with equipment maintained by Functional Genomics and Model Plant Facility in the NIBB. This work was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan [No. 22780007, 25450013 and No. 2654 (part of Joint Research Program implemented at the Institute of Plant Science and Resources, Okayama University in Japan) to K.T.], the NIBB Cooperative Research Program (14-357 to M.M.), Sapporo Bioscience Foundation and the Graduate University for Advanced Studies (Sokendai).
Footnotes
Author Contributions M.H.-T., M.M. and K.T. designed the reserch. M.H.-T. performed most of the experiment. K.T. performed qPCR. M.H.-T., M.M. and K.T. analyzed data and wrote the main manuscript. All authors reviewed the manuscript.
References
- Slotkin R. K. & Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8, 272–285 (2007). [DOI] [PubMed] [Google Scholar]
- Arabidopsis genome initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 (2000). [DOI] [PubMed] [Google Scholar]
- International rice genome sequencing project. The map-based sequence of the rice genome. Nature 436, 793–800 (2005). [DOI] [PubMed] [Google Scholar]
- Wei F. et al. Detailed Analysis of a Contiguous 22-Mb Region of the Maize Genome. PLoS Genet 5, e1000728, 10.1371/journal.pgen.1000728 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H., Tsugane K., Kanno T. & Nishimura T. DNA methylation in plants: relationship to small RNAs and histone modifications, and functions in transposon inactivation. Plant Cell Physiol 53, 766–784 (2012). [DOI] [PubMed] [Google Scholar]
- May B. P. & Martienssen R. A. Transposon mutagenesis in the study of plant development. Crit Rev Plant Sci 22, 1–35 (2003). [Google Scholar]
- Boavida L. C. et al. A collection of Ds insertional mutants associated with defects in male gametophyte development and function in Arabidopsis thaliana. Genetics 181, 1369–1385 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa M., Tsugane K. & Iida S. Effective contribution of the nDart1 transposon-tagging system to rice functional genomics in Advances in Genetics Research (ed Urbano K. V.) 259–272 (Nova Science, 2011).
- Tsugane K. et al. An active DNA transposon nDart causing leaf variegation and mutable dwarfism and its related elements in rice. Plant J 45, 46–57 (2006). [DOI] [PubMed] [Google Scholar]
- Shimatani Z. et al. Characterization of autonomous Dart1 transposons belonging to the hAT superfamily in rice. Mol Genet Genomics 281, 329–344 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johzuka-Hisatomi Y. et al. Homologous recombination-dependent gene targeting and an active DNA transposon nDart-promoted gene tagging for rice functional genomics in Rice Biology in the Genomics Era: Biotechnology in Agriculture and Forestry. Vol. 62 (ed. Hirano H.-Y., Hirai A., Sano Y. & Sasaki T.) 81–94 (Springer, 2008). [Google Scholar]
- Eun C. H. et al. Activation and Epigenetic Regulation of DNA Transposon nDart1 in Rice. Plant Cell Physiol 53, 857–868 (2012). [DOI] [PubMed] [Google Scholar]
- Takagi K., Maekawa M., Tsugane K. & Iida S. Transposition and target preferences of an active nonautonomous DNA transposon nDart1 and its relatives belonging to the hAT superfamily in rice. Mol Genet Genomics 284, 343–355 (2010). [DOI] [PubMed] [Google Scholar]
- Takagi K., Ishikawa N., Maekawa M., Tsugane K. & Iida S. Transposon display for active DNA transposons in rice. Genes Genet Syst 82, 109–122 (2007). [DOI] [PubMed] [Google Scholar]
- Hayashi-Tsugane M. et al. A mutable albino allele in rice reveals that formation of thylakoid membranes requires the SNOW-WHITE LEAF1 gene. Plant Cell Physiol 55, 3–15 (2014). [DOI] [PubMed] [Google Scholar]
- Millar A. A. & Waterhouse P. M. Plant and animal microRNAs: similarities and differences. Functional & integrative genomics 5, 129–135 (2005). [DOI] [PubMed] [Google Scholar]
- Pritchard C. C., Cheng H. H. & Tewari M. MicroRNA profiling: approaches and considerations. Nat rev Genet 13, 358–369 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. & Wang H. The miR156/SPL module, a regulatory hub and versatile toolbox, gears up crops for enhanced agronomic traits. Molecular plant 10.1016/j.molp.2015.01.008 (2015). [DOI] [PubMed] [Google Scholar]
- Miura K. et al. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet 42, 545–549 (2010). [DOI] [PubMed] [Google Scholar]
- Huijser P. & Schmid M. The control of developmental phase transitions in plants. Development 138, 4117–4129 (2011). [DOI] [PubMed] [Google Scholar]
- Chuck G., Cigan A. M., Saeteurn K. & Hake S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet 39, 544–549 (2007). [DOI] [PubMed] [Google Scholar]
- Wang J. W., Schwab R., Czech B., Mica E. & Weigel D. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20, 1231–1243 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y. et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42, 541–544 (2010). [DOI] [PubMed] [Google Scholar]
- Bhogale S. et al. MicroRNA156: a potential graft-transmissible microRNA that modulates plant architecture and tuberization in Solanum tuberosum ssp. andigena. Plant Physiol 164, 1011–1027 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K., Wu C. & Xiong L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol 142, 280–293 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K. et al. Gradual increase of miR156 regulates temporal expression changes of numerous genes during leaf development in rice. Plant Physiol 158, 1382–1394 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J., Stec A. & Gustus C. teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141, 333–346 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer A., Zhao Q., Ross-Ibarra J. & Doebley J. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet 43, 1160–1163 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Zhang J., Yan J. & Song R. Two transposable element insertions are causative mutations for the major domestication gene teosinte branched 1 in modern maize. Cell Res 21, 1267–1270 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda-Kawakatsu K. et al. Expression level of ABERRANT PANICLE ORGANIZATION1 determines rice inflorescence form through control of cell proliferation in the meristem. Plant Physiol 150, 736–747 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Wang H. The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu Rev Plant Biol 65, 505–530 (2014). [DOI] [PubMed] [Google Scholar]
- Dooner H. K, Weil C. Transposon and gene creation in Plant transposons and gene dynamics in evolution (ed. Fedoroff N.) 143–179 (Wiley-Blackwell, 2013).
- Pfaffl M. W. Relative quantification in Real-time PCR. (ed. Dorak M. T.) 63–80 (Taylor & Francis, 2007).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.