Abstract
Background
Uveal melanoma (UM) is the most common primary intraocular malignancy in adults. Early treatment may improve any chances of preventing metastatic disease, but diagnosis of small UM is challenging. Up to 95 % of all UMs carry somatic mutations in the G-coupled proteins GNAQ and GNA11 promoting anchorage-independent growth and proliferation. About 50%of UMs are fatal. Once metastatic, patients have limited options for successful therapy.
Methods
We have developed functionalized gold nanoparticles (AuNPs) to visualize transcripts of mutant GNAQ mRNA in living cells. In addition to their suitability as a specific tool for GNAQ mutation detection, we have developed a novel linker that enables conjugation of siRNAs to AuNPs allowing for greater and more rapid intracellular release of siRNAs compared to previously described approaches.
Results
Binding of modified AuNPs to matching target mRNA leads to conformational changes, resulting in a detectable fluorescent signal that can be used for mutation detection in living cells. Knockdown of GNAQ with siRNA-AuNPs effectively reduced downstream signals and decreased cell viability in GNAQ mutant uveal melanoma cells.
Conclusion
AuNPs may in future be developed to serve as sensors for mutations of vital importance. The new release system for siRNA-AuNP improves previous systems, which conceivably will be useful for future therapeutic gene regulatory approaches.
Keywords: siRNA, GNAQ, Fluorescence, Treatment, Linker
1 Background
Uveal Melanoma (UM) originates from intraocular melanocytes and is the most common primary intraocular malignancy, affecting 5–7 people/million/year (Hu and McCormick 2011; Shields and Shields 2009). More than 90 % of UM involve the choroid, the remainder occurring in the ciliary body or iris (van den Bosch et al. 2010; Singh et al. 2005). Up to one half of patients develop metastasis. Metastatic spread occurs hematogenously, involving the liver in over 90 % of patients with disseminated disease (Bakalian et al. 2008; Diener-West et al. 2005; Scholes et al. 2003).
Most large and medium-sized UMs can be diagnosed clinically. Diagnosis based on clinical findings such as the thickness of the lesion, presence of subretinal fluid and orange pigment, tumor proximity to the optic disk and clinical symptoms such as impairment of vision (Shields et al. 2009). A challenge remains with predicting the biological behaviour of small melanocytic proliferations. Indeterminate pigmented lesions need to be observed and about one third of small tumors show growth in follow-up examinations (Butler et al. 1994). Even though evidence suggests that small lesions can be followed safely and lethal courses are rare, it is still to mention that tumor size and patient outcomes have also been found to correlate (Damato et al. 2009; Kujala et al. 2003; Shields 2009).
The management of patients with UM has recently been revolutionized by the discovery that metastasis almost exclusively occurs in patients with tumors that show chromosome 3 loss or a class 2 gene expression profile (Damato et al. 2010a; Harbour 2014; Scholes et al. 2003). In addition, a growing number of genetic alterations have been shown to correlate with survival (Harbour et al. 2010). There is much scope for (in vivo) genomic analysis of uveal tumors, to predict metastatic disease and identify which asymptomatic tumors can be safely observed and which require therapy.
Several studies have identified mutations in the alpha subunit of the G-coupled protein GNAQ/GNA11 which occur in approximately 90 % of UMs and which predominantly result in the substitution of glutamine by proline (Q209P) or leucine (Q209L) (Van Raamsdonk et al. 2009, 2010). This region is critical for the intrinsic GTPase activity of the protein of which mutations cause constitutive activation, making GNAQ/GNA11 a bona fide oncogene. To date, to our knowledge, it has been impossible to synthetically restore the enzymatic activity of mutant GNAQ or target the permanently active protein. There is a lack of treatment effective for UM metastases. The high prevalence of GNAQ mutations in UM make mutant GNAQ a possibly interesting candidate for new therapeutic approaches (Pópulo et al. 2011).
In this study we sought to answer 2 questions: Can AuNPs be functionalized to detect specific GNAQ mutations? Can AuNPs be modified to achieve GNAQ gene regulation?
First, we provide evidence that functionalized AuNPs can be used to visualize mutant GNAQ mRNA. Upon binding of the fluorophore-labeled oligonucleotides to its complementary sequence, the quenched fluorescein is relocated away from the AuNP core producing a detectable signal in viable cells.
Second, using AuNPs that release distinct small interfering RNAs (siRNAs), we demonstrate target gene regulation, reduced protein expression and decreased cell viability. siRNAs are bound to AuNPs through a novel release system that allows the release of siRNA fast and to a greater extent compared to previous methods once the nanostructure reaches the cytoplasm. The ability of nanoconjugates to prevent siRNA degradation and to allow the delivery into cells without the use of transfection or permeabilization reagents has the potential to improve targeting of hitherto undruggable, driving oncogenes (Conde et al. 2013; Ghosh et al. 2008; Wall and Shi 2003).
2 Results
2.1 GNAQ and GNA11 mutations are frequent in uveal melanomas, indeterminate and benign uveal melanocytic proliferations
GNAQ and GNA11 mutations are frequent in UM. We have sequenced 5 uveal melanomas and found mutations in all 5 tumors. In addition, we also sequenced 4 indeterminate uveal melanocytic proliferations that were initially classified as benign or indeterminate, but progressed to UM over time; interestingly, all tumors harboured mutations in GNAQ orGNA11. Little is known about the mutation status of GNAQ and GNA11 in benign uveal melanocytic proliferations. One out of 4 uveal nevi sequenced in this study carried a mutation in GNAQ, suggesting that these genetic aberrations are not limited to UM. Even though GNAQ mutations seem to be more prevalent in malignant melanocytic tumors sequenced in this study, they might also be present in benign uveal melanocytic proliferations and are therefore of limited value in distinguishing benign from malignant lesions (Table 1).
Table 1.
Mutation status of uveal tumors, initial clinical and final, follow-up diagnosis
| Initial diagnosis | Follow-up diagnosis | Mutation status |
|---|---|---|
| Uveal melanoma | Uveal melanoma | GNA11(Q209L) |
| Uveal melanoma | Uveal melanoma | GNAQ(Q209P) |
| Uveal melanoma | Uveal melanoma | GNAQ(Q209L) |
| Uveal melanoma | Uveal melanoma | GNAQ(Q209P) |
| Uveal melanoma | Uveal melanoma | GNA11(Q209L) |
| Small uveal nevus | Uveal melanoma | GNAQ(Q209R) |
| Small uveal nevus | Uveal melanoma | GNAQ(Q209P) |
| Melanocytoma | Uveal melanoma | GNAQ(Q209P) |
| Melanocytoma | Uveal melanoma | GNAQ(Q209P) |
| Uveal nevus | Uveal nevus | GNAQ(Q209P) |
| Uveal nevus | Uveal nevus | GNAQ(wt) GNA11(wt) |
| Uveal nevus | Uveal nevus | GNAQ(wt) GNA11(wt) |
| Uveal nevus | Uveal nevus | GNAQ(wt) GNA11(wt) |
wt wild type
2.2 Aim 1 – mutation detection. Schematic structure and functionality of modified AuNPs for mutation detection
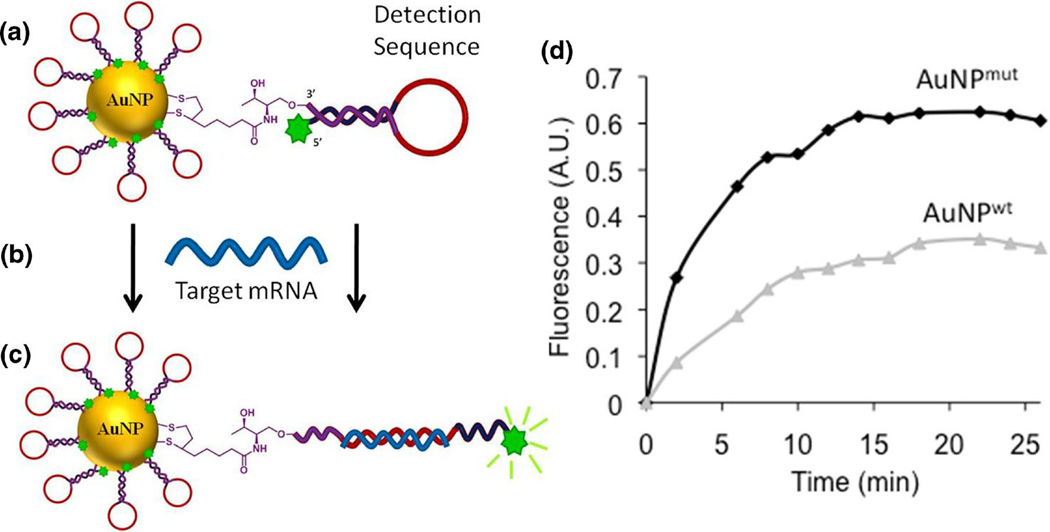
In this study, we used AuNPs modified with oligonucleotides, which were conjugated to a fluorescein derivative and are capable of folding into a hairpin structure. In this conformation, the fluorescent tag is in close proximity to the gold core, which quenches the fluorescent signal (Fig. 1a). After binding to the complementary mRNA transcripts of the gene of interest (Fig. 1b), the hairpin shaped oligonucleotides unfold, shifting the fluorescein derivative away from the gold core (Latorre et al. 2014b). This structural rearrangement leads to a detectable increase in fluorescence (Fig. 1c). To test specificity of modified AuNPs to detect single nucleotide changes, we designed AuNPs recognizing the wild type and the mutant transcript of GNAQ. Characterisation of AuNPs revealed that gold particle designs were stable in liquid solution (PBS). About 4.5 µM of molecular beacons were loaded onto 15 nM of AuNPs and loaded with about 300 molecular beacons per particle (Fig. S1). AuNPs modified to recognize the mutant transcript (AuNPmut) gave a strong fluorescent signal when incubated with synthesised oligonucleotieds of the matching, mutant GNAQ transcript. In contrast, AuNPs bearing the wild type detection sequence only barely showed an increase in fluorescence (Fig. 1d).
Fig. 1.
a Schematic structure of gold nanoparticles. Oligonucleotides are modified with a fluorescein derivate on the 5′ end and a dithiolane moiety on the 3′ end to anchor the oligonucleotide to the AuNP. b, c After binding of the complementary, target mRNA sequence, the hairpin structure unfolds resulting in a fluorescent signal. d AuNPs designed to recognize the mutant transcript of GNAQ (AuNPmut) give a stronger fluorescent signal than AuNPs detecting the wild type sequence (AuNPwt)
2.3 Mutation detection in GNAQ mutant uveal melanoma cells
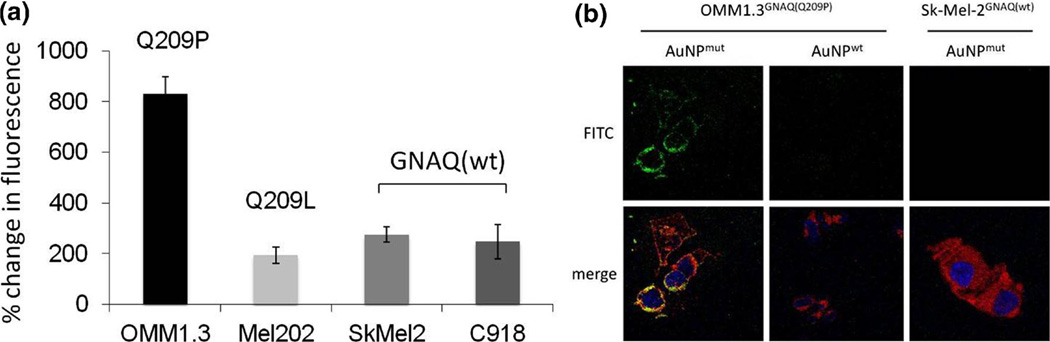
To demonstrate the capacity of modified AuNPs to detect target gene mutations in live cells, we incubated the human uveal melanoma cell lines OMM1.3, Mel202 and C918 and the cutaneous melanoma cell line Sk-Mel-2 with modified AuNPs. We found a near 4-fold increase in fluorescence measured by flow cytometric analysis when GNAQQ209P mutant OMM1.3 cells were incubated with AuNPs that specifically recognize the mutant transcript (Fig. 2a). Non-targeting control AuNPs served as a baseline, negative control. In contrast, Mel202 cells harbouring a different GNAQ mutation (GNAQQ209L) and the GNAQ wild type cell lines Sk-Mel-2 and C918 only gave a marginal increase in signal compared to cells incubated with AuNPs bearing a non-matching control sequence. Confocal microscopy of OMM1.3 cells incubated with AuNPs that specifically recognize the GNAQQ209P mutation (AuNPmut) revealed a significant increase in the fluorescent signal in GNAQ mutant OMM1.3 cells compared to AuNPs bearing the wild type sequence (AuNPwt). Also, GNAQ wild type Sk-Mel-2 cells did not show an increase of fluorescence when incubated with AuNPsmut supporting the specificity of AuNPs for mutation detection (Fig. 2b).
Fig. 2.
a In the GNAQ mutant cell line OMM1.3, gold nanoparticles bearing the exact, mutant GNAQ sequence (AuNPmut) give a strong fluorescent signal, whereas the UM cell line Mel202 harbouring a different GNAQ mutation and the wild type cell lines Sk-Mel-2 and C918 only show a marginal increase in fluorescence. Bars represent the increase of fluorescence compared to untreated controls measured by flow cytometric analysis (FACS). Error bars represent the standard deviation. b Confocal microscopic pictures of the GNAQ mutant UM cell line OMM1.3 and the GNAQ wild type cell line Sk-Mel-2 incubated with the indicated AuNPs. (Green = fluorescein, red = deep red plasma membrane stain, blue = DAPI)
2.4 Aim 2 – targeting un-druggable proteins. A disulphide moiety improves the release of siRNA targeting GNAQ from modified AuNPs
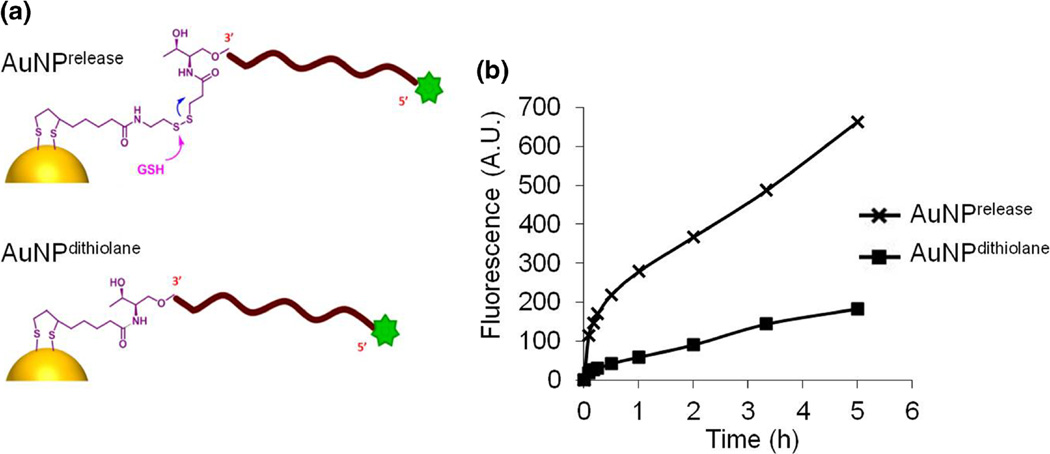
Single AuNPs can be functionalized to serve as a carrier of several active molecules such as siRNAs with the additional benefit of preventing siRNA from being degraded rapidly (Wall and Shi 2003). In most cases, siRNAs are modified with thiol groups to facilitate their conjugation to AuNPs (Conde et al. 2013; Lee et al. 2011). The active molecule is then released in the presence of reducing agents such as glutathione (GSH). This release, however, is slow and inefficient, which reduces the availability of active siRNA for gene regulation and limits resultant knockdown capacity of siRNA-AuNPs. To improve the release of siRNA from AuNPs, we have developed a new approach for the modification and preparation of siRNAs-AuNPs, which increases release-efficacy and is faster and easier to perform than previously reported approaches (Lee et al. 2009). This modification has two major features: i) a dithiolane moiety that facilitates the conjugation with AuNPs and ii) disulphide groups that promote the release of siRNAs in the presence of GSH (Fig. 3a).
Fig. 3.
a Schematic structure of AuNPrelease and AuNPdithiolane. b The new release system (AuNPrelease) improves the release of oligonucleotides in the presence of GSH measured by the increase of fluorescence over time. The active molecule is released faster and to a greater extent compared to AuNPs that lack the disulphide modification (AuNPdithiolane). ([AuNPs]=3 nM; [GSH]=1 mM)
We assessed our novel modification in vitro by comparing AuNPs bearing this new release system (AuNPrelease) with AuNPs bearing only the dithiolane functionality (AuNPdithiolane), but lacking the disulphide moiety. In these experiments, we used DNA stands containing a fluorescein molecule on the 5′ end and the release system modification on the 3′ end. DNA release was monitored over time and measured by the increase of fluorescence emission upon incubation with GSH at 1 mM concentration. Our results demonstrate that AuNPrelease release oligonucleotides significantly faster and to a greater extent than AuNPdithiolane (Fig. 3b).
2.5 AuNPs modified with siRNA targeting GNAQ affect downstream signaling and reduce cell viability in GNAQ mutant cells
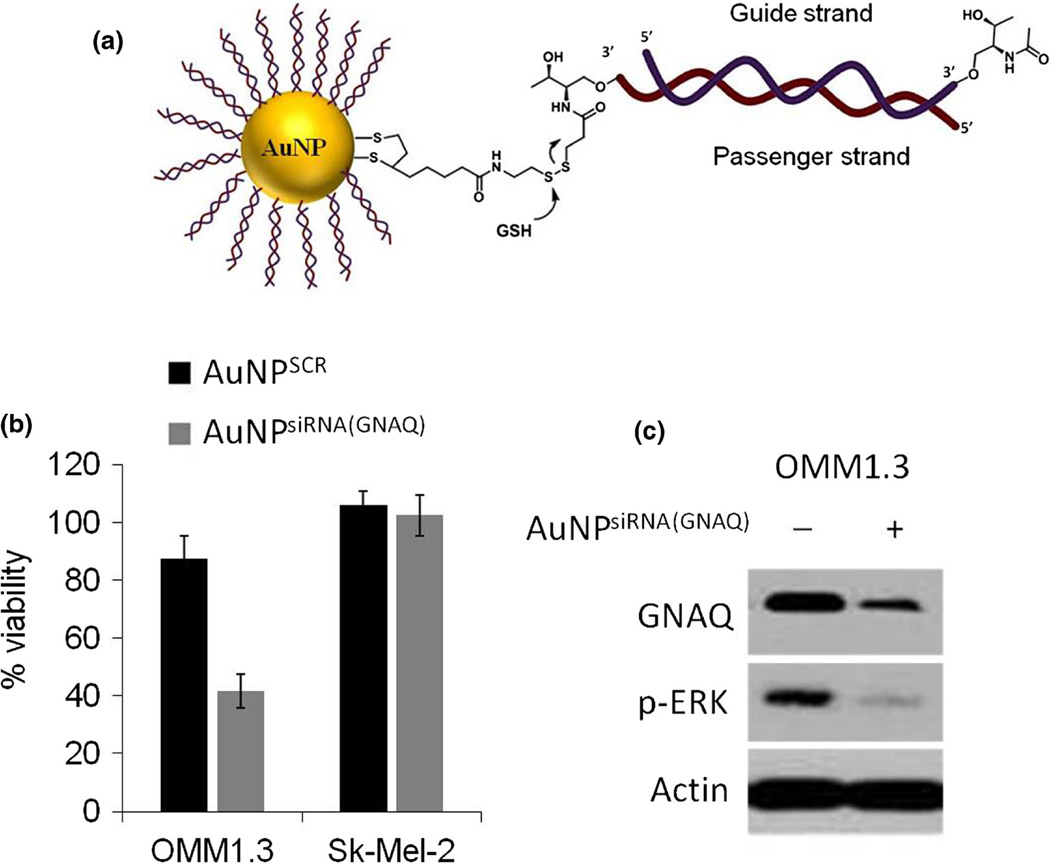
In the next set of experiments, we used our release system to prepare siRNA-AuNPs to target defined GNAQ transcripts (Fig. 4a). Due to high intracellular concentrations of GSH, especially in malignant cells, most siRNA were released intracellularly (Estrela et al. 2006; Latorre et al. 2014a). siRNAs were also modified at the 3′-end of the guide strand with a derivative of threoninol. This modification has improved the stability of siRNAs in serum without reducing the inhibititory activity (Somoza et al. 2010). Using siRNA-AuNPs targeting GNAQ, we sought to modulate GNAQ protein levels, downstream signaling and cell viability in GNAQ mutant cells. Incubation of cells with siRNA-AuNPs for 3 days caused a significant reduction of cell viability in the GNAQ mutant UM cell line OMM1.3. The wild-type cell line Sk-Mel-2 on the other hand remained largely unaffected. Immunoblot analyses showed a reduction of GNAQ protein levels and a decrease of p-ERK levels inOMM1.3 cells confirming the on-target effect of siRNA modified AuNPs compared to AuNPs bearing a non-targeting control sequence (AuNPSCR) (Fig. 4b, c).
Fig. 4.
a Schematic structure of AuNPs for siRNA delivery. siRNA targeting GNAQ was anchored onto the AuNPs with a dithiolane molecule, which also bears a disulphide moiety to allow a fast an efficient release of siRNA after interaction with glutathione (GSH). b The GNAQ mutant cell line OMM1.3, depended on GNAQ signalling, showed a reduction of cell viability with AuNPsiRNA(GNAQ), whereas viability of the GNAQ wild type cell line Sk-Mel-2 remained unchanged. Bars represent the viability of cells after 72 h incubation with the indicated AuNPs. (N=4, error bars represent the standard deviation of the mean) c Immunoblot analyses showed reduction of GNAQ protein and the major downstream effector p-ERK after incubation with AuNPsiRNA(GNAQ)
3 Discussion
Clinical examination of melanocytic lesions in the eye is an important modality for the diagnosis of UM. Although large and medium-sized uveal melanomas can readily be diagnosed, it is often difficult to clinically distinguish small melanomas from benign nevoid proliferations (Damato and Damato 2012; Shields et al. 2004; Shields and Shields 2009). The management of patients with a melanocytic tumor of uncertain malignancy can be difficult, requiring a choice between three options: i) observation, with an uncertain risk of metastatic spread; ii) treatment, which may cause severe visual loss and iii) biopsy, which demands both for the surgeon and the laboratory team, and is at risk of ocular complications.
With advances in our understanding of the genetic background and mutational status of UMs, molecular testing of ocular tumors has become routine in clinical practice, as a means of predicting survival (Damato et al. 2009, 2010a; Höglund et al. 2004; Harbour 2014; Onken et al. 2012; Shields et al. 2011). At present, genetic analyses require tumor material, which is generally acquired by biopsy. Even if lethal genetic abnormalities are excluded, additional aberrations may develop while the tumor is under ophthalmoscopic surveillance, so that sequential genomic analysis may be needed. Minimally invasive tests would therefore be valuable to determine the malignant potential of indeterminate melanocytic uveal tumors and would thus considerably improve patient care.
Several research groups using comparable approaches for AuNP designs have demonstrated that AuNPs can be modified to detect single nucleotide changes. However, a major limitation in the characterization of AuNP efficacy in most studies is that the specificity of the nanostructures was mainly assessed with short (18–20 bases) uniform, synthesized oligonucleotides that only harboured matching versus non-matching target mRNA sequences (Chen et al. 2008; He et al. 2012; Prigodich et al. 2009). We have shown that our functionalized AuNPs selectively detect a specific mutant transcript in living human cells. For this proof-of-principle study we chose GNAQ as our gene of interest and selectively detected mutations in live, GNAQ mutant cells. This approach has the potential to serve as a platform for the development of additional AuNP designs that can be used for the genetic characterization of cells. AuNPs used in this study as well as AuNPs similar in structure targeting other genetic alterations could ultimately be optimized for applications aimed at detecting mutations in vivo.
Regardless of size, the biological behaviour of UMs can vary greatly. Recent research has validated genetic profiles to identify patients that are more likely to develop metastases and who will eventually succumb to their UMs. Molecular tests including gene expression profiling and multiplex ligation dependent probe amplification are useful tools to assess genetic aberrations which help categorizing Ums (Damato et al. 2009, 2010a; Höglund et al. 2004; Harbour 2013, 2014; Onken et al. 2012). The genetic signature of UMs is particularly important for follow-up and treatment decisions: Class I UMs usually grow locally but uncommonly metastasise, whereas class II UMs and UMs with loss of chromosome 3 invariably lead to disseminated disease, which is fatal in more than 70 % of cases. Even though genetic analysis of GNAQ does not distinguish class I from class II UMs, AuNPs can be modified to target other mRNA sequences, such as BAP1, CCND1, BCL-2, AKT3 or detect chromosomal aberrations such as loss of chromosome 3 or gain of chromosome 8q which are associated with poor prognosis (van den Bosch et al. 2010; Damato et al. 2010b; Harbour 2013; Harbour et al. 2010; Onken et al. 2007; Scholes et al. 2003). We envision that in the future functionalized AuNPs may serve as a tool to provide genetic information on tumor cells in vivo, potentially facilitating the classification of UMs without requiring a biopsy.
Once UM has metastasised, treatment rarely prolongs life, so that the median overall survival is only 6–9 months from symptomatic disease (Bedikian et al. 1995; Kujala et al. 2003). Somatic mutations of GNAQ and GNA11 have been shown to drive UM growth and progression. They impair the intrinsic enzymatic activity of the protein, resulting in anchorage-independent GNAQ signalling (Van Raamsdonk et al. 2009, 2010; Sisley et al. 2011). Even though GNAQ provides an interesting target for treatment and drug development, the design of small molecule inhibitors specifically targeting constitutively active GNAQ has thus been unsuccessful. Despite the fact that siRNA mediated GNAQ knockdown has been shown to inhibit UM cell proliferation and to shrink tumors in animal models, significant challenges in the delivery of such GNAQ-targeting oligonucleotides in vitro and in vivo persist (Patel et al. 2011; Van Raamsdonk et al. 2009).
We have modified AuNPs with siRNA directed against GNAQ using a novel linker that enables the conjugation of siRNAs to AuNPs and improves the release of siRNAs in the presence of reducing molecules such as GSH. The cytoplasmatic concentration of GSH exceeds the concentration in the extracellular space significantly, enhancing intracellular release of the active molecule. In addition, we have previously shown that malignant cells have higher concentrations of GSH compared to non-malignant cells thereby increasing the delivery of therapeutics with AuNPs in cancer cells (Latorre et al. 2014c). We found that incubation with siRNA-AuNPs reduced GNAQ protein levels and disrupted the mitogen activated protein kinase signalling pathway (MAPK) downstream of GNAQ (Fig. 4). As expected, we also observed that knockdown of mutant GNAQ with siRNA-AuNPs results in a significant decrease of cell viability consistent with the established role of the MAPK pathway in maintaining cell viability. In contrast, wild type cells remain largely unaffected.
AuNPs can be functionalized with any siRNAs that specifically interfere with diseases propagating transcripts for the improved treatment of a given diseases. The unique properties of AuNPs hold particular promise for medical applications; they i) function as carriers of active molecules, such as antisense RNAs and therapeutics such as doxorubicin or AZD8055; ii) protect active molecules from degradation and iii) enable cellular uptake without the need of transfection or permeabilization reagents (Conde et al. 2012; Ding et al. 2014; Ghosh et al. 2008; Latorre et al. 2014c; Lee et al. 2009, 2011; Prigodich et al. 2009). Functionalized AuNPs individually modified with therapeutics relevant to the mutation and signaling characteristics of individual patients, would be a valuable addition to current local or systemic treatment modalities with the potential to target molecules such as GNAQ that are to date considered un-druggable.
4 Conclusion
Our study provides evidence that modified AuNPs can be functionalized to detect single nucleotide changes in cells. Furthermore, we have developed a novel linker that has the potential to increase the efficacy of siRNA-AuNP mediated gene regulation. Both AuNP designs are conceivably useful for future diagnostic and therapeutic applications.
5 Material and methods
Oligonucleotide synthesis
Oligonucleotides were prepared using a MerMade4 DNA and RNA Synthesizer using phosphoramidites (Link Technologies). DNA oligonucleotides were prepared at 1 µMol scale. After solid-phase synthesis, the solid support was transferred to a screw-cap glass vial and incubated at 55 °C for 4 h with 2 mL of ammonia solution (33 %). After the vial was cooled on ice, the supernatant was transferred by pipette to microcentrifuge tubes and the solid support and vial were rinsed with water. The combined solutions were evaporated to dryness using an evaporating centrifuge. RNA samples were further treated with TBAF to remove TBDMS groups following the procedure suggested by Link Technologies. Then, samples were purified by 20 % polyacrylamide gel electrophoresis and the oligonucleotides were eluted from gel fractions using an elutrap system. The solutions were desalted using a NAP-10 column and concentrated in an evaporating centrifuge.
Preparation of modified AuNPs
Gold nanoparticles (AuNPs) were prepared by the Turkevich method, where HAuCl4 and sodium citrate are mixed in boiling water (Kimling et al. 2006). This procedure affords AuNPs of 13 nm at around 12 nM concentration as a stable reddish dispersion where the gold metallic core is decorated with citrate groups. Subsequently, AuNPs were modified using an excess of oligonucleotides with a dithiolane group, which allows for rapid interaction to yield stable modified AuNPs. The particles showed a characteristic peak at 520 nm in the UV–vis spectra, and also revealed a peak fluorescence emission, recorded at 518 nm after 20 min incubation with the matching target sequence (exCitation at 470 nm) (Fig. S1a,b). The UV/VIS spectra were recorded at room temperature with the Synergy H4 microplate reader. In order to quantify loading of oligonucleotides onto the nanoparticles, DNA strands were labeled with a fluorescein molecule at the 5′ end. After conjugation with the nanostructures, fluorescence of the supernatant was quantified (λexc=470 nm, λem=520 nm) to determine loading efficacy. A detailed description of all chemical structures including the syntehsis of modified solid supports, sequences of oligonucleotides as well as NMR spectra is given in the electronic SI 2–6.
Cell lines and cell culture
Human melanoma cell lines OMM1.3, Sk-Mel-2, C918 and Mel202 were a generous gift from Boris Bastian at the University of California San Francisco. All cell lines were maintained in RPMI 1640 medium supplemented with 10 % FBS and incubated at 37 °C under 5 % CO2.
Flow cytometry
Cells were incubated with medium containing AuNPs at a concentration of 5nM for 6 h. Then the medium was replaced, cells were washed with PBS and analyzed with the AccuriC6 Flow Cytometer® using the CFlow® software (Version 1.0.227.4). Results represent the average of three independent experiments with error bars indicating the standard deviation. Non-modified AuNPs served as controls.
Imaging
Cells were grown in 2-well glass chamber slides at sub-confluent levels and incubated with 5nM GNPs. Cells were stained with CellMask Deep Red Plasma membrane Stain (Life Technologies, C10046) and DAPI (Vector, H-1200). Images were taken with the Zeiss Confocal Laser-scanning Microscope (LSM 510) using a 63× oil immersion objective with a 1.4 NA. The filter sets and laser lines used were: CellMask Deep Red Plasma membrane Stain: excitation 633 nm, BP 650–710; fluorescein: excitation 470 nm BP 500–610; DAPI: excitation 800 nm, BP390-465.
Cell viability assay
All viability assays were at least carried out in triplicates. The relative number of viable cells was calculated using CellTiter-Glo® (Promega, G7570) after 72 h of incubation with the indicated conditions. Total luminescence was measured on the SynergyHT plate reader (BioTek) using Gen5 software (Version 1.11.5). Cells were treated with the indicated AuNPs in concentrations ranging from 1.5 µM to 375 nM.
Sequencing
Archival left over tissue from patients with UM were generously provided by Devron Char at the California Pacific Medical Center San Francisco. Formalin fixed and paraffin embedded tissue was cut in 7 µM sections. Tumor cells were extracted by hand micro dissection under a 10× magnifying dissection microscope. A haematoxylin and eosin stained reference slide was used for orientation. DNA was extracted using the QIAmp DNA FFPE tissue kit (Qiagen 56404) according to the manufacturer’s recommendations. The following M13-PCR primers were used: GNAQ (F) 5′-TGTAAAACGACGGCCAGTAATCCATTG C-3′ GNAQ (R) 5′-AGCGGATAACAATTTCACACAGGTTTTCC-3′ GNA11 (F) 5′-TGTAAAACGACGGCCAGTCGTCCTGG GA-3′ GNA11 (R) AGCGGATAACAATTTCACACAGGC TTGG-3′. Sanger sequencing was carried out after clean-up of PCR products with exonuclease I and Shrimp alkaline phosphatase. Sequences were then analyzed with the Mutation Surveyor Version 4.0.9 (Softgenetics).
Supplementary Material
Acknowledgments
We thank Bertil Damato and James E. Cleaver for their helpful advice in the elaboration of this manuscript. We further thank Boris Bastian for providing the cell lines used in this study. This work was supported by, the National Institute of Health (1K08CA155035-01A1), the Melanoma Research Alliance, the UCSF Catalyst Program, the Max Kade Foundation, the Verein für Dermatologie Krankenanstalt Rudolfstiftung and the René Touraine Foundation the Ministry of Economy and competitiveness (SAF2010-15440) and IMDEA Nanociencia. We further acknowledge T. Dattels for his generous support.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10544-014-9908-7) contains supplementary material, which is available to authorized users.
Coflict of interests The authors have no conflict of interest.
Elements of the structure of gold nanoparticles used in this study are patented: 61/933,881 (US); EP14382033.0 (EU).
Contributor Information
Christian Posch, Email: cposch81@gmail.com, Department of Dermatology, Mount Zion Cancer Research Center, University of California San Francisco, 2340 Sutter Street N461, 94115 San Francisco, CA, USA; Department of Dermatology, The Rudolfstiftung Hospital, Juchgasse 25, 1030 Vienna, Austria.
Alfonso Latorre, Instituto Madrileño de Estudios Avanzados en Nanociencia (IMDEA Nanociencia), & CNB-CSIC-IMDEA Nanociencia Associated Unit “Unidad de Nanobiotecnología”, 28049 Madrid, Spain.
Michelle B. Crosby, North County Eye Surgery Associates, 320 Santa Fe Drive Suite 104, Encinitas 92024, USA
Anna Celli, Department of Dermatology, Mount Zion Cancer Research Center, University of California San Francisco, 2340 Sutter Street N461, 94115 San Francisco, CA, USA.
Ana Latorre, Instituto Madrileño de Estudios Avanzados en Nanociencia (IMDEA Nanociencia), & CNB-CSIC-IMDEA Nanociencia Associated Unit “Unidad de Nanobiotecnología”, 28049 Madrid, Spain.
Igor Vujic, Department of Dermatology, Mount Zion Cancer Research Center, University of California San Francisco, 2340 Sutter Street N461, 94115 San Francisco, CA, USA; Department of Dermatology, The Rudolfstiftung Hospital, Juchgasse 25, 1030 Vienna, Austria.
Martina Sanlorenzo, Department of Dermatology, Mount Zion Cancer Research Center, University of California San Francisco, 2340 Sutter Street N461, 94115 San Francisco, CA, USA; Department of Medical Sciences, Section of Dermatology, University of Turin, Via Cherasco 23, 10100 Torino, Italy.
Gary A. Green, Department of Dermatology, Mount Zion Cancer Research Center, University of California San Francisco, 2340 Sutter Street N461, 94115 San Francisco, CA, USA
Jingly Weier, Department of Dermatology, Mount Zion Cancer Research Center, University of California San Francisco, 2340 Sutter Street N461, 94115 San Francisco, CA, USA.
Mitchell Zekhtser, Department of Dermatology, Mount Zion Cancer Research Center, University of California San Francisco, 2340 Sutter Street N461, 94115 San Francisco, CA, USA.
Jeffrey Ma, Department of Dermatology, Mount Zion Cancer Research Center, University of California San Francisco, 2340 Sutter Street N461, 94115 San Francisco, CA, USA.
Gabriela Monico, Department of Dermatology, Mount Zion Cancer Research Center, University of California San Francisco, 2340 Sutter Street N461, 94115 San Francisco, CA, USA.
Devron H. Char, Departments of Ophthalmology, Stanford University, University of California San Francisco and California Pacific Medical Center, 45 Castro Street, 94114 San Francisco, USA
Denis Jusufbegovic, Departments of Ophthalmology, Stanford University, University of California San Francisco and California Pacific Medical Center, 45 Castro Street, 94114 San Francisco, USA.
Klemens Rappersberger, Department of Dermatology, The Rudolfstiftung Hospital, Juchgasse 25, 1030 Vienna, Austria.
Álvaro Somoza, Department of Dermatology, The Rudolfstiftung Hospital, Juchgasse 25, 1030 Vienna, Austria.
Susana Ortiz-Urda, Department of Dermatology, Mount Zion Cancer Research Center, University of California San Francisco, 2340 Sutter Street N461, 94115 San Francisco, CA, USA.
References
- Bakalian S, Marshall J-C, Logan P, Faingold D, Maloney S, Di Cesare S, Martins C, Fernandes BF, Burnier MN., Jr Clin Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008;14:951. doi: 10.1158/1078-0432.CCR-06-2630. [DOI] [PubMed] [Google Scholar]
- Bedikian AY, Legha SS, Mavligit G, Carrasco CH, Khorana S, Plager C, Papadopoulos N, Benjamin RS. Cancer. 1995;76:1665. doi: 10.1002/1097-0142(19951101)76:9<1665::aid-cncr2820760925>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Butler P, Char DH, Zarbin M, Kroll S. Ophthalmology. 1994;101:710. doi: 10.1016/s0161-6420(94)31274-7. [DOI] [PubMed] [Google Scholar]
- Chen Y-T, C-L Hsu, Hou S-Y. Anal. Biochem. 2008;375:299. doi: 10.1016/j.ab.2007.12.036. [DOI] [PubMed] [Google Scholar]
- Conde J, Ambrosone A, Sanz V, Hernandez Y, Marchesano V, Tian F, Child H, Berry CC, Ibarra MR, Baptista PV, Tortiglione C, de la Fuente JM. ACS Nano. 2012;6:8316. doi: 10.1021/nn3030223. [DOI] [PubMed] [Google Scholar]
- Conde J, Tian F, Hernández Y, Bao C, Cui D, Janssen K-P, Ibarra MR, Baptista PV, Stoeger T, de la Fuente JM. Biomaterials. 2013;34:7744. doi: 10.1016/j.biomaterials.2013.06.041. [DOI] [PubMed] [Google Scholar]
- Damato B, Dopierala JA, Coupland SE. Clin. Cancer Res. 2010a;16:6083. doi: 10.1158/1078-0432.CCR-10-2076. [DOI] [PubMed] [Google Scholar]
- Damato B, Dopierala JA, Coupland SE. Clin. Cancer Res. Off. J. Am. Assoc Cancer Res. 2010b;16:6083. doi: 10.1158/1078-0432.CCR-10-2076. [DOI] [PubMed] [Google Scholar]
- Damato B, Dopierala J, Klaasen A, van Dijk M, Sibbring J, Coupland SE. Invest. Ophthalmol. Vis. Sci. 2009;50:3048. doi: 10.1167/iovs.08-3165. [DOI] [PubMed] [Google Scholar]
- Damato EM, Damato BE. Ophthalmology. 2012;119:1582. doi: 10.1016/j.ophtha.2012.01.048. [DOI] [PubMed] [Google Scholar]
- Diener-West M, Reynolds SM, Agugliaro DJ, Caldwell R, Cumming K, Earle JD, Hawkins BS, Hayman JA, Jaiyesimi I, Jampol LM, Kirkwood JM, Koh W-J, Robertson DM, Shaw JM, Straatsma BR, Thoma J, Collaborative Ocular Melanoma Study Group Arch. Ophthalmol. 2005;123:1639. doi: 10.1001/archopht.123.12.1639. [DOI] [PubMed] [Google Scholar]
- Ding Y, Jiang Z, Saha K, Kim CS, Kim ST, Landis RF, Rotello VM. Mol. Ther. J. Am. Soc. Gene Ther. 2014 doi: 10.1038/mt.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrela JM, Ortega A, Obrador E. Crit. Rev. Clin. Lab. Sci. 2006;43:143. doi: 10.1080/10408360500523878. [DOI] [PubMed] [Google Scholar]
- Ghosh P, Han G, De M, Kim CK, Rotello VM. Adv. Drug Deliv. Rev. 2008;60:1307. doi: 10.1016/j.addr.2008.03.016. [DOI] [PubMed] [Google Scholar]
- Höglund M, Gisselsson D, Hansen GB, White VA, Säll T, Mitelman F, Horsman D. Int. J. Cancer. 2004;108:57. doi: 10.1002/ijc.11558. [DOI] [PubMed] [Google Scholar]
- Harbour JW. Am. Soc. Clin. Oncol. Educ. Book ASCO Am. Soc. Clin. Oncol. Meet. 2013;388 doi: 10.1200/EdBook_AM.2013.33.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour JW. Methods Mol. Biol. Clifton NJ. 2014;1102:427. doi: 10.1007/978-1-62703-727-3_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour JW, Onken MD, Roberson EDO, Duan S, Cao L, Worley LA, Council ML, Matatall KA, Helms C, Bowcock AM. Science. 2010;330:1410. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Zhang X, Zhang S, Kris MKL, Man FC, Kawde A-N, Liu G. Biosens. Bioelectron. 2012;34:37. doi: 10.1016/j.bios.2011.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, McCormick SA. Eye Sci. 2011;26:18. doi: 10.3969/j.issn.1000-4432.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Kimling J, Maier M, Okenve B, Kotaidis V, Ballot H, Plech A. J. Phys. Chem. B. 2006;110:15700. doi: 10.1021/jp061667w. [DOI] [PubMed] [Google Scholar]
- Kujala E, Mäkitie T, Kivelä T. Invest. Ophthalmol. Vis. Sci. 2003;44:4651. doi: 10.1167/iovs.03-0538. [DOI] [PubMed] [Google Scholar]
- Latorre A, Posch C, Garcimartín Y, Celli A, Sanlorenzo M, Vujic I, Ma J, Zekhtser M, Rappersberger K, Ortiz-Urda S, Somoza A. Nanoscale. 2014a doi: 10.1039/c4nr00019f. [DOI] [PubMed] [Google Scholar]
- Latorre A, Posch C, Garcimartín Y, Ortiz-Urda S, Somoza Á. Chem. Commun. Camb. Engl. 2014b;50:3018. doi: 10.1039/c3cc47862a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre A, Posch C, Gracimartin Y, Celli A, Sanlorenzo M, Vujic I, Ma J, Zekhtser M, Rappersberger K, Ortiz-Urda S, Somoza A. Nanoscale. 2014c doi: 10.1039/c4nr00019f. [DOI] [PubMed] [Google Scholar]
- Lee J-S, Green JJ, Love KT, Sunshine J, Langer R, Anderson DG. Nano Lett. 2009;9:2402. doi: 10.1021/nl9009793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Han MS, Asokan S, Tung C-H. Small Weinh. Bergstr. Ger. 2011;7:364. doi: 10.1002/smll.201001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onken MD, Worley LA, Char DH, Augsburger JJ, Correa ZM, Nudleman E, Aaberg TM, Jr, Altaweel MM, Bardenstein DS, Finger PT, Gallie BL, Harocopos GJ, Hovland PG, McGowan HD, Milman T, Mruthyunjaya P, Simpson ER, Smith ME, Wilson DJ, Wirostko WJ, Harbour JW. Ophthalmology. 2012;119:1596. doi: 10.1016/j.ophtha.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onken MD, Worley LA, Person E, Char DH, Bowcock AM, Harbour JW. Clin. Cancer Res. Off. J. Am. Assoc Cancer Res. 2007;13:2923. doi: 10.1158/1078-0432.CCR-06-2383. [DOI] [PubMed] [Google Scholar]
- Patel M, Smyth E, Chapman PB, Wolchok JD, Schwartz GK, Abramson DH, Carvajal RD. Clin. Cancer Res. Off. J. Am. Assoc Cancer Res. 2011;17:2087. doi: 10.1158/1078-0432.CCR-10-3169. [DOI] [PubMed] [Google Scholar]
- Pópulo H, Vinagre J, Lopes JM, Soares P. Br. J. Ophthalmol. 2011;95:715. doi: 10.1136/bjo.2009.174417. [DOI] [PubMed] [Google Scholar]
- Prigodich AE, Seferos DS, Massich MD, Giljohann DA, Lane BC, Mirkin CA. ACS Nano. 2009;3:2147. doi: 10.1021/nn9003814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes AGM, Damato BE, Nunn J, Hiscott P, Grierson I, Field JK. Invest. Ophthalmol. Vis. Sci. 2003;44:1008. doi: 10.1167/iovs.02-0159. [DOI] [PubMed] [Google Scholar]
- Shields CL. Arch. Ophthalmol. 2009;127:989. doi: 10.1001/archophthalmol.2009.208. [DOI] [PubMed] [Google Scholar]
- Shields CL, Demirci H, Materin MA, Marr BP, Mashayekhi A, Shields JA. Can. J. Ophthalmol. J. Can. Ophtalmol. 2004;39:351. doi: 10.1016/s0008-4182(04)80005-x. [DOI] [PubMed] [Google Scholar]
- Shields CL, Furuta M, Thangappan A, Nagori S, Mashayekhi A, Lally DR, Kelly CC, Rudich DS, Nagori AV, Wakade OA, Mehta S, Forte L, Long A, Dellacava EF, Kaplan B, Shields JA. Arch. Ophthalmol. 2009;127:989. doi: 10.1001/archophthalmol.2009.208. [DOI] [PubMed] [Google Scholar]
- Shields CL, Ganguly A, Bianciotto CG, Turaka K, Tavallali A, Shields JA. Ophthalmology. 2011;118:396. doi: 10.1016/j.ophtha.2010.05.023. [DOI] [PubMed] [Google Scholar]
- Shields CL, Shields JA. Clin. Dermatol. 2009;27:122. doi: 10.1016/j.clindermatol.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Singh AD, Bergman L, Seregard S. Ophthalmol. Clin. N. Am. 2005;18:75. doi: 10.1016/j.ohc.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Sisley K, Doherty R, Cross NA. Br. J. Ophthalmol. 2011;95:620. doi: 10.1136/bjo.2010.182097. [DOI] [PubMed] [Google Scholar]
- Somoza A, Terrazas M, Eritja R. Chem. Commun. Camb. Engl. 2010;46:4270. doi: 10.1039/c003221b. [DOI] [PubMed] [Google Scholar]
- van den Bosch T, Kilic E, Paridaens D, de Klein A. Dermatol. Res. Pract. 2010;2010:360136. doi: 10.1155/2010/360136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O’Brien JM, Simpson EM, Barsh GS, Bastian BC. Nature. 2009;457:599. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, Obenauf AC, Wackernagel W, Green G, Bouvier N, Sozen MM, Baimukanova G, Roy R, Heguy A, Dolgalev I, Khanin R, Busam K, Speicher MR, O’Brien J, Bastian BC, Engl N. J. Med. 2010;363:2191. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall NR, Shi Y. Lancet. 2003;362:1401. doi: 10.1016/S0140-6736(03)14637-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.