Abstract
Persistent hepatitis B virus (HBV) infection relies on the stable maintenance and proper functioning of a nuclear episomal form of the viral genome called covalently closed circular (ccc) DNA. One of the major reasons for the failure of currently available antiviral therapeutics to achieve a cure of chronic HBV infection is their inability to eradicate or inactivate cccDNA. In this review article, we summarize our current understanding of cccDNA metabolism in hepatocytes and the modulation of cccDNA by host pathophysiological and immunological cues. Perspectives on the future investigation of cccDNA biology, as well as strategies and progress in therapeutic elimination and/or transcriptional silencing of cccDNA through rational design and phenotypic screenings, are also discussed. This article forms part of a symposium in Antiviral Research on “An unfinished story: from the discovery of the Australia antigen to the development of new curative therapies for hepatitis B.”
1. Introduction
Despite the availability of effective vaccines for more than three decades, an estimated 240 million individuals are still chronically infected with hepatitis B virus (HBV), and HBV-related diseases account for 600,000 deaths per year (Ott et al., 2012). Seven drugs are currently approved for the treatment of chronic hepatitis B, which include two formulations of alpha-interferon (standard and pegylated) that modulate the host antiviral immune responses and five nucleos(t)ide analogues (lamivudine, adefovir, entecavir, telbivudine, and tenofovir) that inhibit HBV DNA polymerase (Keeffe et al., 2008; Zoulim and Locarnini, 2009). Pegylated IFN-α therapy is effective in achieving a sustained virological response -- defined as hepatitis e antigen (HBeAg) seroconversion and/or reduction in HBV DNA levels to below 20,000 copies/mL at 6 months after the completion of a 48-week course of therapy -- in only 30% of HBeAg-positive and 40% of HBeAg-negative cases (Janssen et al., 2005; Lau et al., 2005; Perrillo, 2009). Nucleos(t)ide analogues profoundly reduce the viral load in vast majority of treated patients, which is associated with significant improvement of liver function and reduced incidence of liver failure and hepatocellular carcinoma (Liaw, 2013). However, even prolonged treatment rarely cures chronic HBV infection, and virological relapse is common after treatment cessation (Zoulim and Locarnini, 2009).
Accumulating evidence suggests that the inability to eliminate the nuclear episomal form of the viral genome, the covalently closed circular (ccc) DNA, by currently available therapeutics, is one of the critical reasons for failure to achieve a functional cure (Bowden et al., 2015; Sung et al., 2005; Werle-Lapostolle et al., 2004; Wursthorn et al., 2006). Unraveling the molecular mechanism of cccDNA metabolism and regulation by host antiviral immune responses could facilitate the discovery and development of antiviral agents and immunotherapeutics that would effectively induce more durable suppression of HBV replication following the cessation of treatment and ideally achieve a functional cure (Block et al., 2013).
2. Molecular pathway of cccDNA biosynthesis
HBV is a small DNA virus with a relaxed circular (rc) partially double-stranded 3.2kb genome (Summers et al., 1975). Unique structural properties of the genome include the asymmetry in length of the two strands of DNA and the covalent linkage of the viral DNA polymerase protein to the 5’-terminus of minus-strand DNA (Bartenschlager and Schaller, 1988; Gerlich and Robinson, 1980; Molnar-Kimber et al., 1983). These structural features provided early clues for uncovering HBV DNA replication via a mechanism other than semiconservative replication (Mason et al., 1982), i.e., polymerase protein-primed reverse transcription of an RNA intermediate called pregenomic (pg) RNA (Summers and Mason, 1982; Wang and Seeger, 1992). However, distinct from retroviruses, the integration of HBV genomic DNA into host cellular chromosomes is not an obligatory step in the life cycle of HBV (Seeger and Mason, 2015). Instead, upon entry into the cytoplasm of hepatocyte, rcDNA in the nucleocapsid is transported into the nucleus and converted into the episomal cccDNA, which serves as the template for the transcription of viral mRNAs (Summers and Mason, 1982).
In addition to being the template for reverse transcription, the pgRNA is translated to produce both the core protein and HBV DNA polymerase. The DNA polymerase binds to a stem-loop structure (termed epsilon) within the 5’ portion of the pgRNA to prime viral DNA synthesis and initiate nucleocapsid assembly (Tavis and Ganem, 1995; Tavis and Ganem, 1996; Wang and Seeger, 1992, 1993). The encapsidated pgRNA is then reverse transcribed to form the minus-strand viral DNA by viral DNA polymerase. The minus-strand serves as the template for the plus-strand DNA synthesis. The nucleocapsid matures as rcDNA is formed and can be either enveloped and secreted out of the cell as a virion particle or redirected into the nucleus to amplify the cccDNA pool (Tuttleman et al., 1986; Wu et al., 1990).
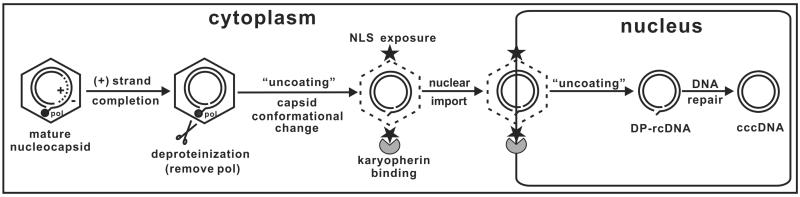
The essential steps in the biosynthesis of cccDNA from the incoming virion or from the progeny cytoplasmic nucleocapsid rcDNA include rcDNA uncoating (nucleocapsid disassembly), nuclear transportation and conversion into cccDNA. The molecular mechanisms involved in rcDNA uncoating have been investigated by different approaches. Studies from Kann's group demonstrated that the cytoplasmic HBV capsids can freely pass the nuclear pore complex (NPC) and are captured in the nuclear basket on the nuclear side of the NPC, through interaction with nucleoporin 153 (Nup153). However, only the capsids containing a mature rcDNA disassemble in the nuclear baskets and release rcDNA into the nucleus (Kann et al., 1999; Rabe et al., 2003; Schmitz et al., 2010). On the other hand, results from our studies on the production and function of deproteinized rcDNA support a model that the completion of plus-strand DNA synthesis triggers the removal of viral DNA polymerase covalently linked to the 5’-end of minus-strand DNA. Plus-strand DNA synthesis also triggers a structural change in nucleocapsids in the cytoplasm, which leads to the translocation of the carboxyl termini of core protein to the exterior of the capsids.
Exposure of the nuclear localization signal (NLS) located at the carboxyl terminus of the core protein allows the matured capsids to interact with host karyopherins and import the deproteinized rcDNA into the nuclei for cccDNA formation. In this regard, the deproteinized rcDNA is a functional precursor of cccDNA (Fig. 1) (Guo et al., 2007; Guo et al., 2010). In agreement with the notion that maturation of nucleocapsids triggers their structural change or partial disassembly, biochemical and biophysical analyses of cytoplasmic nucleocapsids by another group revealed that the maturation of rcDNA was associated with nucleocapsid destabilization, as indicated by increased protease and nuclease sensitivity, altered sedimentation during sucrose gradient centrifugation, and slower mobility in native agarose gel electrophoresis (Cui et al., 2013).
Fig 1. Proposed model for the molecular pathway of HBV cccDNA formation.
In the cytoplasm, completion of viral plus-strand DNA synthesis inside the nucleocapsid triggers the removal of the viral polymerase from minus-strand DNA. This deproteinization reaction is associated with a nucleocapsid conformational change (“uncoating” step 1), resulting in exposure of the nuclear localization signal (NLS) at the C-terminus of the capsid protein on the nucleocapsid surface, followed by binding of cellular karyopherins and nuclear import of the DP-rcDNA-containing capsid. In the nucleus, DP-rcDNA is released from the capsid (“uncoating” step 2) and converted into cccDNA by employing the host DNA repair machinery.
Biochemically, conversion of rcDNA to cccDNA requires the removal of the polymerase protein and RNA primers from the 5’-terminus of minus-strand and plus-strand DNA, respectively, as well as trimming the DNA ends, filling in the gaps and ligation of both strand of DNA. Although it is speculated that all those reactions are most probably catalyzed by host cellular DNA repair machinery, the exact DNA repair proteins involved in those reactions remain to be identified. Tyrosyl-DNA phosphodiesterase-2 (Tdp2) was recently identified as the enzyme responsible for cleavage of tyrosyl-5′ DNA linkages formed between topoisomerase II and cellular DNA (Cortes Ledesma et al., 2009). Tdp-2 also specifically cleaves a unique protein-RNA covalent linkage generated for priming the viral genomic RNA replication during a picornavirus infection (Virgen-Slane et al., 2012). Since the hepadnaviral DNA polymerase-DNA linkage is also a 5′ DNA-phosphotyrosyl bond, it is plausible that Tdp2 might be responsible for the removal of DNA-linked viral polymerase and essential for cccDNA synthesis. However, although two independent studies convincingly demonstrated that Tdp2 is indeed able to cleave the tyrosyl-minus strand DNA linkage, using authentic rcDNA isolated from nucleocapsids as substrate, in vitro, the role of this enzyme in hepadnaviral cccDNA formation in virus-replicating cells remains controversial (Cui et al., 2015b; Koniger et al., 2014). Our studies do not favor a role of Tdp2 in HBV cccDNA formation either, as stably knocking down of Tdp2 in an inducible HBV stable cell line upregulated transgene-based viral RNA transcription and subsequent DNA replication, and the levels of deproteinzed rcDNA and cccDNA were also proportionally increased (D. Cai and H. Guo, unpublished data).
While it is generally believed that multiple DNA repair proteins/pathways recognize the two gaps in rcDNA as DNA breaks and directly repair the rcDNA into cccDNA (Guo et al., 2012; Sohn et al., 2009), it is also postulated that rcDNA may first be converted into a double-stranded linear DNA containing terminal repeats (TR-dsl DNA) through extension of both plus- and minus- strands over the cohesive end region by viral or host DNA polymerases, and cccDNA is subsequently formed via intra-molecular homologous recombination of the TR-dsl DNA (Yang et al., 1996). Although the episomal TR-dsl DNA has not been detected in virally- infected hepatocytes by conventional hybridization methods, sequence analysis of cccDNA recombinant joints and virus-host DNA junctions in the livers of DHBV-infected ducks and WHV-infected woodchucks suggested that the putative TR-dsl DNA did exist and could be converted into cccDNA and integrated into host chromosome by nonhomologous end joining (NHEJ) DNA repair pathway (Bill and Summers, 2004; Yang et al., 1996; Yang and Summers, 1995, 1999).
It is also worth noting that in addition to rcDNA, approximately 5 to 10% of hepadnaviral genomic DNA is double-stranded linear (dsl) DNA (Staprans et al., 1991). HBV plus-strand DNA synthesis in the cytoplasmic nucleocapsids is primed by a RNA oligomer derived from the 5’ terminal 17-18 ribonucleotides of pgRNA, which is translocated from the 3’ end of minus- strand DNA to pair with the DR2 sequence near the 5’ end of minus strand DNA to initiate plus-strand synthesis (Lien et al., 1986). The subsequent template switch circularizes viral DNA to yield a rcDNA (Liu et al., 2003). However, occasional failure of primer translocation results in in situ priming of plus-strand DNA synthesis at the 3’ end of minus-strand DNA to produce a dslDNA (Staprans et al., 1991). The dslDNA and TR-dslDNA have been shown to be the direct precursors of integrated HBV in host cellular chromosome (Bill and Summers, 2004; Yang and Summers, 1999).
Interestingly, a recent clinical study suggested that serum dslDNA proportions progressively increase during the development of HBV-related liver diseases. The dslDNA proportion can be regulated by inflammatory cytokines, suggesting an association between inflammation, increased production of HBV dslDNA and development of HCC (Zhao et al., 2015). It has been previously shown that cccDNA can be formed from both rcDNA and dslDNA in hepadnavirus-infected cells (Tuttleman et al., 1986; Yang and Summers, 1995). However, while rcDNA is faithfully repaired to form wild-type cccDNA, the cccDNA derived from dslDNA carries deletions or insertions around the site of end-joining (Yang and Summers, 1995). These observations suggest that rcDNA and dslDNA are converted into cccDNA via different DNA repair pathways. Indeed, using a panel of skin fibroblast and CHO cell-derived lines deficient in specific DNA repair genes, we demonstrated that Ku80, a component of NHEJ DNA repair pathway, was essential for the synthesis of cccDNA from dslDNA, but not rcDNA (Guo et al., 2012). The advent of novel gene editing technologies will facilitate the systematic identification of host cellular genes required for cccDNA biosynthesis from rcDNA (Pyzocha et al., 2014; Zhang et al., 2014a).
3. Regulation of cccDNA formation by viral and host factors
The copy number of cccDNA varies among individual hepatocytes and fluctuates under different pathobiological conditions. Studies from WHV-infected woodchucks indicated that the copy numbers of cccDNA in hepatocytes range from 1 to 50 (Dandri et al., 2000; Kajino et al., 1994). A single cell-based study revealed that in DHBV-infected ducks, cccDNA copy number differed significantly from cell to cell and at different time points post-infection. While 90% of hepatocytes contained 1 to 17 copies of cccDNA, the remaining 10% contained more copies (Zhang et al., 2003). Clinical studies showed that the median level of intrahepatic cccDNA in chronic hepatitis B patients was 1.05 copies per cell, ranging from 0.035 to 195 copies per cell; and the levels of cccDNA decreased after HBeAg seroconversion (Laras et al., 2006; Wursthorn et al., 2006).
Although superinfection exclusion has not been firmly excluded for HBV-infected hepatocytes, the numbers of cccDNA in a virus-infected hepatocyte is presumably controlled by the efficiency of the intracellular cccDNA amplification pathway. For many animal viruses, genome uncoating strictly relies on extracellular virion phase, because the nucleocapsids of these viruses become mature only during the budding and secretion process (Ganser-Pornillos et al., 2008; Mettenleiter et al., 2009), or require interaction with viral receptors or an acid bath in endosomes during entry into host cells to trigger disassembly (Alain et al., 2007; Day et al., 2003; Smith et al., 2008). The uncoating of progeny nuclocapsids in infected cells is a unique feature of hepadnaviruses (Tuttleman et al., 1986; Wu et al., 1990). Considering that the large amounts of viral rcDNA, the precursor of cccDNA formation, exist in the cytoplasm and relatively small numbers of cccDNA accumulate in the nucleus, mature nucleocapsid uncoating and cccDNA formation must be tightly controlled by host and/or viral factors. In addition, because untimely uncoating and release of viral DNA in the cytoplasm may result in recognition of HBV infection by host cellular innate immune sensors to induce an antiviral response (Rasaiyaah et al., 2013), deliberating control of HBV genome uncoating is not only important for cccDNA formation, but also critical for the virus to evade host restriction (Le Sage et al., 2014).
A series of elegant studies by Summers and colleagues suggested that the DHBV large envelope protein regulated intracellular HBV DNA traffic and cccDNA formation (Lenhoff and Summers, 1994; Summers et al., 1990). When the level of large envelope protein is low in the early phase of infection, nuclear delivery of mature nucleocapsid is favored, which allows rapid establishment of the cccDNA pool and ensures that infected cells will be stably colonized. When cccDNA is amplified to reach a level that supports the synthesis of sufficient amount of large envelope proteins for virion assembly, the mature nucleocapsids will be primarily enveloped and secreted as virions. The tight control of DHBV cccDNA amplification by large envelope protein is demonstrated in primary duck hepatocytes and DHBV-infected ducks in vivo. For instance, infection of envelope protein-deficient recombinant DHBV results in approximately 20-fold more cccDNA accumulation and ultimately death of the infected hepatocytes (Lenhoff et al., 1999; Summers et al., 1990; Summers et al., 1991). However, investigations in human hepatoma cells demonstrated that the deficiency of HBV envelope proteins increased the level of cccDNA by only two-fold, but resulted in a dramatic accumulation of the deproteinized rcDNA (Gao and Hu, 2007; Guo et al., 2007; Lentz and Loeb, 2011). The modest impact of HBV envelope proteins on cccDNA amplification suggests that other viral and host cellular factors regulate HBV cccDNA synthesis. Identification of the viral and host factors that determine the set points of cccDNA in HBV-infected hepatocytes may provide therapeutic targets for elimination of cccDNA.
DHBV cccDNA can be efficiently accumulated in many different types of human and animal cells (Guo et al., 2012), but formation of HBV cccDNA has only been consistently observed in infected human hepatocytes and selected human hepatoma cell lines. Particularly, HBV fails to synthesize cccDNA in the hepatocytes of HBV transgenic mice, despite the existence of large amount of rcDNA-containing nucleocapsids (Guidotti et al., 1995). Moreover, transgenic expression of human sodium taurocholate cotransporting polypeptide (NTCP) in mouse hepatocytes, the bona fide receptor of HBV and its satellite hepatitis D virus (HDV) (Yan et al., 2012), confers HDV infection susceptibility to mouse hepatocytes, but not HBV-infection susceptibility (He et al., 2015). This finding suggests that besides the lack of a functional receptor, failure to support efficient cccDNA formation might also account for the nonsusceptibility of mouse hepatocytes to HBV infection. Interestingly, trace amount of cccDNA can be detected in the livers of HNF-1α-null HBV transgenic mice, suggesting that host factors regulate cccDNA formation and/or accumulation in mouse hepatocytes (Raney et al., 2001).
It was reported recently by Cui and colleagues that HBV cccDNA could be detected in a TGF-α-immortalized mouse hepatocyte cell line AML12HBV10, and that AML12HBV10 cells support a tetracycline-inducible HBV replication (Xu et al., 2010), where mature nucleocapsids are not stable and rcDNA are efficiently uncoated for cccDNA formation (Cui et al., 2015a). This work thus suggests that disassembly of mature nucleocapsids is subject to host cellular regulation and could thus be one of the critical determinants of host range and/or cell tropism of HBV. Further investigation into the molecular mechanism regulating cccDNA formation in mouse hepatocytes could establish intellectual basis for the development of mouse models supporting efficient HBV infection.
4. Structure and function of cccDNA minichromosome
Like other nuclear viral episomal DNA, hepadnaviral cccDNA molecules in the nuclei of infected cells associate with nucleosomes and assemble into host cellular chromatin-like structures, designated as minichromosomes (Bock et al., 1994; Newbold et al., 1995). Mapping the distribution of nucleosomes along DHBV cccDNA in the livers of infected ducks showed that nucleosomes are not randomly positioned on cccDNA. Particularly, several nucleosome-protected sites in the region of the DHBV genome [nucleotides (nt) 2000 to 2700], known to harbor various cis transcription regulatory elements, were consistently identified in all DHBV-infected livers. These observations suggest that, like host cellular chromatin, nucleosome positioning and histone modification of the minichromosomes may play an important role in regulation of cccDNA transcription (Shi et al., 2012). Indeed, HBV replication activity in the liver of patients correlated with the acetylation status of HBV cccDNA-bound H3 and H4 histones (Pollicino et al., 2006). We also found that certain histone deacetylase (HDAC) inhibitors potently inhibited DHBV and HBV cccDNA transcription, and IFN-α suppressed DHBV cccDNA transcription by reducing the acetylation of cccDNA-associated histone H3 lysine 9 (H3K9) and 27 (H3K27) (Liu et al., 2013).
In addition to nucleosomes, host cellular transcription factors, co-activators, corepressors, chromatin-modifying enzymes, as well as viral proteins, such as core and HBx proteins, have also been demonstrated to be recruited to cccDNA minichromoses under certain conditions to regulate cccDNA transcription activity (Levrero et al., 2009; Nassal, 2015). WHV X protein has been shown to be essential for the virus to establish infection in vivo (Zoulim et al., 1994). Infection of primary human hepatocyte and differentiated HepaRG cultures with HBV virions containing genomes-deficient in HBx expression resulted in equal amounts of cccDNA formation as in cells infected with HBV virions containing wild-type genomes. However, HBV RNA was only detectable in cells infected with wild-type HBV virions (Lucifora et al., 2011; Riviere et al., 2015). Interestingly, trans-complementation of HBx was able to rescue HBV mRNA transcription in cells infected with HBx-deficient virions, even weeks after initial infection (Lucifora et al., 2011). Further epigenetic analyses indicated that HBx was recruited to the cccDNA minichromosomes and modulated histone modifications, which promoted histone acetylation of cccDNA minichromosome and thus favored the establishment of a transcriptionally active chromatin structure (Belloni et al., 2009). In addition, HBx was also demonstrated to enhance cccDNA transcription by counteracting the activity of transcriptional repressor(s) that silence the virus transcription. For instance, it was reported recently that Spindlin1, a cellular Tudor-domain protein, was more efficiently recruited to HBV cccDNA in the absence of HBx and suppressed the transcription activity of cccDNA. Silencing of Spindlin1 expression restored cccDNA transcription in the absence of HBx expression, which correlated with increased H3K4me3 at cccDNA minichromosomes. On the contrary, expression of HBx reduced Spindlin1 recruitment to cccDNA and activated its transcription activity (Ducroux et al., 2014). Moreover, a recent study also showed that HBx protein helped overcome SETDB1-mediated H3K9me3 and HP1 induced condensation of cccDNA minichromosomes to activate cccDNA transcription (Riviere et al., 2015).
Several lines of evidence from independent research groups have suggested that HBV capsid protein (HBcAg) is also associated with cccDNA minichromosomes (Bock et al., 2001; Guo et al., 2011; Lucifora et al., 2014). Interestingly, it was reported that the primary duck hepatocytes infected with DHBV containing genomes deficient in capsid protein expression accumulated similar amounts of cccDNA, but significantly less amounts of viral mRNA, compared to the cells infected with wild-type DHBV in the presence of foscarnet to block viral DNA replication and intracellular amplification of cccDNA (Schultz et al., 1999). This finding suggests that DHBV capsid protein may regulate cccDNA transcription and/or viral RNA turnover. However, we found that HepG2 cells transfected with either wild-type or capsid-deficient unit-length HBV DNA accumulated similar amounts of HBV mRNA, indicating that capsid protein did not regulate HBV transcription or viral mRNA turnover under our experimental conditions (Zhang et al., 2014b). Thus, the possible role of HBV capsid protein in cccDNA metabolism and function awaits further investigation in more biologically relevant systems.
HBV cccDNA contains three major CpG islands that can potentially be methylated (Zhang et al., 2013). Accumulating evidence suggests that variable degrees of CpG methylation were present in cccDNA from patients with chronic HBV infection. Old age and low viral loads were significantly associated with elevated cccDNA methylation (Kim et al., 2011). Our recent profiling of cccDNA methylation in chronic HBV infected patients revealed that while CpG island I was rarely methylated, methylation of CpG islands II and III was associated with low viral loads. Moreover, cell-based assays also demonstrated that CpG island II methylation significantly reduced the transcriptional activity of cccDNA (Zhang et al., 2014b).
Obviously, the minichromosomal structure and epigenetic modifications critically regulate the maintenance and transcriptional activity of cccDNA. Further understanding the epigenetic regulation and its relationship with cccDNA stability and function may reveal molecular targets for development of therapeutic agents to destabilize cccDNA or specifically silence its transcription.
5. Metabolism of cccDNA and mechanism of the cure of infected hepatocytes
Unlike the covalently closed circular form of nuclear episomal DNA from other DNA viruses, such as polyomaviruses, papillomaviruses and γ-herpesviruses, that can semiconservatively replicate in the nuclei of host cells, HBV cccDNA cannot replicate itself, but can only be made from cytoplasmic capsid DNA (Wu et al., 1990). Maintenance of cccDNA in a persistently infected hepatocyte thus relies on the intricate stability of cccDNA minichromosmes and/or the efficient replenishment of cccDNA through intracellular amplification pathway. If the turnover of cccDNA molecules does occur in infected hepatocytes, complete inhibition of viral DNA replication with antiviral agents inhibiting any step of viral DNA replication should ultimately deplete the precursors of cccDNA and consequentially, eradicate cccDNA. Accordingly, answers to the following three related questions are critical to understand the metabolism of cccDNA and mechanism of the cure of infected hepatocytes: (i) How stable are cccDNA minichromosomes in non-dividing cells? (ii) Could cccDNA molecules survive cell division? (iii) How are cccDNA molecules cleared during the resolution of HBV infections?
5.1 Longevity of cccDNA
Many attempts have been made to determine the half-life of HBV and other hepadnavirus cccDNA under various conditions. It has been shown that WHV cccDNA is stably maintained for months in non-dividing primary woodchuck hepatocytes (Moraleda et al., 1997). HBV cccDNA in confluent HepG2-derived cell lines, such as HepAD38 and HepDES19 cells, is also apparently very stable (Cai et al., 2012; Zhou et al., 2006). These observations corroborate the long half-life of hepadnavirus cccDNA, observed in HBV-infected chimpanzees (Wieland et al., 2004), WHV-infected woodchucks (Zhu et al., 2001) and DHBV-infected ducks (Addison et al., 2002) receiving viral polymerase inhibitor therapies, which were 9-14, 33-50 and 35-57 days, respectively. Nevertheless, in all those experimental conditions, it is difficult to conclude whether the observed cccDNA turnover is due to the decay of cccDNA in living cells, loss and/or dilution of cccDNA during cell division, or death of infected cells.
5.2 Fate of cccDNA during cell division
HBV is not a cytopathic virus, and infected cells continue to grow and divide (Chisari and Ferrari, 1995). Even though hepatocytes may divide only once in half a year in the normal liver (MacDonald, 1961; Magami et al., 2002), the proliferation rate is greatly enhanced during viral infection, due to inflammation and regeneration (Guo et al., 2000). cccDNA minichromosomes do not have centromeres and therefore can be lost during cell division (Calos, 1998). However, work reported by others and us has suggested that DHBV cccDNA could be passed into daughter cells (Guo et al., 2003; Lutgehetmann et al., 2010; Reaiche-Miller et al., 2013). Therefore, the open questions are: how cccDNA molecules survive mitosis and whether cccDNA are randomly or equally distributed into daughter cells.
The same questions apply to the nuclear episomal DNA of other viruses. In fact, papillomaviruses and γ-herpesviruses do establish latent infections as episomes in the nuclei of dividing cells. These viruses attach their episomal genomes to mitotic cellular chromosomes non-covalently via a viral protein for effective preservation and partitioning during cell division (Ballestas et al., 1999; Hung et al., 2001; Lehman and Botchan, 1998; You et al., 2004; You et al., 2006). So far, it is not known if hepadnavirus cccDNA is distributed into progeny cells via a similar mechanism, but obviously, disruption of cccDNA partitioning into daughter cells should be an effective way to accelerate the decay of cccDNA.
5.3 Non-cytolytic cure of HBV-infected hepatocytes
In regard to the mechanism of resolution of acute HBV infection, one possibility is that all the infected hepatocytes are killed during the recovery phase and replaced by the proliferation of uninfected hepatocytes. However, studies in WHV-infected woodchucks, DHBV-infected ducks and HBV-infected chimpanzees demonstrated that at the peak of transient infection, almost 100% of hepatocytes were infected (Guidotti et al., 1999; Jilbert et al., 1992; Kajino et al., 1994). Despite the fact that killing of hepatocytes does occur in the recovery phase, the total amount of hepatocyte death and turnover are significantly less than required for resolution of the infection under the condition that cccDNA are partitioned into daughter cells during cell division and every cccDNA-containing hepatocytes are killed (Guidotti et al., 1999; Wieland et al., 2004). Instead, experimental data support the hypothesis that during the clearing phase of HBV infection, host immune responses non-cytopathically inhibit viral replication and greatly shorten the half-life of cccDNA, thereby limiting the extent to which cytopathic T-cell effector functions and tissue destruction are required to terminate the infection (Murray et al., 2005).
Intriguingly, using integrated viral DNA as a genetic marker of virally infected hepatocytes, Summers and colleagues demonstrated that the integrated viral DNA persist in liver tissues from recovered animals at essentially undiminished levels of 1 viral genome per 1,000-3,000 liver cells (Summers et al., 2003). This finding unambiguously supports the notion that the hepatocytes in the recovered liver were derived primarily from the infected cell population and non-cytolytic eradication of cccDNA from infected hepatocytes does occur during the resolution of HBV infection. One of the possible mechanisms to eradicate cccDNA from hepatocytes is that cytokine-induced antiviral responses either lead to the degradation of cccDNA or accelerate the loss of cccDNA during cell division. The latter hypothesis predicts that the clearance of cccDNA requires infected hepatocytes to divide at least once during the recovery phase.
Interestingly, it was reported recently that the treatment of HBV-infected human primary hepatocytes or HepaRG cells with IFN-α or a lymphotoxin-β receptor (LTβR) agonist induced the expression of cytidine deaminase APOBEC3A and 3B, which were subsequently recruited to cccDNA minichromosomes by HBV core protein to edit cccDNA on the plus strand. Deaminated cccDNA was subjected to DNA repair and was ultimately degraded via a mechanism involving apurinic/apyrimidinic (AP) endonuclease (APE-1), (Lucifora et al., 2014). It is thus of interest to know whether IFN-α induces cccDNA decay via a similar mechanism in human hepatocytes, in vivo, and why IFN-α therapy only cures a small fraction of treated patients (Chisari et al., 2014; Shlomai and Rice, 2014).
Taken together, it appears that cccDNA is stable, but can be degradated by innate and adaptive immune response-induced intracellular antiviral programs in stationary cells. Like many other viral episomal DNAs, cccDNA can also be partitioned into daughter cells during cell division. Theoretically, cccDNA-free hepatocytes can be produced by single or multiple rounds of cell division due to unequal distribution and/or dilution of cccDNA. Nevertheless, our recent studies indicate that IFN-α does not significantly accelerate DHBV cccDNA loss during cell division (F. Liu and J.-T. Guo, unpublished observations).
6. Therapeutic elimination or silencing of cccDNA
6.1 Sequence-specific cleavage of cccDNA
Selective elimination of cccDNA relies on targeting its unique molecular features. Obviously, the most distinct molecular feature of cccDNA that differentiates it from host cellular chromatin is its unique DNA sequence. Therefore, several sequence-specific DNA targeting technologies, such as zinc finger proteins (Hoeksema and Tyrrell, 2010; Weber et al., 2014; Zimmerman et al., 2008), transcription activator-like effector nuclease (TALENs) (Bloom et al., 2013; Chen et al., 2014), and the clustered regularly interspaced short palindromic repeats (CRISPR)/CAS9 (Dong et al., 2015; Lin et al., 2014; Liu et al., 2015; Ramanan et al., 2015; Ren et al., 2015; Seeger and Sohn, 2014), have been evaluated in cultured cells and in mouse models. These approaches have demonstrated specific cleavage of cccDNA or inhibition of cccDNA transcription.
It is particularly interesting that CRISPR/CAS9 with different HBV-specific guide RNAs can be efficiently recruited to cccDNA minichromosomes, formed in HBV-infected, NTCP-expressing HepG2 cells, to induce mutations and deletions in cccDNA similar to those observed with chromosomal DNA cleaved by Cas9 and repaired by NHEJ pathway (Seeger and Sohn, 2014). However, efficient delivery of these gene-editing molecules into all HBV-infected cells in vivo is a major challenge for their clinical application.
6.2 Potential therapeutic targets for elimination or silencing of cccDNA
Recent findings about the immunological control of cccDNA by interferons and lymphotoxin raise exciting possibilities to target cccDNA via pharmacological activation or augmentation of the host intrinsic antiviral pathways (Liu et al., 2013; Lucifora et al., 2014). In addition, investigation of the mechanisms of HBx and core protein in cccDNA metabolism and function may reveal viral-host interactions for selective elimination or silencing of cccDNA (Lucifora et al., 2011; Riviere et al., 2015).
It is generally claimed that failure to eliminate cccDNA underlines the inability of current therapeutics to cure HBV infection, but the persistence of cccDNA can occasionally be detected in the livers of individuals with resolved HBV infections, and HBV replication can be reactivated in these individuals while suffering from systemic immune suppression or receiving anti-cancer chemotherapy (Pattullo, 2015; Rehermann et al., 1996). These observations indicate that the restoration of a functional antiviral immune response against HBV, but not necessarily the sterilized clearance of HBV cccDNA, might be more important for achieving a functional cure for HBV infection. Investigation to understand how immune system purges cccDNA and keeps the residual HBV (cccDNA) under surveillance will shed light into the development of immunotherapeutics to durably control HBV infection. In particular, reactivation of occult or immune-controlled HBV infection by anti-TNF-α therapy suggests that the cytokine plays a critical role in immune surveillance of HBV infection (Cassano et al., 2011).
It will be interesting to know if the immune system controls residual HBV replication by suppressing cccDNA transcription or via posttranscriptional mechanisms.
As we learned from studying the mechanism of IFN-α suppression of DHBV cccDNA transcription in cultured cells, IFN-α or possibly other cytokines may induce epigenetic remodeling of cccDNA minichromosome to induce a prolonged suppression of its transcription (Liu et al., 2013). It can thus be speculated that the immune surveillance of residual HBV replication may work through cytokine-induced transcriptional silencing of cccDNA function. Pharmacological activation of such intracellular responses in virus-infected cells might be able to durably control HBV replication.
6.3 Discovery of cccDNA-targeting antivirals through phenotypic screening
Despite being a reasonable approach, screening for anti-cccDNA agents has not been vigorously conducted, due to the lack of efficient in vitro infection models and a practical approach to measure cccDNA in medium- to high-throughput formats. Although there are primary human hepatocytes (PHH), HepaRG cells, and the recently established HepG2-NTCP cell culture systems available to support cccDNA-dependent HBV replication, the efficiency of HBV infection in vitro remains low, even with large virus inocula (Gripon et al., 2002; Iwamoto et al., 2014; Ni et al., 2014; Schulze-Bergkamen et al., 2003; Seeger and Sohn, 2014; Yan et al., 2014). In addition, there is a high degree of variability of HBV infectivity in primary hepatocytes from donor to donor, with some batches proving to be uninfectable, and the HepaRG system requires much manipulation for experimental setup (Gripon et al., 2002). These characteristics restrict their utility for screening compound libraries.
Alternatively, cccDNA formation can be achieved through the intracellular amplification pathway in stably-transfected HBV cell cultures that constitutively or conditionally replicate HBV genome (Chou et al., 2005; Guo et al., 2007; Ladner et al., 1997; Sells et al., 1988), as represented by HepG2.2.15 line (Schmidt and Korba, 2000; Sells et al., 1987). However, the direct detection of cccDNA from HBV cell lines by either Southern blot hybridization or real-time PCR assays would not be amenable to high throughput screening due to their limited sensitivity and specificity, respectively. On the other hand, there is no suitable surrogate marker for cccDNA in HepG2.2.15 cells since the majority of viral products are derived from integrated viral transgene, which is indistinguishable from cccDNA contributions.
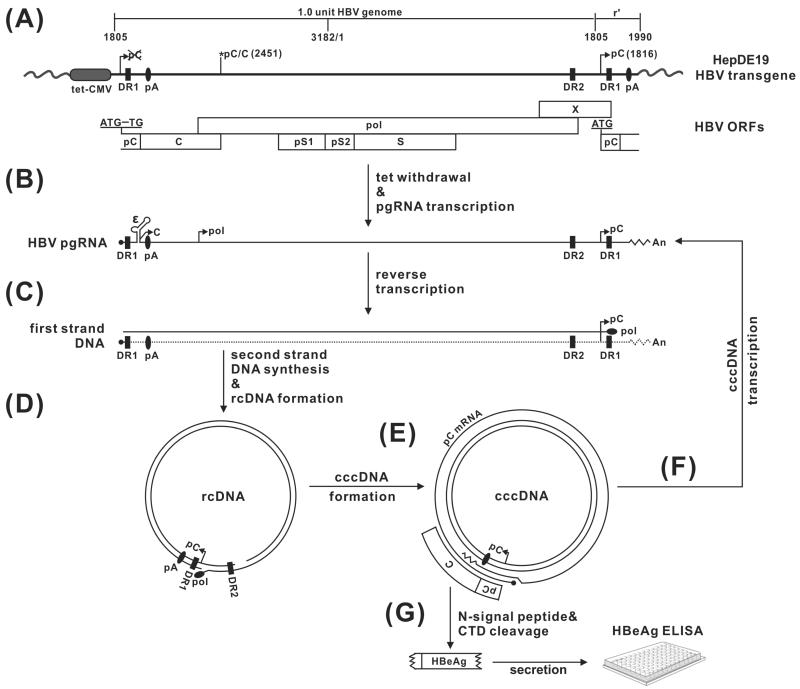
To surmount this obstacle, we previously established a HepDE19 stable cell line that expresses HBeAg in a cccDNA-dependent manner (Fig. 2). HBeAg is a secreted viral protein dispensable for HBV DNA replication and cccDNA formation. The strategy is based on the principle that the HBV cccDNA can be made from a pgRNA transcript alone, along with its translation products (core and pol) via the intracellular amplification pathway. The ORF of HBeAg is divided between the two termini of pgRNA and is only reconstituted when the pgRNA is reverse transcribed and circularized into cccDNA. HepDE19 cells contain an integrated HBV genome spanning the entire viral pgRNA and precore mRNA region under the control of a tetracycline-regulated (tet-off) CMV-IE promoter. A point mutation of the precore start codon at the 5’ end of the transgene was introduced to prevent expression of precore from the transgene. Upon induction, the transcribed pgRNA expresses viral core protein and polymerase and initiates reverse transcription of pgRNA to generate rcDNA, resulting in cccDNA formation. Meanwhile, the start codon of the precore open reading frame (ORF) at the 3’ end of pgRNA is copied into a viral DNA sequence, and the ORF of precore is restored during rcDNA conversion into cccDNA. Thus, authentic precore mRNA can be transcribed only from cccDNA. The precore protein product is further processed and secreted as HBeAg, which serves as a surrogate marker for cccDNA and can be conveniently detected by ELISA. HepDE19 cell line provided the first cccDNA reporter system for screening inhibitors targeting HBV cccDNA establishment or maintenance. By making use of the HepDE19 system, we previously screened a small-molecule compound library and identified two disubstituted sulfonamide (DSS) compounds that act as inhibitors of cccDNA formation by blocking rcDNA deproteinization. DSS compounds serve as proof-of-concept drug candidates for development into therapeutics, to eliminate cccDNA from chronically HBV infected patients. (Cai et al., 2012).
Fig 2. HBV nucleic acid metabolism in HepDE19 cells, with the rationale for design of a cccDNA-dependent HBeAg assay.
(A) The transgene in HepDE19 cells contains a 1.1 overlength HBV genome (genotype D, subtype ayw), starting from nt 1805, under the control of a tet-CMV promoter (tet-off), in which the start codon (AUG) of the precore (pC) ORF has been mutated to UG at the 5’ end of HBV DNA, with the second one at the 3’ redundancy (r’) left unchanged. C, pol, pS1, pS2, S and X represent ORF for core, polymerase, preS1, preS2 and S domain of HBV surface antigen, and X protein, respectively. DR represents the identical direct repeat sequences 1 and 2. pA is polyadenylation site. (B) Upon tetracycline withdrawal, HBV pgRNA is transcribed and the viral core and polymerase are produced, resulting in pgRNA encapsidation, (C) reverse transcription of pgRNA to minus-strand DNA, and (D) sequential plus-strand DNA synthesis and circularization into rcDNA. (E) rcDNA is converted to the cccDNA template, in which the precore ORF is restored, giving rise to authentic precore mRNA, and (F) pgRNA. (G) cccDNA-derived precore mRNA is translated into the precore protein, which is further processed into secreted HBeAg through proteolytic cleavage of the N-terminal signal peptide and C-terminal domain (CTD). The secreted HBeAg in the supernatant of HepDE19 cells can be detected by ELISA assay and serves as a surrogate marker for intracellular cccDNA.
However, the existing assay is not ideal for high-throughput screening, because the HBeAg ELISA cross-reacts with an HBeAg homologue, the core antigen (HBcAg). HBcAg is an abundant, secreted protein, produced largely from the transgene template in HepDE19 cells (Guo et al., 2007). The HBeAg shares 149 amino acid (aa) sequence homology with HBcAg (Block et al., 2007), and a highly specific HBeAb or HBeAg ELISA is not available, resulting in a low HBeAg signal-to-noise ratio (tet- vs. tet-/3TC) in HepDE19 cell-based assay. To further improve the specificity and sensitivity of the assay, we have embarked upon generating the “second-generation” cccDNA reporter cell systems by incorporating an epitope tag in the precore region or replacing the precore ORF with a luminescent or colorimetric reporter gene. We have recently established a new HBV cell line expressing human influenza hemagglutinin (HA) epitope-tagged HBeAg in cccDNA-dependent manner. The HA-HBeAg ELISA assay with HA antibody serving as the capture antibody and anti-HBe antibody serving as detection antibody, completely eliminated the contaminating signal from HBcAg (D. Cai and H. Guo, unpublished data). The novel reporter systems provide powerful tools for phenotypic screening of cccDNA-targeting antiviral substances, as well as host factors that regulate cccDNA metabolism and function.
7. Conclusions and perspectives
Our knowledge of cccDNA biosynthesis, transcription regulation and turnover in HBV-infected hepatocytes is incomplete, and we have no therapeutic means to reliably control cccDNA. Future investigation towards
-
(i)
understanding the molecular pathway of cccDNA formation,
-
(ii)
knowing how cccDNA survives cell division,
-
(iii)
revealing the mechanism of immunologic elimination and silencing of cccDNA and
-
(iv)
understanding how HBx and core protein control cccDNA transcription
should not only advance our knowledge of hepatitis B pathobiology, but also provide clues or validate targets for the development of therapeutics to eradicate or suppress cccDNA. Further development and optimization of cccDNA-reporter cells and NTCP-based HBV infection systems should greatly improve our ability to investigate cccDNA metabolism and functional regulation, as well as the discovery of drug candidates for durable control of HBV infection.
Highlights.
Hepatitis B virus (HBV) persistence relies on the stable maintenance and proper functioning of cccDNA.
cccDNA is synthesized from viral genomic DNA of the infecting virion or from the progeny mature nucleocapsid.
Copy numbers of cccDNA in the nuclei of infected hepatocytes are controlled by both viral and host cellular factors.
Eradication or transcriptional silencing of cccDNA is essential for a cure of HBV infection.
High-throughput cccDNA quantification and functional assays are needed for discovery of cccDNA-targeting antivirals.
Acknowledgements
We thank Dr. Siddhartha Rawat for critical reading of the manuscript. Ju-Tao Guo is supported by NIH grant (R01AI113267) and the Hepatitis B Foundation, through an appropriation from the Commonwealth of Pennsylvania. Haitao Guo is supported by NIH grant (R01AI094474 and R21AI103838) and the Trustees of Indiana University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Addison WR, Walters KA, Wong WW, Wilson JS, Madej D, Jewell LD, Tyrrell DL. Half-life of the duck hepatitis B virus covalently closed circular DNA pool in vivo following inhibition of viral replication. Journal of virology. 2002;76:6356–6363. doi: 10.1128/JVI.76.12.6356-6363.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alain T, Kim TS, Lun X, Liacini A, Schiff LA, Senger DL, Forsyth PA. Proteolytic disassembly is a critical determinant for reovirus oncolysis. Mol Ther. 2007;15:1512–1521. doi: 10.1038/sj.mt.6300207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballestas ME, Chatis PA, Kaye KM. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- Bartenschlager R, Schaller H. The amino-terminal domain of the hepadnaviral P-gene encodes the terminal protein (genome-linked protein) believed to prime reverse transcription. Embo J. 1988;7:4185–4192. doi: 10.1002/j.1460-2075.1988.tb03315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belloni L, Pollicino T, De Nicola F, Guerrieri F, Raffa G, Fanciulli M, Raimondo G, Levrero M. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19975–19979. doi: 10.1073/pnas.0908365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bill CA, Summers J. Genomic DNA double-strand breaks are targets for hepadnaviral DNA integration. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11135–11140. doi: 10.1073/pnas.0403925101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block TM, Gish R, Guo H, Mehta A, Cuconati A, Thomas London W, Guo JT. Chronic hepatitis B: what should be the goal for new therapies? Antiviral research. 2013;98:27–34. doi: 10.1016/j.antiviral.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block TM, Guo H, Guo JT. Molecular virology of hepatitis B virus for clinicians. Clinics in liver disease. 2007;11:685–706. vii. doi: 10.1016/j.cld.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom K, Ely A, Mussolino C, Cathomen T, Arbuthnot P. Inactivation of hepatitis B virus replication in cultured cells and in vivo with engineered transcription activator-like effector nucleases. Mol Ther. 2013;21:1889–1897. doi: 10.1038/mt.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock CT, Schranz P, Schroder CH, Zentgraf H. Hepatitis B virus genome is organized into nucleosomes in the nucleus of the infected cell. Virus Genes. 1994;8:215–229. doi: 10.1007/BF01703079. [DOI] [PubMed] [Google Scholar]
- Bock CT, Schwinn S, Locarnini S, Fyfe J, Manns MP, Trautwein C, Zentgraf H. Structural organization of the hepatitis B virus minichromosome. Journal of molecular biology. 2001;307:183–196. doi: 10.1006/jmbi.2000.4481. [DOI] [PubMed] [Google Scholar]
- Bowden S, Locarnini S, Chang TT, Chao YC, Han KH, Gish RG, de Man RA, Yu M, Llamoso C, Tang H. Covalently closed-circular hepatitis B virus DNA reduction with entecavir or lamivudine. World journal of gastroenterology : WJG. 2015;21:4644–4651. doi: 10.3748/wjg.v21.i15.4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Mills C, Yu W, Yan R, Aldrich CE, Saputelli JR, Mason WS, Xu X, Guo JT, Block TM, Cuconati A, Guo H. Identification of disubstituted sulfonamide compounds as specific inhibitors of hepatitis B virus covalently closed circular DNA formation. Antimicrobial agents and chemotherapy. 2012;56:4277–4288. doi: 10.1128/AAC.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calos MP. Stability without a centromere. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:4084–4085. doi: 10.1073/pnas.95.8.4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassano N, Mastrandrea V, Principi M, Loconsole F, De Tullio N, Di Leo A, Vena GA. Anti-tumor necrosis factor treatment in occult hepatitis B virus infection: a retrospective analysis of 62 patients with psoriatic disease. J Biol Regul Homeost Agents. 2011;25:285–289. [PubMed] [Google Scholar]
- Chen J, Zhang W, Lin J, Wang F, Wu M, Chen C, Zheng Y, Peng X, Li J, Yuan Z. An efficient antiviral strategy for targeting hepatitis B virus genome using transcription activator-like effector nucleases. Mol Ther. 2014;22:303–311. doi: 10.1038/mt.2013.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisari FV, Ferrari C. Hepatitis B virus immunopathology. Springer seminars in immunopathology. 1995;17:261–281. doi: 10.1007/BF00196169. [DOI] [PubMed] [Google Scholar]
- Chisari FV, Mason WS, Seeger C. Virology. Comment on “Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA”. Science. 2014;344:1237. doi: 10.1126/science.1254082. [DOI] [PubMed] [Google Scholar]
- Chou YC, Jeng KS, Chen ML, Liu HH, Liu TL, Chen YL, Liu YC, Hu CP, Chang C. Evaluation of transcriptional efficiency of hepatitis B virus covalently closed circular DNA by reverse transcription-PCR combined with the restriction enzyme digestion method. Journal of virology. 2005;79:1813–1823. doi: 10.1128/JVI.79.3.1813-1823.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, Caldecott KW. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461:674–678. doi: 10.1038/nature08444. [DOI] [PubMed] [Google Scholar]
- Cui X, Guo JT, Hu J. Hepatitis B Virus Covalently Closed Circular DNA Formation in Immortalized Mouse Hepatocytes Associated with Nucleocapsid Destabilization. Journal of virology. 2015a doi: 10.1128/JVI.01261-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Ludgate L, Ning X, Hu J. Maturation-associated destabilization of hepatitis B virus nucleocapsid. Journal of virology. 2013;87:11494–11503. doi: 10.1128/JVI.01912-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, McAllister R, Boregowda R, Sohn JA, Ledesma FC, Caldecott KW, Seeger C, Hu J. Does Tyrosyl DNA Phosphodiesterase-2 Play a Role in Hepatitis B Virus Genome Repair? PloS one. 2015b;10:e0128401. doi: 10.1371/journal.pone.0128401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandri M, Burda MR, Will H, Petersen J. Increased hepatocyte turnover and inhibition of woodchuck hepatitis B virus replication by adefovir in vitro do not lead to reduction of the closed circular DNA. Hepatology. 2000;32:139–146. doi: 10.1053/jhep.2000.8701. [DOI] [PubMed] [Google Scholar]
- Day PM, Lowy DR, Schiller JT. Papillomaviruses infect cells via a clathrin-dependent pathway. Virology. 2003;307:1–11. doi: 10.1016/s0042-6822(02)00143-5. [DOI] [PubMed] [Google Scholar]
- Dong C, Qu L, Wang H, Wei L, Dong Y, Xiong S. Targeting hepatitis B virus cccDNA by CRISPR/Cas9 nuclease efficiently inhibits viral replication. Antiviral research. 2015;118:110–117. doi: 10.1016/j.antiviral.2015.03.015. [DOI] [PubMed] [Google Scholar]
- Ducroux A, Benhenda S, Riviere L, Semmes OJ, Benkirane M, Neuveut C. The Tudor domain protein Spindlin1 is involved in intrinsic antiviral defense against incoming hepatitis B Virus and herpes simplex virus type 1. PLoS pathogens. 2014;10:e1004343. doi: 10.1371/journal.ppat.1004343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, Yeager M, Sundquist WI. The structural biology of HIV assembly. Curr Opin Struct Biol. 2008;18:203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Hu J. Formation of Hepatitis B Virus Covalently Closed Circular DNA: Removal of Genome-linked Protein. Journal of virology. 2007 doi: 10.1128/JVI.02721-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich WH, Robinson WS. Hepatitis B virus contains protein attached to the 5′ terminus of its complete DNA strand. Cell. 1980;21:801–809. doi: 10.1016/0092-8674(80)90443-2. [DOI] [PubMed] [Google Scholar]
- Gripon P, Rumin S, Urban S, Le Seyec J, Glaise D, Cannie I, Guyomard C, Lucas J, Trepo C, Guguen-Guillouzo C. Infection of a human hepatoma cell line by hepatitis B virus. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15655–15660. doi: 10.1073/pnas.232137699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti LG, Matzke B, Schaller H, Chisari FV. High-level hepatitis B virus replication in transgenic mice. Journal of virology. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti LG, Rochford R, Chung J, Shapiro M, Purcell R, Chisari FV. Viral clearance without destruction of infected cells during acute HBV infection [In Process Citation]. Science. 1999;284:825–829. doi: 10.1126/science.284.5415.825. [DOI] [PubMed] [Google Scholar]
- Guo H, Jiang D, Zhou T, Cuconati A, Block TM, Guo JT. Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation. Journal of virology. 2007;81:12472–12484. doi: 10.1128/JVI.01123-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Mao R, Block TM, Guo JT. Production and function of the cytoplasmic deproteinized relaxed circular DNA of hepadnaviruses. Journal of virology. 2010;84:387–396. doi: 10.1128/JVI.01921-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Xu C, Zhou T, Block TM, Guo JT. Characterization of the Host Factors Required for Hepadnavirus Covalently Closed Circular (ccc) DNA Formation. PloS one. 2012;7:e43270. doi: 10.1371/journal.pone.0043270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J-T, Zhou H, Liu C, Aldrich C, Saputelli J, Whitaker T, Barrasa MI, Mason WS, Seeger C. Apoptosis and regeneration of hepatocytes during recovery from transient hepadnavirus infection. J. Virology. 2000;74:1495–1505. doi: 10.1128/jvi.74.3.1495-1505.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JT, Pryce M, Wang X, Barrasa MI, Hu J, Seeger C. Conditional replication of duck hepatitis B virus in hepatoma cells. Journal of virology. 2003;77:1885–1893. doi: 10.1128/JVI.77.3.1885-1893.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YH, Li YN, Zhao JR, Zhang J, Yan Z. HBc binds to the CpG islands of HBV cccDNA and promotes an epigenetic permissive state. Epigenetics : official journal of the DNA Methylation Society. 2011;6:720–726. doi: 10.4161/epi.6.6.15815. [DOI] [PubMed] [Google Scholar]
- He W, Ren B, Mao F, Jing Z, Li Y, Liu Y, Peng B, Yan H, Qi Y, Sun Y, Guo JT, Sui J, Wang F, Li W. Hepatitis D Virus Infection of Mice Expressing Human Sodium Taurocholate Co-transporting Polypeptide. PLoS pathogens. 2015;11:e1004840. doi: 10.1371/journal.ppat.1004840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeksema KA, Tyrrell DL. Inhibition of viral transcription using designed zinc finger proteins. Methods in molecular biology. 2010;649:97–116. doi: 10.1007/978-1-60761-753-2_6. [DOI] [PubMed] [Google Scholar]
- Hung SC, Kang MS, Kieff E. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:1865–1870. doi: 10.1073/pnas.031584698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M, Watashi K, Tsukuda S, Aly HH, Fukasawa M, Fujimoto A, Suzuki R, Aizaki H, Ito T, Koiwai O, Kusuhara H, Wakita T. Evaluation and identification of hepatitis B virus entry inhibitors using HepG2 cells overexpressing a membrane transporter NTCP. Biochemical and biophysical research communications. 2014;443:808–813. doi: 10.1016/j.bbrc.2013.12.052. [DOI] [PubMed] [Google Scholar]
- Janssen HL, van Zonneveld M, Senturk H, Zeuzem S, Akarca US, Cakaloglu Y, Simon C, So TM, Gerken G, de Man RA, Niesters HG, Zondervan P, Hansen B, Schalm SW. Pegylated interferon alfa-2b alone or in combination with lamivudine for HBeAg-positive chronic hepatitis B: a randomised trial. Lancet. 2005;365:123–129. doi: 10.1016/S0140-6736(05)17701-0. [DOI] [PubMed] [Google Scholar]
- Jilbert AR, Wu TT, England JM, Hall PM, Carp NZ, O'Connell AP, Mason WS. Rapid resolution of duck hepatitis B virus infections occurs after massive hepatocellular involvement. Journal of virology. 1992;66:1377–1388. doi: 10.1128/jvi.66.3.1377-1388.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajino K, Jilbert AR, Saputelli J, Aldrich CE, Cullen J, Mason WS. Woodchuck hepatitis virus infections: very rapid recovery after a prolonged viremia and infection of virtually every hepatocyte. Journal of virology. 1994;68:5792–5803. doi: 10.1128/jvi.68.9.5792-5803.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kann M, Sodeik B, Vlachou A, Gerlich WH, Helenius A. Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J Cell Biol. 1999;145:45–55. doi: 10.1083/jcb.145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeffe EB, Dieterich DT, Han SH, Jacobson IM, Martin P, Schiff ER, Tobias H. A treatment algorithm for the management of chronic hepatitis B virus infection in the United States: 2008 update. Clin Gastroenterol Hepatol. 2008;6:1315–1341. doi: 10.1016/j.cgh.2008.08.021. quiz 1286. [DOI] [PubMed] [Google Scholar]
- Kim JW, Lee SH, Park YS, Hwang JH, Jeong SH, Kim N, Lee DH. Replicative activity of hepatitis B virus is negatively associated with methylation of covalently closed circular DNA in advanced hepatitis B virus infection. Intervirology. 2011;54:316–325. doi: 10.1159/000321450. [DOI] [PubMed] [Google Scholar]
- Koniger C, Wingert I, Marsmann M, Rosler C, Beck J, Nassal M. Involvement of the host DNA-repair enzyme TDP2 in formation of the covalently closed circular DNA persistence reservoir of hepatitis B viruses. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E4244–4253. doi: 10.1073/pnas.1409986111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner SK, Otto MJ, Barker CS, Zaifert K, Wang GH, Guo JT, Seeger C, King RW. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrobial agents and chemotherapy. 1997;41:1715–1720. doi: 10.1128/aac.41.8.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laras A, Koskinas J, Dimou E, Kostamena A, Hadziyannis SJ. Intrahepatic levels and replicative activity of covalently closed circular hepatitis B virus DNA in chronically infected patients. Hepatology. 2006;44:694–702. doi: 10.1002/hep.21299. [DOI] [PubMed] [Google Scholar]
- Lau GK, Piratvisuth T, Luo KX, Marcellin P, Thongsawat S, Cooksley G, Gane E, Fried MW, Chow WC, Paik SW, Chang WY, Berg T, Flisiak R, McCloud P, Pluck N. Peginterferon Alfa-2a, lamivudine, and the combination for HBeAg-positive chronic hepatitis B. N Engl J Med. 2005;352:2682–2695. doi: 10.1056/NEJMoa043470. [DOI] [PubMed] [Google Scholar]
- Le Sage V, Mouland AJ, Valiente-Echeverria F. Roles of HIV-1 capsid in viral replication and immune evasion. Virus research. 2014;193:116–129. doi: 10.1016/j.virusres.2014.07.010. [DOI] [PubMed] [Google Scholar]
- Lehman CW, Botchan MR. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:4338–4343. doi: 10.1073/pnas.95.8.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhoff RJ, Luscombe CA, Summers J. Acute liver injury following infection with a cytopathic strain of duck hepatitis B virus. Hepatology. 1999;29:563–571. doi: 10.1002/hep.510290236. [DOI] [PubMed] [Google Scholar]
- Lenhoff RJ, Summers J. Coordinate regulation of replication and virus assembly by the large envelope protein of an avian hepadnavirus. J.Virol. 1994;68:4565–4571. doi: 10.1128/jvi.68.7.4565-4571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz TB, Loeb DD. Roles of the envelope proteins in the amplification of covalently closed circular DNA and completion of synthesis of the plus-strand DNA in hepatitis B virus. Journal of virology. 2011;85:11916–11927. doi: 10.1128/JVI.05373-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levrero M, Pollicino T, Petersen J, Belloni L, Raimondo G, Dandri M. Control of cccDNA function in hepatitis B virus infection. Journal of hepatology. 2009;51:581–592. doi: 10.1016/j.jhep.2009.05.022. [DOI] [PubMed] [Google Scholar]
- Liaw YF. Impact of therapy on the outcome of chronic hepatitis B. Liver international : official journal of the International Association for the Study of the Liver. 2013;33(Suppl 1):111–115. doi: 10.1111/liv.12057. [DOI] [PubMed] [Google Scholar]
- Lien JM, Aldrich CE, Mason WS. Evidence that a capped oligoribonucleotide is the primer for duck hepatitis B virus plus-strand DNA synthesis. Journal of virology. 1986;57:229–236. doi: 10.1128/jvi.57.1.229-236.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SR, Yang HC, Kuo YT, Liu CJ, Yang TY, Sung KC, Lin YY, Wang HY, Wang CC, Shen YC, Wu FY, Kao JH, Chen DS, Chen PJ. The CRISPR/Cas9 System Facilitates Clearance of the Intrahepatic HBV Templates In Vivo. Molecular therapy. Nucleic acids. 2014;3:e186. doi: 10.1038/mtna.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Campagna M, Qi Y, Zhao X, Guo F, Xu C, Li S, Li W, Block TM, Chang J, Guo JT. Alpha-interferon suppresses hepadnavirus transcription by altering epigenetic modification of cccDNA minichromosomes. PLoS pathogens. 2013;9:e1003613. doi: 10.1371/journal.ppat.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Tian R, Loeb DD. Base pairing among three cis-acting sequences contributes to template switching during hepadnavirus reverse transcription. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:1984–1989. doi: 10.1073/pnas.0436218100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hao R, Chen S, Guo D, Chen Y. Inhibition of Hepatitis B Virus by CRISPR/Cas9 System via Targeting the Conserved Regions of Viral Genome. The Journal of general virology. 2015 doi: 10.1099/vir.0.000159. [DOI] [PubMed] [Google Scholar]
- Lucifora J, Arzberger S, Durantel D, Belloni L, Strubin M, Levrero M, Zoulim F, Hantz O, Protzer U. Hepatitis B virus X protein is essential to initiate and maintain virus replication after infection. Journal of hepatology. 2011;55:996–1003. doi: 10.1016/j.jhep.2011.02.015. [DOI] [PubMed] [Google Scholar]
- Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T, Remouchamps C, Chou WM, Thasler WE, Huser N, Durantel D, Liang TJ, Munk C, Heim MH, Browning JL, Dejardin E, Dandri M, Schindler M, Heikenwalder M, Protzer U. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 2014;343:1221–1228. doi: 10.1126/science.1243462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutgehetmann M, Volz T, Kopke A, Broja T, Tigges E, Lohse AW, Fuchs E, Murray JM, Petersen J, Dandri M. In vivo proliferation of hepadnavirus-infected hepatocytes induces loss of covalently closed circular DNA in mice. Hepatology. 2010;52:16–24. doi: 10.1002/hep.23611. [DOI] [PubMed] [Google Scholar]
- MacDonald RA. “Lifespan” of liver cells. Autoradio-graphic study using tritiated thymidine in normal, cirrhotic, and partially hepatectomized rats. Arch. Intern. Med. 1961;107:335–343. doi: 10.1001/archinte.1961.03620030023003. [DOI] [PubMed] [Google Scholar]
- Magami Y, Azuma T, Inokuchi H, Kokuno S, Moriyasu F, Kawai K, Hattori T. Cell proliferation and renewal of normal hepatocytes and bile duct cells in adult mouse liver. Liver. 2002;22:419–425. doi: 10.1034/j.1600-0676.2002.01702.x. [DOI] [PubMed] [Google Scholar]
- Mason WS, Aldrich C, Summers J, Taylor JM. Asymmetric replication of duck hepatitis B virus DNA in liver cells: Free minus-strand DNA. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:3997–4001. doi: 10.1073/pnas.79.13.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettenleiter TC, Klupp BG, Granzow H. Herpesvirus assembly: An update. Virus research. 2009 doi: 10.1016/j.virusres.2009.03.018. [DOI] [PubMed] [Google Scholar]
- Molnar-Kimber KL, Summers J, Taylor JM, Mason WS. Protein covalently bound to minus-strand DNA intermediates of duck hepatitis B virus. Journal of virology. 1983;45:165–172. doi: 10.1128/jvi.45.1.165-172.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraleda G, Saputelli J, Aldrich CE, Averett D, Condreay L, Mason WS. Lack of effect of antiviral therapy in nondividing hepatocyte cultures on the closed circular DNA of woodchuck hepatitis virus. Journal of virology. 1997;71:9392–9399. doi: 10.1128/jvi.71.12.9392-9399.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JM, Wieland SF, Purcell RH, Chisari FV. Dynamics of hepatitis B virus clearance in chimpanzees. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17780–17785. doi: 10.1073/pnas.0508913102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015 doi: 10.1136/gutjnl-2015-309809. [DOI] [PubMed] [Google Scholar]
- Newbold JE, Xin H, Tencza M, Sherman G, Dean J, Bowden S, Locarnini S. The covalently closed duplex form of the hepadnavirus genome exists in situ as a heterogeneous population of viral minichromosomes. Journal of virology. 1995;69:3350–3357. doi: 10.1128/jvi.69.6.3350-3357.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Y, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, Falth M, Stindt J, Koniger C, Nassal M, Kubitz R, Sultmann H, Urban S. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology. 2014;146:1070–1083. doi: 10.1053/j.gastro.2013.12.024. [DOI] [PubMed] [Google Scholar]
- Ott JJ, Stevens GA, Groeger J, Wiersma ST. Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine. 2012;30:2212–2219. doi: 10.1016/j.vaccine.2011.12.116. [DOI] [PubMed] [Google Scholar]
- Pattullo V. Hepatitis B reactivation in the setting of chemotherapy and immunosuppression - prevention is better than cure. World journal of hepatology. 2015;7:954–967. doi: 10.4254/wjh.v7.i7.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrillo R. Benefits and risks of interferon therapy for hepatitis B. Hepatology. 2009;49:S103–111. doi: 10.1002/hep.22956. [DOI] [PubMed] [Google Scholar]
- Pollicino T, Belloni L, Raffa G, Pediconi N, Squadrito G, Raimondo G, Levrero M. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology. 2006;130:823–837. doi: 10.1053/j.gastro.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Pyzocha NK, Ran FA, Hsu PD, Zhang F. RNA-guided genome editing of mammalian cells. Methods in molecular biology. 2014;1114:269–277. doi: 10.1007/978-1-62703-761-7_17. [DOI] [PubMed] [Google Scholar]
- Rabe B, Vlachou A, Pante N, Helenius A, Kann M. Nuclear import of hepatitis B virus capsids and release of the viral genome. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9849–9854. doi: 10.1073/pnas.1730940100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan V, Shlomai A, Cox DB, Schwartz RE, Michailidis E, Bhatta A, Scott DA, Zhang F, Rice CM, Bhatia SN. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Scientific reports. 2015;5:10833. doi: 10.1038/srep10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raney AK, Eggers CM, Kline EF, Guidotti LG, Pontoglio M, Yaniv M, McLachlan A. Nuclear covalently closed circular viral genomic DNA in the liver of hepatocyte nuclear factor 1 alpha-null hepatitis B virus transgenic mice. Journal of virology. 2001;75:2900–2911. doi: 10.1128/JVI.75.6.2900-2911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasaiyaah J, Tan CP, Fletcher AJ, Price AJ, Blondeau C, Hilditch L, Jacques DA, Selwood DL, James LC, Noursadeghi M, Towers GJ. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature. 2013;503:402–405. doi: 10.1038/nature12769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaiche-Miller GY, Thorpe M, Low HC, Qiao Q, Scougall CA, Mason WS, Litwin S, Jilbert AR. Duck hepatitis B virus covalently closed circular DNA appears to survive hepatocyte mitosis in the growing liver. Virology. 2013;446:357–364. doi: 10.1016/j.virol.2013.08.014. [DOI] [PubMed] [Google Scholar]
- Rehermann B, Ferrari C, Pasquinelli C, Chisari FV. The hepatitis B virus persists for decades after patients' recovery from acute viral hepatitis despite active maintenance of a cytotoxic T- lymphocyte response. Nature medicine. 1996;2:1104–1108. doi: 10.1038/nm1096-1104. [DOI] [PubMed] [Google Scholar]
- Ren Q, Li C, Yuan P, Cai C, Zhang L, Luo GG, Wei W. A Dual-reporter system for real-time monitoring and high-throughput CRISPR/Cas9 library screening of the hepatitis C virus. Scientific reports. 2015;5:8865. doi: 10.1038/srep08865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere L, Gerossier L, Ducroux A, Dion S, Deng Q, Michel ML, Buendia MA, Hantz O, Neuveut C. HBx relieves chromatin-mediated transcriptional repression of hepatitis B viral cccDNA involving SETDB1 histone methyltransferase. Journal of hepatology. 2015 doi: 10.1016/j.jhep.2015.06.023. [DOI] [PubMed] [Google Scholar]
- Schmidt K, Korba B. Hepatitis B virus cell culture assays for antiviral activity. Methods Mol Med. 2000;24:51–67. doi: 10.1385/1-59259-245-7:51. [DOI] [PubMed] [Google Scholar]
- Schmitz A, Schwarz A, Foss M, Zhou L, Rabe B, Hoellenriegel J, Stoeber M, Pante N, Kann M. Nucleoporin 153 arrests the nuclear import of hepatitis B virus capsids in the nuclear basket. PLoS pathogens. 2010;6:e1000741. doi: 10.1371/journal.ppat.1000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz U, Summers J, Staeheli P, Chisari FV. Elimination of duck hepatitis B virus RNA-containing capsids in duck interferon-alpha-treated hepatocytes. Journal of virology. 1999;73:5459–5465. doi: 10.1128/jvi.73.7.5459-5465.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Bergkamen H, Untergasser A, Dax A, Vogel H, Buchler P, Klar E, Lehnert T, Friess H, Buchler MW, Kirschfink M, Stremmel W, Krammer PH, Muller M, Protzer U. Primary human hepatocytes--a valuable tool for investigation of apoptosis and hepatitis B virus infection. Journal of hepatology. 2003;38:736–744. doi: 10.1016/s0168-8278(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Seeger C, Mason WS. Molecular biology of hepatitis B virus infection. Virology. 2015;479-480C:672–686. doi: 10.1016/j.virol.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger C, Sohn JA. Targeting Hepatitis B Virus With CRISPR/Cas9. Molecular therapy. Nucleic acids. 2014;3:e216. doi: 10.1038/mtna.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sells MA, Chen M, Acs G. Production of hepatitis B virus particles in hepG2 cells transfected with cloned hepatitis B virus DNA. Proc. Natl. Acad. Sci. USA. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sells MA, Zelent AZ, Shvartsman M, Acs G. Replicative intermediates of hepatitis B virus in HepG2 cells that produce infectious virions. Journal of virology. 1988;62:2836–2844. doi: 10.1128/jvi.62.8.2836-2844.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Li S, Shen F, Li H, Qian S, Lee DH, Wu JZ, Yang W. Characterization of nucleosome positioning in hepadnaviral covalently closed circular DNA minichromosomes. Journal of virology. 2012;86:10059–10069. doi: 10.1128/JVI.00535-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomai A, Rice CM. Virology. Getting rid of a persistent troublemaker to cure hepatitis. Science. 2014;343:1212–1213. doi: 10.1126/science.1252186. [DOI] [PubMed] [Google Scholar]
- Smith JL, Campos SK, Wandinger-Ness A, Ozbun MA. Caveolin-1-dependent infectious entry of human papillomavirus type 31 in human keratinocytes proceeds to the endosomal pathway for pH-dependent uncoating. Journal of virology. 2008;82:9505–9512. doi: 10.1128/JVI.01014-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn JA, Litwin S, Seeger C. Mechanism for CCC DNA synthesis in hepadnaviruses. PloS one. 2009;4:e8093. doi: 10.1371/journal.pone.0008093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staprans S, Loeb DD, Ganem D. Mutations affecting hepadnavirus plus-strand DNA synthesis dissociate primer cleavage from translocation and reveal the origin of linear viral DNA. Journal of virology. 1991;65:1255–1262. doi: 10.1128/jvi.65.3.1255-1262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers J, Jilbert AR, Yang W, Aldrich CE, Saputelli J, Litwin S, Toll E, Mason WS. Hepatocyte turnover during resolution of a transient hepadnaviral infection. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:11652–11659. doi: 10.1073/pnas.1635109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers J, Mason WS. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- Summers J, O'Connell A, Millman I. Genome of hepatitis B virus: restriction enzyme cleavage and structure of DNA extracted from Dane particles. Proc. Natl. Acad. Sci. USA. 1975;72:4597–4601. doi: 10.1073/pnas.72.11.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers J, Smith PM, Horwich AL. Hepadnavirus envelope proteins regulate covalently closed circular DNA amplification. Journal of virology. 1990;64:2819–2824. doi: 10.1128/jvi.64.6.2819-2824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers J, Smith PM, Huang MJ, Yu MS. Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. Journal of virology. 1991;65:1310–1317. doi: 10.1128/jvi.65.3.1310-1317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung JJ, Wong ML, Bowden S, Liew CT, Hui AY, Wong VW, Leung NW, Locarnini S, Chan HL. Intrahepatic hepatitis B virus covalently closed circular DNA can be a predictor of sustained response to therapy. Gastroenterology. 2005;128:1890–1897. doi: 10.1053/j.gastro.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Tavis J, Ganem D. RNA sequences controlling the initiation of duck hepatits B virus minus-strand DNA. J. Virol. 1995;69:4283–4291. doi: 10.1128/jvi.69.7.4283-4291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavis J, Ganem D. Evidence for activation of the hepatitis B virus polymerase by binding of its RNA template. J. Virol. 1996;70:5741–5750. doi: 10.1128/jvi.70.9.5741-5750.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttleman JS, Pourcel C, Summers J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell. 1986;47:451–460. doi: 10.1016/0092-8674(86)90602-1. [DOI] [PubMed] [Google Scholar]
- Virgen-Slane R, Rozovics JM, Fitzgerald KD, Ngo T, Chou W, van der Heden van Noort GJ, Filippov DV, Gershon PD, Semler BL. An RNA virus hijacks an incognito function of a DNA repair enzyme. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:14634–14639. doi: 10.1073/pnas.1208096109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GH, Seeger C. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell. 1992;71:663–670. doi: 10.1016/0092-8674(92)90599-8. [DOI] [PubMed] [Google Scholar]
- Wang GH, Seeger C. Novel mechanism for reverse transcription in hepatitis B viruses. Journal of virology. 1993;67:6507–6512. doi: 10.1128/jvi.67.11.6507-6512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber ND, Stone D, Sedlak RH, De Silva Feelixge HS, Roychoudhury P, Schiffer JT, Aubert M, Jerome KR. AAV-mediated delivery of zinc finger nucleases targeting hepatitis B virus inhibits active replication. PloS one. 2014;9:e97579. doi: 10.1371/journal.pone.0097579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werle-Lapostolle B, Bowden S, Locarnini S, Wursthorn K, Petersen J, Lau G, Trepo C, Marcellin P, Goodman Z, Delaney W.E.t., Xiong S, Brosgart CL, Chen SS, Gibbs CS, Zoulim F. Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology. 2004;126:1750–1758. doi: 10.1053/j.gastro.2004.03.018. [DOI] [PubMed] [Google Scholar]
- Wieland SF, Spangenberg HC, Thimme R, Purcell RH, Chisari FV. Expansion and contraction of the hepatitis B virus transcriptional template in infected chimpanzees. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2129–2134. doi: 10.1073/pnas.0308478100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TT, Coates L, Aldrich CE, Summers J, Mason WS. In hepatocytes infected with duck hepatitis B virus, the template for viral RNA synthesis is amplified by an intracellular pathway. Virology. 1990;175:255–261. doi: 10.1016/0042-6822(90)90206-7. [DOI] [PubMed] [Google Scholar]
- Wursthorn K, Lutgehetmann M, Dandri M, Volz T, Buggisch P, Zollner B, Longerich T, Schirmacher P, Metzler F, Zankel M, Fischer C, Currie G, Brosgart C, Petersen J. Peginterferon alpha-2b plus adefovir induce strong cccDNA decline and HBsAg reduction in patients with chronic hepatitis B. Hepatology. 2006;44:675–684. doi: 10.1002/hep.21282. [DOI] [PubMed] [Google Scholar]
- Xu C, Guo H, Pan XB, Mao R, Yu W, Xu X, Wei L, Chang J, Block TM, Guo JT. Interferons accelerate decay of replication-competent nucleocapsids of hepatitis B virus. Journal of virology. 2010;84:9332–9340. doi: 10.1128/JVI.00918-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Peng B, Liu Y, Xu G, He W, Ren B, Jing Z, Sui J, Li W. Viral entry of hepatitis B and D viruses and bile salts transportation share common molecular determinants on sodium taurocholate cotransporting polypeptide. Journal of virology. 2014;88:3273–3284. doi: 10.1128/JVI.03478-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife. 2012;1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Mason WS, Summers J. Covalently closed circular viral DNA formed from two types of linear DNA in woodchuck hepatitis virus-infected liver. Journal of virology. 1996;70:4567–4575. doi: 10.1128/jvi.70.7.4567-4575.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Summers J. Illegitimate replication of linear hepadnavirus DNA through nonhomologous recombination. Journal of virology. 1995;69:4029–4036. doi: 10.1128/jvi.69.7.4029-4036.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Summers J. Integration of hepadnavirus DNA in infected liver: evidence for a linear precursor. Journal of virology. 1999;73:9710–9717. doi: 10.1128/jvi.73.12.9710-9717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J, Croyle JL, Nishimura A, Ozato K, Howley PM. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell. 2004;117:349–360. doi: 10.1016/s0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- You J, Srinivasan V, Denis GV, Harrington WJ, Jr., Ballestas ME, Kaye KM, Howley PM. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes. Journal of virology. 2006;80:8909–8919. doi: 10.1128/JVI.00502-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wen Y, Guo X. CRISPR/Cas9 for genome editing: progress, implications and challenges. Human molecular genetics. 2014a;23:R40–46. doi: 10.1093/hmg/ddu125. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li C, Zhang Y, Zhu H, Kang Y, Liu H, Wang J, Qin Y, Mao R, Xie Y, Huang Y, Zhang J. Comparative analysis of CpG islands among HBV genotypes. PloS one. 2013;8:e56711. doi: 10.1371/journal.pone.0056711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Mao R, Yan R, Cai D, Zhang Y, Zhu H, Kang Y, Liu H, Wang J, Qin Y, Huang Y, Guo H, Zhang J. Transcription of hepatitis B virus covalently closed circular DNA is regulated by CpG methylation during chronic infection. PloS one. 2014b;9:e110442. doi: 10.1371/journal.pone.0110442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YY, Zhang BH, Theele D, Litwin S, Toll E, Summers J. Single-cell analysis of covalently closed circular DNA copy numbers in a hepadnavirus-infected liver. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12372–12377. doi: 10.1073/pnas.2033898100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XL, Yang JR, Lin SZ, Ma H, Guo F, Yang RF, Zhang HH, Han JC, Wei L, Pan XB. Serum viral duplex-linear DNA proportion increases with the progression of liver disease in patients infected with HBV. Gut. 2015 doi: 10.1136/gutjnl-2014-308989. [DOI] [PubMed] [Google Scholar]
- Zhou T, Guo H, Guo JT, Cuconati A, Mehta A, Block TM. Hepatitis B virus e antigen production is dependent upon covalently closed circular (ccc) DNA in HepAD38 cell cultures and may serve as a cccDNA surrogate in antiviral screening assays. Antiviral research. 2006;72:116–124. doi: 10.1016/j.antiviral.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Yamamoto T, Cullen J, Saputelli J, Aldrich CE, Miller DS, Litwin S, Furman PA, Jilbert AR, Mason WS. Kinetics of hepadnavirus loss from the liver during inhibition of viral DNA synthesis. Journal of virology. 2001;75:311–322. doi: 10.1128/JVI.75.1.311-322.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman KA, Fischer KP, Joyce MA, Tyrrell DL. Zinc finger proteins designed to specifically target duck hepatitis B virus covalently closed circular DNA inhibit viral transcription in tissue culture. Journal of virology. 2008;82:8013–8021. doi: 10.1128/JVI.00366-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoulim F, Locarnini S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology. 2009;137:1593–1608. e1591–1592. doi: 10.1053/j.gastro.2009.08.063. [DOI] [PubMed] [Google Scholar]
- Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. Journal of virology. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]