Abstract
The dismal outcome of gastric cancer patients highlights the need for diagnostic biomarkers and effective therapeutic targets, such as microRNAs. We sought to discover microRNAs involved in gastric cancer, and to elucidate their downstream target mechanisms. Both cultured gastric epithelial cells (HFE145 and NCI-N87) and primary human gastric tissues (31 non-neoplastic stomach (NS) and 25 gastric carcinomas (GC)) were studied. MicroRNA microarrays and quantitative RT–PCR were applied to discover and verify differentially expressed microRNAs. in vitro cell migration and invasion, cell proliferation, cell cycle and apoptosis assays were executed to elucidate biological effects of micro-RNA-192 and -215. Western blotting and luciferase assays were performed to confirm direct messenger RNA targeting by microRNA-192 and -215. MicroRNA microarray analyses revealed that 25 and 20 microRNAs were upregulated and downregulated in GC vs NS, respectively. Expression levels of both microRNA-192 and -215 were significantly higher in GC than in NS (P < 0.05). Luciferase assays suggested that microRNA-215 inhibits activated leukocyte cell adhesion molecule (ALCAM) expression at the posttranscriptional level. In addition, expression levels of ALCAM were significantly lower in GC than in NS. Mimics and inhibitors, respectively, of microRNA-192 or -215 exerted no effect on cell cycle or apoptosis in the immortalized normal gastric cell line HFE145 or the gastric cancer cell line NCI-N87. However, mimics of microRNA-192 or -215 significantly increased growth rates in HFE145 cells, whereas inhibitors of microRNA-192 or -215 caused significant decreases in growth rates in NCI-N87 cells. ALCAM knockdown by an ALCAM-specific siRNA significantly increased cell growth in HFE145 cells. Both transfection of mimics of microRNA-192 or -215 and ALCAM knockdown by an ALCAM-specific siRNA significantly increased the migration of HFE145 cells. In conclusion, in gastric cancer, both microRNA-192 and -215 are overexpressed in vivo and exert cell growth and migration-promoting effects in vitro, thus representing potential microRNAs with a role in cancer in the human stomach.
Keywords: microRNA-192 and -215, gastric cancer, ALCAM
Introduction
Gastric cancer (GC) remains one of the most lethal malignancies and a major cause of cancer death worldwide. Estimated new cases of GC in the United States numbered 21 500 in 2008, with deaths estimated at 10 880 (Jemal et al., 2006). Overall 5-year survival from GC is below 20%, largely as a consequence of late detection (Jemal et al., 2007). Failure patterns include both local recurrence and systemic spread (including peritoneal metastasis), particularly in patients with serosal invasion or lymph node metastasis (Lnmeta) (Khushalani, 2008). Thus, to improve clinical care and the early detection of gastric cancer, the identification of novel molecular biomarkers based on a comprehensive understanding of molecular pathogenesis would be very helpful.
MicroRNAs are a class of abundant, approximately 21–25-nucleotide non-coding RNAs that mediate posttranscriptional regulation of messenger RNA (mRNA) targets by interfering with mRNA stability or protein translation (Bartel, 2004), and dysregulation of micro-RNAs is a hallmark of cancer development and progression (Shi et al., 2008). Growing evidence reveals that many microRNAs are involved in tumorigenesis and/or tumor progression, including GC (Li et al., 2006; Volinia et al., 2006; Motoyama et al., 2008; Petrocca et al., 2008; Wang et al., 2008; Xia et al., 2008; Ando et al., 2009; Guo et al., 2009; Kim et al., 2009). Xia et al. reported that microRNA-15b and -16 are involved in the development of multidrug resistance in gastric cancer cells, at least in part by modulating apoptosis via targeting of BCL2 (Xia et al., 2008). Petrocca et al. found that the microRNA-106b-25 cluster is involved in E2F1 posttranscriptional regulation and has a function in the development of transforming growth factor-β resistance in gastric cancer (Petrocca et al., 2008). MicroRNA regulatory mechanisms in cancer progression are now essential for understanding the complete molecular genetic underpinning of GC. However, thus far there have been few reports describing microRNA expression or microRNA targets in GC.
In the current study, we investigated microRNA expression profiles in human gastric cancer using microRNA microarrays, and then validated the expression levels of the microRNA-192 and -215 using real-time quantitative RT–PCR (qRT-PCR). We found that microRNA-192 and -215 are upregulated in GC in vivo and suppress activated leukocyte cell adhesion molecule (ALCAM, synonym CD166) expression in vitro. In addition, our results indicated that the expression of ALCAM is significantly downregulated in human gastric cancer tissues in vivo. Finally, functional studies presented herein suggested that microRNA-192 and -215 function as microRNAs with a role in cancer in the human stomach.
Results
MicroRNA microarrays
MicroRNA microarray analyses were performed on three non-neoplastic gastric tissues (non-neoplastic stomach (NS)) and seven GCs. MicroRNAs that were at least two-fold up- or downregulated with a P-value of <0.05 in GC vs NS are shown in Supplementary Table 1. Both microRNA-192 and -215 were significantly upregulated in GC vs NS by micro-RNA microarray.
Quantitative reverse-transcriptase PCR (qRT–PCR) for microRNA-192 and -215 expression in human gastric tissues and cell lines
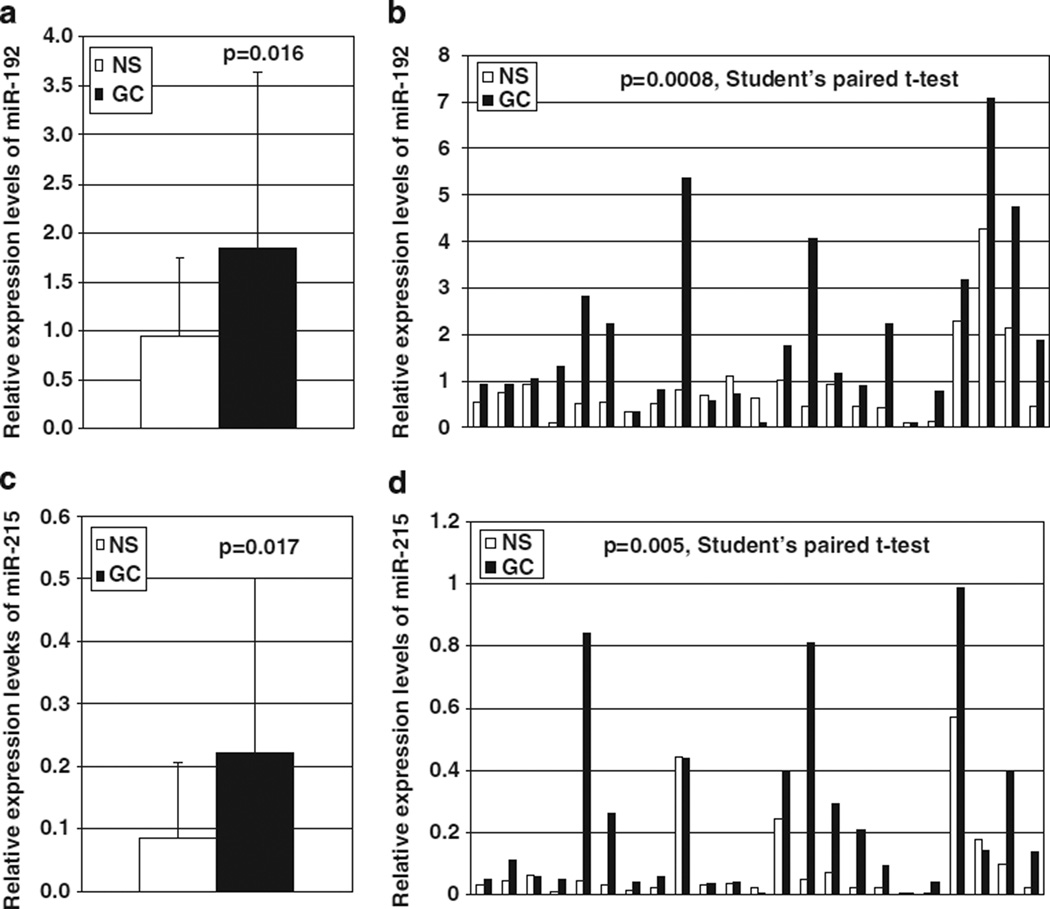
Both microRNA-192 and -215 were significantly more highly expressed in GC than in NS (1.838 vs 0.937, P = 0.016; 0.222 vs 0.083, P = 0.017, respectively; Figures 1a and c). Moreover, expression levels of both microRNA-192 and -215 in GCs (1.955 and 0.238, respectively) were significantly higher than in matching NS (0.867 and 0.090, respectively) in 23 cases with matching NS and GC (P = 0.0008 and 0.005, respectively; Student’s paired t-test; Figures 1b and d).
Figure 1.
MicroRNA-192 and -215 expression in human gastric tissues by qRT–PCR. Expression levels of both microRNA-192 and -215 were significantly higher in GC than in NS (a and c, respectively). Moreover, in 23 cases with matching NS and GC, expression levels of both microRNA-192 and -215 in GC were significantly higher than in paired NS (b and d, respectively). NS, non-neoplastic stomach; GC, gastric cancer.
Furthermore, we determined expression levels of microRNA-192 and -215 in one immortalized normal human gastric epithelial cell line (HFE145) and six gastric cancer-derived cell lines (NCI-N87, KATO III, RF-48, AGS, MKN28 and RF-1). Expression levels of both microRNA-192 and -215 were considerably higher (fold change >5) in NCI-N87, KATO III, RF-48 and AGS than in HFE145 cells, whereas the expression levels of these in MKN28 and RF-1 cells were similar to those in HFE145 cells (Supplementary Figure 1).
Cell proliferation assays
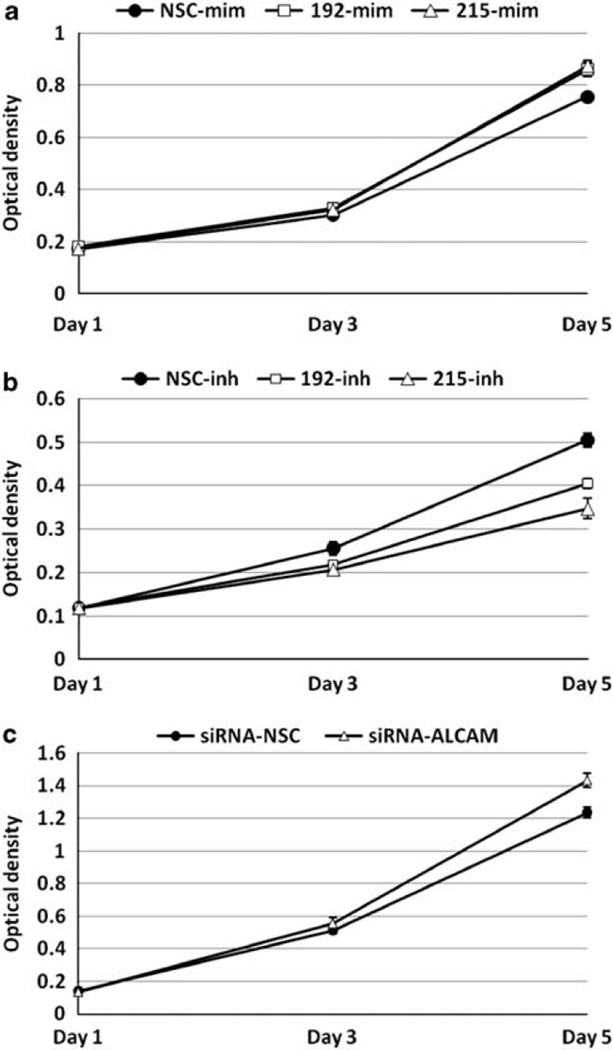
To investigate potential cancer-relevant biological effects exerted by microRNA-192 and -215, we first performed cell proliferation assays using WST-1 reagent. Transfection of mimics of microRNA-192 or -215 induced significantly increased growth rates in HFE145 cells, whereas transfection of inhibitors of microRNA-192 or -215 caused significant decreases in growth rates in NCI-N87 cells (Figures 2a and b). Interestingly, knockdown of ALCAM (a predicted target of micro-RNA-192 and -215; see below) by an ALCAM-specific siRNA significantly increased cell growth in HFE145 cells (Figure 2c), suggesting that microRNA-192 and -215 promote cell growth by downregulating ALCAM in gastric epithelial cells.
Figure 2.
Cell proliferation assay by WST-1 in HFE145 and NCI-N87 cells. Both microRNA-192 and -215 mimics induced significantly increased growth rates in HFE145 cells (a; P < 0.01 and P < 0.01, respectively), and inhibitors of both microRNA-192 and -215 caused a significant decrease in cell growth in NCI-N87 gastric cancer cells (b; P < 0.01 and P < 0.01, respectively). ALCAM knockdown by an ALCAM-specific siRNA significantly increased the growth rates of HFE145 cells (c; P < 0.01). Experiments were repeated at least three times, with triplicates performed in each independent experiment. NSC, non-specific control; mim, mimic; inh, inhibitor.
Cell cycle and apoptosis assays
Next, we performed experiments to assess the effects of these two microRNAs on cell-cycle progression and programmed cell death. Transfection of mimics of microRNA-192 or -215 and inhibitors of microRNA-192 or -215 had no effect on cell-cycle progression and apoptosis in HFE145 cells or NCI-N87 cells, respectively (Supplementary Figures 2 and 3).
Target searches for microRNA-192/-215, and ALCAM protein and mRNA expression following microRNA mimic transfections
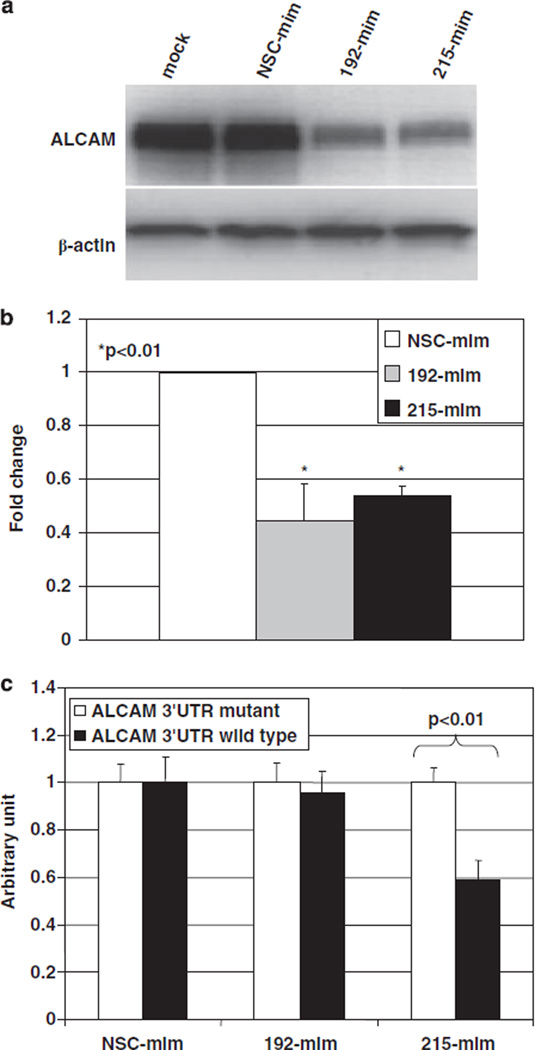
We next focused our attention on potential mRNA targets of microRNA-192 and -215. To identify these targets, we performed database searches in microRNA target prediction engines, including miR-Base (http://microrna.sanger.ac.uk/cgi-bin/targets/v4/search.pl), TargetScan (http://www.targetscan.org/vert_40/), PicTar (http://pictar.bio.nyu.edu/) and microRNA.org (http://www.microrna.org/microrna/home.do). We ultimately identified ALCAM as a putative target of microRNA-192 and -215. Transfecting mimics of microRNA-192 or -215 decreased ALCAM protein expression in HFE145 cells (Figure 3a). In addition, transfecting mimics of microRNA-192 or -215 significantly decreased ALCAM mRNA levels in HFE145 cells (Figure 3b).
Figure 3.
ALCAM protein and mRNA expression after micro-RNA mimics transfection, and luciferase reporter assay in HFE145 cells. Western blotting showed that mimics of microRNA-192 or -215 decreased ALCAM expression in HFE145 cells (a). qRT–PCR assays showed that transfection of mimics of microRNA-192 or -215 significantly decreased ALCAM mRNA levels (P < 0.01 and P < 0.01, respectively) in normal immortalized HFE145 gastric cells (b). Luciferase reporter assays were repeated three times, with triplicates performed in each independent experiment. Luciferase reporter activity was significantly reduced by a mimic of micro-RNA-215 (P < 0.01), but not by a mimic of microRNA-192 (c). NSC, non-specific control; mim, mimic; inh, inhibitor.
Luciferase reporter assays
In order to assess direct binding to and silencing of ALCAM by microRNA-192 and -215, we performed luciferase assays. In HFE145 cells, luciferase reporter activity was significantly reduced by a mimic of microRNA-215, but not by a mimic of microRNA-192 (Figure 3c).
Cell migration/invasion assays
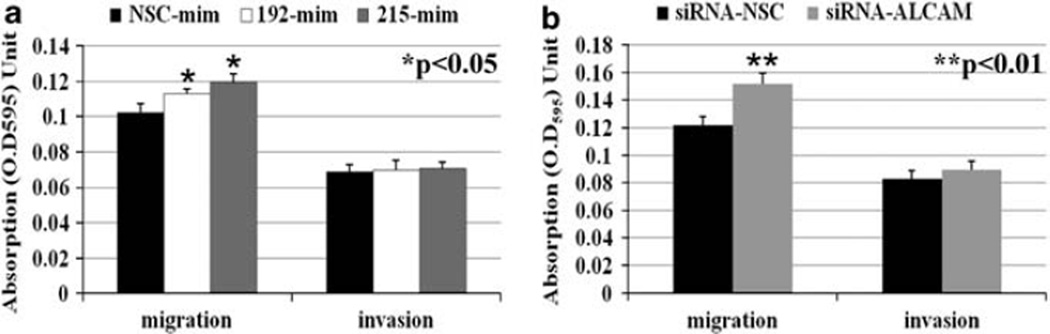
As ALCAM is known to be involved in cell–cell adhesion, we next investigated whether microRNA-192 and -215 or ALCAM influence cell motility or invasiveness. As shown in Figure 4, both transfection of mimics of microRNA-192 or -215 and ALCAM knockdown by an ALCAM-specific siRNA significantly increased the migration of HFE145 cells, but had no effect on the invasive potential of these cells. We normalized Matrigel chamber data to Transwell chamber data. The ratios for NSC-mim, 192-mim and 215-mim were 0.67, 0.62 and 0.59, respectively (P = 0.20 and P = 0.10, respectively). The ratios for siRNA-NSC and siRNA-ALCAM were 0.68 and 0.59, respectively (P = 0.16).
Figure 4.
Migration and invasion assays after transfection of microRNA mimics or ALCAM siRNA in HFE145 cells. Both transfection of mimics of microRNA-192 or -215 (a) and ALCAM knockdown by an ALCAM-specific siRNA (b) significantly increased the migration of HFE145 cells (P < 0.05 and P < 0.01, respectively), but had no effect on the invasive potential of these cells. NSC, non-specific control; mim, mimic; siRNA, small interfering RNA.
ALCAM mRNA and protein expression in human gastric tissues and cell lines
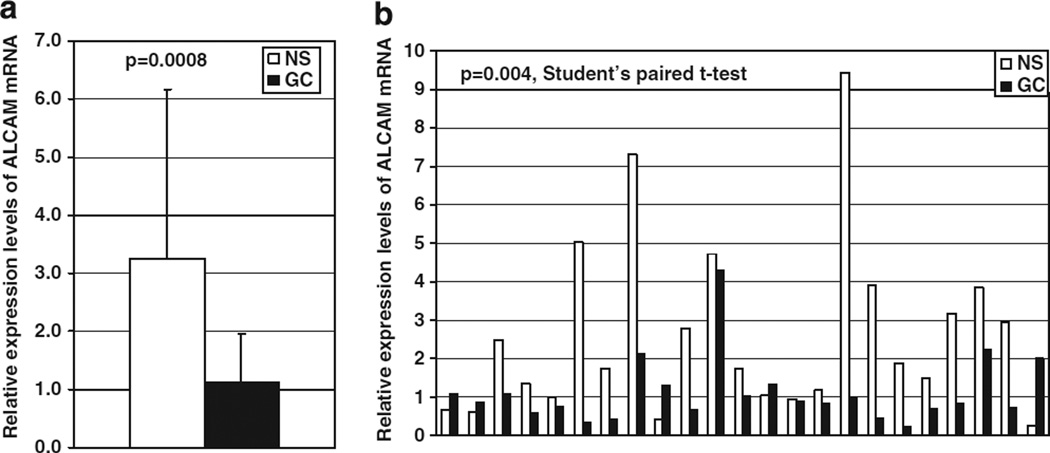
ALCAM mRNA expression levels were significantly lower in GC (mean = 1.120) than in NS (mean = 3.257; P = 0.0008, Figure 5a). Moreover, ALCAM expression levels in GC (mean = 1.117) were significantly lower than in matching NS (mean = 2.608) in 23 cases with corresponding NS and GC (P = 0.004, Student’s paired t-test; Figure 5b). Although it did not achieve statistical significance, there was a negative correlation between ALCAM mRNA levels and expression of microRNA-192 or -215 (Pearson’s correlation coefficient, r = −0.0621, P = 0.6491 and r = −0.2043, P = 0.1309, respectively; Supplementary Figure 4).
Figure 5.
ALCAM mRNA expression levels in human gastric tissues by qRT–PCR. ALCAM mRNA expression levels were significantly lower in GC than in NS (a). In 23 cases with paired NS and GC, ALCAM levels in GC were significantly lower than in matching NS (b). NS, non-neoplastic stomach; GC, gastric cancer.
We further tested ALCAM protein expression and localization by immunohistochemical staining in a tissue microarray containing 13 cases with matched NS, GC and LNmetas). ALCAM protein was localized to both the membrane and cytoplasm in NS, but only to the cytoplasm in GC and LNmeta (Supplementary Figure 5). The frequency of ALCAM membranous staining was significantly higher in NS (100%) than in GC (15.4%) or LNmeta (7.7%; P < 0.0001 and P < 0.0001, respectively; Fisher’s exact test, Table 1).
Table 1.
ALCAM expression by immunohistochemistry in human gastric tissue
| Case no. | Membrane | Cytoplasm | Histology of GC |
|||||
|---|---|---|---|---|---|---|---|---|
| NS | GC | LN meta | NS | GC | LNmeta | |||
| TMA1 | + | − | − | + | + | + | Mod | |
| TMA2 | + | − | − | + | + | + | Poor | |
| TMA3 | + | − | − | + | + | + | Poor | |
| TMA4 | + | − | − | + | + | + | Poor | |
| TMA5 | + | + | − | + | + | + | Poor | |
| TMA6 | + | − | − | + | + | + | Poor | |
| TMA7 | + | + | + | + | + | + | Mod- | |
| −poor | ||||||||
| TMA8 | + | − | − | + | + | + | Mod | |
| TMA9 | + | − | − | + | + | + | Poor | |
| TMA10 | + | − | − | + | + | + | Poor | |
| TMA11 | + | − | − | + | + | + | Mod | |
| TMA12 | + | − | − | + | + | + | Poor | |
| TMA13 | + | − | − | + | − | − | Poor | |
| Total | P-value | |||||||
| + | 13 | 2 | 1 | */$P<0.01 | 13 | 12 | 12 | |
| − | 0 | 11 | 12 | 0 | 1 | 1 | ||
Abbreviations: GC, gastric cancer; LNmeta, lymph node metastasis; Mod, moderately differentiated adenocarcinoma; NS, non–neoplastic stomach; poor, poorly differentiated adenocarcinoma.
, GC vs NS;
LNmeta vs NS.
Discussion
In the current study, we systematically investigated the expression of microRNA-192 and -215 in primary human gastric tissues in vivo and their cancer-related functions in gastric cells in vitro. Our results demonstrate that both microRNA-192 and -215 are upregulated in human gastric cancer. Notably, we discovered that mRNA encoding the adhesion molecule ALCAM is targeted by both microRNA-192 and -215 and downregulated at both the mRNA and protein levels in human gastric cancer. We found a negative correlation between ALCAM mRNA levels and expression of microRNA-192 and -215, implying that downregulation of ALCAM in GC may be due, at least in part, to overexpression of microRNA-192 and -215. In addition, overexpression of microRNA-192 or -215 significantly increased growth rates in immortalized normal HFE145 cells, whereas downregulation of microRNA-192 or -215 significantly decreased growth rates in NCI-N87 gastric cancer cells, and ALCAM knockdown by an ALCAM-specific siRNA significantly increased cell growth in HFE145 cells. Furthermore, we demonstrated that both overexpression of microRNA-192 or -215 and knockdown of ALCAM by siRNA significantly increased the migration of gastric epithelial cells, although there was no effect on the invasive potential of these cells, suggesting involvement of both microRNA-192/-215 and ALCAM in the control of gastric cell migration, which is a critical step in cancer metastasis. Taken together, these findings suggest that microRNA-192 and -215 are potentially important in the initiation and/or progression of GCs by functioning as micro-RNAs with a role in cancer, likely via suppression of ALCAM.
During preparation of the current manuscript, it was reported that microRNA-192 and -215 are downregulated in colon cancers (Braun et al., 2008; Song et al., 2008). In contrast to colon cancers, however, these two microRNAs were both upregulated in gastric cancers in the current study. MicroRNA-192 and -215 have the same 8-mer seed sequence, and only two nucleotides differ between them, with both microRNAs belonging to the miR-192 family. MicroRNA-192 and -215 are located on chromosomes 11q13.1 and 1q41, respectively. Chromosome 11q and 1q gains have been reported in gastric cancers by several different groups (Noguchi et al., 2001; Kimura et al., 2004; Tsukamoto et al., 2008), suggesting that the upregulation of microRNA-192 and -215 may be due to genomic DNA amplification in human gastric cancer.
In colon cancer, microRNA-192 and -215 contributed to enhanced CDKN1A/p21 levels, colony suppression, cell-cycle arrest and cell detachment from a solid support. These effects were partially dependent on the presence of wild-type p53. MicroRNA-192 and -215 function as both effectors and regulators of p53; they appear to suppress carcinogenesis via p21 accumulation and cell-cycle arrest (Braun et al., 2008). Another study showed that microRNA-192 is involved in the p53 tumor suppressor network, with significant effects on cell-cycle control in colon cancer (Song et al., 2008). Still a third study demonstrated that the activation of microRNA-192 and -215 induces cell-cycle arrest in HCT116 DICERex5 cells (Georges et al., 2008). In the current study, both microRNA-192 and -215 showed no effect on cell-cycle progression in either HFE145 or NCI-N87 cells. It is of note that the HFE-145 cells were immortalized by SV40 large-T antigen, which inactivates p53 (Carbone et al., 1997), and NCI-N87 cells carry a p53 mutation (Kim et al., 1991). Thus, the absence of an effect of microRNA-192 and -215 on the cell cycle in our study may have been due to the inactivation of p53 in these two cell lines.
ALCAM is a cell adhesion molecule expressed by epithelial cells in several organs and mediates both heterophilic (ALCAM–CD6) and homophilic (ALCAM–ALCAM) cell–cell interactions (Swart, 2002). ALCAM expression has been described in a number of tumor types, although these data are somewhat inconsistent. ALCAM was found to be upregulated in melanoma, colon cancer, bladder cancer and esophageal squamous cell carcinoma, but downregulated in breast cancer (Ofori-Acquah and King, 2008). In addition, variable levels of ALCAM expression have been reported at different stages of tumor development, even within the same type of malignancy. In prostate carcinoma, ALCAM was upregulated in low-grade tumors, but downregulated in high-grade tumors (Kristiansen et al., 2003). Clearly, ALCAM is involved in oncogenesis, but its mechanism of action is still elusive. ALCAM has been suggested to have a function in cell motility and neoplastic progression. In primary breast carcinoma, high levels of ALCAM significantly correlated with small tumor diameter and low tumor grade, whereas ALCAM/MMP-2 ratios were significantly higher in cancers characterized by small tumor size and low tumor grade, suggesting that degradation of the cellular network established by adhesion molecules, such as ALCAM causes tumor tissue relaxation and increases metastasis (Jezierska et al., 2006; Jezierska and Motyl, 2009). In human melanoma cells, stable transfection of a transmembrane, amino-terminally truncated ALCAM variant diminished cell clustering mediated by wild-type ALCAM, promoted cell motility in vitro, and accelerated spontaneous lung metastases in vivo (van Kempen et al., 2004). Inhibition of ADAM17 (a disintegrin and metalloprotease), which is a proteolytic sheddase of ALCAM, inhibited cell motility, whereas a recombinant antibody blocking ALCAM’s adhesive functions enhanced cell motility in epithelial ovarian cancer cells (Rosso et al., 2007). Interestingly, it has been reported that micro-RNA-192 and -215 were capable of inducing cell detachment in HCT116 colon cancer cells (Braun et al., 2008). In the current study, we discovered that ALCAM was downregulated and targeted by both microRNA-192 and -215 in human gastric cancer. Furthermore, both transfection of mimics of micro-RNA-192 or -215 and knockdown of ALCAM by an ALCAM-specific siRNA significantly increased the migration of HFE145 cells. Considering our own and these previous published findings, it is tempting to speculate that microRNA-192/-215-dependent changes in ALCAM could contribute to the invasion and metastasis of cancer cells.
Most studies have found that microRNAs function through the 3-untranslated regions (UTRs) of target mRNAs; however, recent studies have shown that microRNAs can also target the 5′-UTR or coding DNA sequence (Lytle et al., 2007; Orom et al., 2008; Tsai et al., 2009; Zhou et al., 2009). It has been reported that luciferase translation was repressed after introducing the complementary site of let-7 into the 5′-UTR of luciferase in a gene expression system in HeLa cells (Lytle et al., 2007). Human miR-10a targets at the 5′-UTR of ribosome protein mRNAs and stimulates their translation (Orom et al., 2008). In our study, although microRNA-192 mimics suppressed both ALCAM mRNA and protein expression, they failed to inhibit ALCAM 3′-UTR luciferase reporter activity in HFE145 cells. We considered the following possible explanations for these results: (1) there are other target sites of microRNA-192 within the ALCAM 3′-UTR; (2) micro-RNA-192 may regulate ALCAM expression by targeting the ALCAM 5′-UTR or coding DNA sequence; or (3) regulation of ALCAM expression by microRNA-192 may constitute a secondary (that is, indirect) effect.
In conclusion, in gastric cancer, both microRNA-192 and -215 are overexpressed in vivo and participate in the regulation of ALCAM, which is downregulated in primary human gastric cancer. Moreover, both overexpression of microRNA-192 or -215 and knockdown of ALCAM by an siRNA significantly increased the proliferation and migration of gastric epithelial cells. Thus, microRNA-192 and -215 represent potential gastric microRNAs with a role in cancer, and their suppression may ultimately be explored in novel strategies in gastric cancer treatment.
Materials and methods
Tissue samples
In the current study, 31 non-neoplastic gastric tissues (NS), 25 gastric cancers (GCs) (23 of which had corresponding NS) were examined. All patients provided written informed consent under a protocol approved by the Institutional Review Board at the Johns Hopkins University School of Medicine. All specimens were stored in liquid nitrogen before RNA extraction. Clinicopathological characteristics are summarized in Supplementary Table 2.
Cell lines
Immortalized human normal gastric epithelial cells (HFE145) and gastric cancer cells (NCI-N87) were obtained from collaborator at Howard University (Dr Duane T Smoot) and the American Type Culture Collection, respectively. These cells were cultured in 90% DMEM supplemented with 10% fetal bovine serum.
RNA extraction
Total RNAs were extracted from tissues and cultured cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNAs were stored at −80 °C before analysis.
MicroRNA microarrays
MicroRNA Labeling Reagent and Hybridization Kits (Agilent, Santa Clara, CA, USA) and Human microRNA Microarray Kits (V2, Agilent), each containing 723 human microRNAs and 76 human viral microRNAs, were used to generate global microRNA expression profiles. This platform is designed to ensure extremely high data fidelity and robustness. Each microRNA is represented by 30 probes on the array (that is, 15 replicates of two distinct probes hybridize to each microRNA). Furthermore, these 30 probes are evenly distributed across the array in order to minimize positional bias. 100 ng of total RNA from three NS and seven GC tissues was phosphatase treated and then labeled with cyanine 3-pCp. The labeled RNA was purified using Micro Bio-spin columns (Bio-Rad, Hercules, CA, USA) and subsequently hybridized to a human microRNA microarray slide at 55 °C for 20 h. After hybridization, the slides were washed with Gene Expression Wash Buffer (Agilent) and scanned on an Agilent Microarray Scanner (Agilent) using Agilent’s Scan Control, version A.7.0.1 software. Raw data were further automatically processed in Microsoft Excel. Hybridization signals that failed to exceed the average background value by more than three standard deviations were excluded from analysis.
Quantitative reverse-transcriptase PCR (qRT–PCR) for microRNA and ALCAM expression
TaqMan MicroRNA Assays, Human (Applied Biosystems, Foster City, CA, USA) were used to confirm microRNA expression changes identified on microRNA microarrays according to the manufacturer’s protocol. U6 small nuclear RNA TaqMan RT–PCR amplicon (RNU6B TaqMan micro-RNA Assay kit, Applied Biosystems) was used as an internal control.
To detect ALCAM mRNA expression, isolated RNA was reverse-transcribed using iScript cDNA Synthesis kit (Bio-Rad) according to the manufacturer’s instructions. iQ SYBR Green Supermix (Bio-Rad) was used for real-time PCR applications. The primer set for ALCAM qRT–PCR was 5′-AAGTTGGGTGACTGCATTTC-3′ (forward) and 5′-ATTATGACCACCGCTCCTTC-3′ (reverse). β-Actin was used to normalize data. PCR conditions were as follows: 2 min at 95 °C followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s and 72 °C for 30 s.
Transfection of microRNA mimics and inhibitors
Synthesized RNA duplexes of microRNA mimics (agomiRs) and microRNA inhibitors (antagomiRs) were purchased from Dharmacon (Lafayette, CO, USA). 30 ~ 50% confluent cells were transfected with 60nm of each agomiR or antagomiR using Lipofectamine RNAi MAX (Invitrogen). Both RNA and protein were harvested after 48 h of transfection. Nonspecific controls for both mimics and inhibitors were used as negative controls.
Cell proliferation assays
Cells were plated at a density of 1000 cells per well onto 96-well plates at day 0 (24 h after microRNA mimic or inhibitor transfection). At every other day until day 5, Cell Proliferation Reagent WST-1 (Roche, Mannheim, Germany) was added to each well, and then incubated at 37 °C for 1 h. Optical density was measured at 660 nm (background) and 440 nm (signal) using a plate reader (Molecular Devices, Sunnyvale, CA, USA) at the incubation time of 1 h.
Cell-cycle assays
5 × 105 HFE145 and NCI-N87 cells were transfected with 60 nm mimics or inhibitors of microRNA-192, -215 or nonspecific controls, respectively. After 48 h, cells were stained with propidium iodide (Cellular DNA Flow Cytometric Analysis Reagent Set, Roche) and fluorescence intensity was measured using a flow cytometer to assess DNA content.
Apoptosis assays
5 × 105 HFE145 and NCI-N87 cells were transfected with 60 nm mimics or inhibitors of microRNA-192, -215 or nonspecific controls, respectively. After 48 h, cells were doubly stained with propidium iodide and annexin-V (Vybrant Apoptosis Assay Kit #3, Invitrogen). Fluorescence intensity was measured using a flow cytometer to assess early apoptotic cells, defined as those staining only with annexin-V.
Cell migration/invasion assays
Cell motility and invasiveness were determined by a Transwell chamber and a Matrigel chamber plate (24-well format, BD Biosciences, St Louis, MO, USA), as described previously (Adiseshaiah et al., 2007; Agarwal et al., 2009). Cells (5 × 104) were seeded onto a Transwell or a Matrigel insert membrane with 8µm pores on day 2 after transfection. Growth medium containing 20% fetal bovine serum was used as a chemoattractant. After incubation at 37 °C for 24 h, cells that did not migrate or invade through the pores of the Transwell inserts were manually removed with a cotton swab. Cells present at the bottom of the membrane were fixed in cold methanol for 10 min and then stained with 0.01% crystal violet in 20% ethanol. After 10 min incubation, the filters were washed thoroughly in water and suspended in 200 ml of 5% acetic acid and 5% methanol. Colorimetric readings were taken at OD 595 nm.
Western blotting
Cells were lysed in Laemmli sample buffer (Bio-Rad) with a protease inhibitor, complete EDTA free (Roche). Protein concentration was estimated using a BCA Protein Assay kit (Pierce, Rockford, MA, USA). Cell lysates (20 ug) were electrophoresed on 4–20% Linear Gradient polyacrylamide gels (Bio-Rad) and transferred to Immobilon-PSQ polyvinylidene difluoride membranes (Millipore, Bedford, MA, USA). The membranes were blocked with TBS containing 5% skim milk and 0.1% Tween-20, then, incubated with the primary antibody. The primary antibodies used for the analysis included mouse anti-ALCAM monoclonal antibody (1:400, Novocastra, Newcastle, UK) and mouse anti-β-actin mono-clonal antibody (1:25 000, Sigma-Aldrich, Bedford, MA, USA).
Immunohistochemical staining
Immunohistochemical staining was performed on a 4-µm thick gastric tissue microarray, which was containing 13 matched NS, GC and LNmeta, using an Envision kit (Dako Cytomation, Glostrup, Denmark). Previous studies have reported both nuclear and membranous ALCAM staining to be potentially important to the clinical prognosis of human malignancies (Ofori-Acquah and King, 2008) and thus, the staining reaction was separately evaluated in tumor cell nuclei and cell membranes. A positive result was defined as the presence of staining in > 10% of the cells.
Luciferase reporter assays
The full-length ALCAM 3′-UTR, containing flanking predicted microRNA-responsive sites, was amplified from HFE145 gastric cells using linker primers containing XbaI restriction sites. Amplicons were cut by XbaI and cloned into an XbaI site just downstream of the firefly luciferase structural gene in vector pGL4.13 (Promega, Madison, WI, USA). After sequence verification, we obtained plasmid clones pGL4.13 (Promega) containing correctly oriented inserts (pGL4.13-ALCAMUTR). We also constructed separate plasmids containing the ALCAM 3′-UTR with mutated seed regions for the predicted microRNA-192 and microRNA-215 binding sites (pGL4.13-mutALCAMUTR) as negative controls. In total, 6000 cells per well were seeded onto 96-well plates on the day before transfection, then transfected with microRNA mimics or inhibitors as described above. The constructed pGL4.13 vector and an internal control pRL-CMV (Renilla luciferase) vector were cotransfected 24 h after microRNA mimic or inhibitor transfection. Around 24 h after plasmid vector transfection, the luciferase reporter assay was performed using a Dual-Glo luciferase assay kit (Promega). Luminescence intensity was measured by VICTOR2 fluorometry (Perkin Elmer, Waltham, MA, USA), and the luminescence intensity of firefly luciferase was normalized to that of Renilla luciferase.
Statistical analyses
For all statistical tests, Statistica (version 6.1; StatSoft Inc., Tulsa, OK, USA) was used. Differences with P < 0.05 were considered significant.
Footnotes
Conflict of interest
The authors declare no conflicts of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Adiseshaiah P, Lindner DJ, Kalvakolanu DV, Reddy SP. FRA-1 proto-oncogene induces lung epithelial cell invasion and anchorage-independent growth in vitro, but is insufficient to promote tumor growth in vivo. Cancer Res. 2007;67:6204–6211. doi: 10.1158/0008-5472.CAN-06-4687. [DOI] [PubMed] [Google Scholar]
- Agarwal R, Mori Y, Cheng Y, Jin Z, Olaru AV, Hamilton JP, et al. Silencing of claudin-11 is associated with increased invasiveness of gastric cancer cells. PLoS One. 2009;4:e8002. doi: 10.1371/journal.pone.0008002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando T, Yoshida T, Enomoto S, Asada K, Tatematsu M, Ichinose M, et al. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int J Cancer. 2009;124:2367–2374. doi: 10.1002/ijc.24219. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, et al. p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 2008;68:10094–10104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone M, Rizzo P, Grimley PM, Procopio A, Mew DJ, Shridhar V, et al. Simian virus-40 large-T antigen binds p53 in human mesotheliomas. Nat Med. 1997;3:908–912. doi: 10.1038/nm0897-908. [DOI] [PubMed] [Google Scholar]
- Georges SA, Biery MC, Kim SY, Schelter JM, Guo J, Chang AN, et al. Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008;68:10105–10112. doi: 10.1158/0008-5472.CAN-08-1846. [DOI] [PubMed] [Google Scholar]
- Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, et al. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24:652–657. doi: 10.1111/j.1440-1746.2008.05666.x. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, et al. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- Jezierska A, Motyl T. Matrix Metalloproteinase-2 involvement in breast cancer progression: a mini-review. Med Sci Monit. 2009;15:RA32–RA40. [PubMed] [Google Scholar]
- Jezierska A, Olszewski WP, Pietruszkiewicz J, Olszewski W, Matysiak W, Motyl T. Activated Leukocyte Cell Adhesion Molecule (ALCAM) is associated with suppression of breast cancer cells invasion. Med Sci Monit. 2006;12:BR245–BR256. [PubMed] [Google Scholar]
- Khushalani N. Cancer of the esophagus and stomach. Mayo Clin Proc. 2008;83:712–722. [PubMed] [Google Scholar]
- Kim JH, Takahashi T, Chiba I, Park JG, Birrer MJ, Roh JK, et al. Occurrence of p53 gene abnormalities in gastric carcinoma tumors and cell lines. J Natl Cancer Inst. 1991;83:938–943. doi: 10.1093/jnci/83.13.938. [DOI] [PubMed] [Google Scholar]
- Kim YK, Yu J, Han TS, Park SY, Namkoong B, Kim DH, et al. Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009;37:1672–1681. doi: 10.1093/nar/gkp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Noguchi T, Kawahara K, Kashima K, Daa T, Yokoyama S. Genetic alterations in 102 primary gastric cancers by comparative genomic hybridization: gain of 20q and loss of 18q are associated with tumor progression. Mod Pathol. 2004;17:1328–1337. doi: 10.1038/modpathol.3800180. [DOI] [PubMed] [Google Scholar]
- Kristiansen G, Pilarsky C, Wissmann C, Stephan C, Weissbach L, Loy V, et al. ALCAM/CD166 is up-regulated in low-grade prostate cancer and progressively lost in high-grade lesions. Prostate. 2003;54:34–43. doi: 10.1002/pros.10161. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhan W, Wang Z, Zhu B, He Y, Peng J, et al. Inhibition of PRL-3 gene expression in gastric cancer cell line SGC7901 via microRNA suppressed reduces peritoneal metastasis. Biochem Biophys Res Commun. 2006;348:229–237. doi: 10.1016/j.bbrc.2006.07.043. [DOI] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motoyama K, Inoue H, Nakamura Y, Uetake H, Sugihara K, Mori M. Clinical significance of high mobility group A2 in human gastric cancer and its relationship to let-7 microRNA family. Clin Cancer Res. 2008;14:2334–2340. doi: 10.1158/1078-0432.CCR-07-4667. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Wirtz HC, Michaelis S, Gabbert HE, Mueller W. Chromosomal imbalances in gastric cancer. Correlation with histologic subtypes and tumor progression. Am J Clin Pathol. 2001;115:828–834. doi: 10.1309/2Q9E-3EP5-KYPK-VFGQ. [DOI] [PubMed] [Google Scholar]
- Ofori-Acquah SF, King JA. Activated leukocyte cell adhesion molecule: a new paradox in cancer. Transl Res. 2008;151:122–128. doi: 10.1016/j.trsl.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Rosso O, Piazza T, Bongarzone I, Rossello A, Mezzanzanica D, Canevari S, et al. The ALCAM shedding by the metalloprotease ADAM17/TACE is involved in motility of ovarian carcinoma cells. Mol Cancer Res. 2007;5:1246–1253. doi: 10.1158/1541-7786.MCR-07-0060. [DOI] [PubMed] [Google Scholar]
- Shi XB, Tepper CG, White RW. Cancerous miRNAs and their regulation. Cell Cycle. 2008;7:1529–1538. doi: 10.4161/cc.7.11.5977. [DOI] [PubMed] [Google Scholar]
- Song B, Wang Y, Kudo K, Gavin EJ, Xi Y, Ju J. miR-192 Regulates dihydrofolate reductase and cellular proliferation through the p53-microRNA circuit. Clin Cancer Res. 2008;14:8080–8086. doi: 10.1158/1078-0432.CCR-08-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart GW. Activated leukocyte cell adhesion molecule (CD166/ALCAM): developmental and mechanistic aspects of cell clustering and cell migration. Eur J Cell Biol. 2002;81:313–321. doi: 10.1078/0171-9335-00256. [DOI] [PubMed] [Google Scholar]
- Tsai NP, Lin YL, Wei LN. MicroRNA mir-346 targets the 50-untranslated region of receptor-interacting protein 140 (RIP140) mRNA and up-regulates its protein expression. Biochem J. 2009;424:411–418. doi: 10.1042/BJ20090915. [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y, Uchida T, Karnan S, Noguchi T, Nguyen LT, Tanigawa M, et al. Genome-wide analysis of DNA copy number alterations and gene expression in gastric cancer. J Pathol. 2008;216:471–482. doi: 10.1002/path.2424. [DOI] [PubMed] [Google Scholar]
- van Kempen LC, Meier F, Egeblad M, Kersten-Niessen MJ, Garbe C, Weidle UH, et al. Truncation of activated leukocyte cell adhesion molecule: a gateway to melanoma metastasis. J Invest Dermatol. 2004;122:1293–1301. doi: 10.1111/j.0022-202X.2004.22531.x. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, He YL, Cai SR, Zhan WH, Li ZR, Zhu BH, et al. Expression and prognostic impact of PRL-3 in lymph node metastasis of gastric cancer: its molecular mechanism was investigated using artificial microRNA interference. Int J Cancer. 2008;123:1439–1447. doi: 10.1002/ijc.23643. [DOI] [PubMed] [Google Scholar]
- Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, et al. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372–379. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- Zhou X, Duan X, Qian J, Li F. Abundant conserved microRNA target sites in the 5′-untranslated region and coding sequence. Genetica. 2009;137:159–164. doi: 10.1007/s10709-009-9378-7. [DOI] [PubMed] [Google Scholar]