Abstract
Introduction
Liver enzyme levels and total serum bilirubin are under genetic control and in recent years genome-wide population-based association studies have identified different susceptibility loci for these traits. We conducted a genome-wide association study in European ancestry participants from the Electronic Medical Records and Genomics (eMERGE) Network dataset of patient medical records with available genotyping data in order to identify genetic contributors to variability in serum bilirubin levels and other liver function tests and to compare the effects between adult and pediatric populations.
Methods
The process of whole genome imputation of eMERGE samples with standard quality control measures have been described previously. After removing missing data and outliers based on principal components (PC) analyses, 3294 samples from European ancestry were used for the GWAS study. The association between each single nucleotide polymorphism (SNP) and total serum bilirubin and other liver function tests was tested using linear regression, adjusting for age, gender, site, platform and ancestry principal components (PC).
Results
Consistent with previous results, a strong association signal has been detected for UGT1A gene cluster (best SNP rs887829, beta = 0.15, p = 1.30x10-118) for total serum bilirubin level. Indeed, in this region more than 176 SNPs (or indels) had p<10−8 spanning 150Kb on the long arm of chromosome 2q37.1. In addition, we found a similar level of magnitude in a pediatric group (p = 8.26x10-47, beta = 0.17). Further imputation using sequencing data as a reference panel revealed association of other markers including known TA7 repeat indels (rs8175347) (p = 9.78x10-117) and rs111741722 (p = 5.41x10-119) which were in proxy (r2 = 0.99) with rs887829. Among rare variants, two Asian subjects homozygous for coding SNP rs4148323 (G71R) were identified. Additional known effects for total serum bilirubin were also confirmed including organic anion transporters SLCO1B1-SLCO1B3, TDRP and ZMYND8 at FDR<0.05 with no gene-gene interaction effects. Phenome-wide association studies (PheWAS) suggest a protective effect of TA7 repeat against cerebrovascular disease in an adult cohort (OR = 0.75, p = 0.0008). Among other liver function tests, we also confirmed the previous effect of the ABO blood group locus for variation in serum alkaline phosphatase (rs579459, p = 9.44x10-15).
Conclusions
Taken together, our data present interesting findings with strong confirmation of previous effects by simply using the eMERGE electronic health record phenotyping. In addition, our findings indicate that similar to the adult population, the UGT1A1 is the main locus responsible for normal variation of serum bilirubin in pediatric populations.
Introduction
Bilirubin is the ultimate product of catabolism of heme with complex interplay of different enzymes. Levels of serum bilirubin are significantly elevated in a number of diseases associated with jaundice and hemolytic blood disorders. Bilirubin levels have a large variability in normal population and bilirubin concentration is under strong genetic regulation with a heritability estimate of roughly 0.50 [1]. In particular, polymorphism TATA box promoter motif in the UGT1A1 gene promoter (UGT1A1*28, rs8175347) is associated with UGT1A1 expression [2]; the mean bilirubin levels of TA7 homozygotes are approximately double those of TA6 homozygotes [3–5].
This homozygosity predisposes individuals not only to Gilbert syndrome which is a benign form of episodic jaundice [2] but also to hyperbilirubinemia induced by several drugs, such as Indinavir, an HIV protease inhibitor, that competitively inhibits UGT enzymatic activity [6]. In addition, TA7 homozygote individuals are also susceptible to severe neutropenia following the administration of Irinotecan, an anti-cancer drug mainly used in metastatic colorectal cancer patients [7]. In this instance, there is a decrease in the inactivation process of the active metabolite SN-38 by glucuronidation to SN-38 glucuronide (SN-38G) [7]. Apart from classical TA repeat insertion, recent data have also shown that other common variants can contribute to mild hyperbilirubinemia in an additive manner with TA repeat, in particular rs4124874 and a coding SNP in UGT1A1 rs4148323 (G71R), with an influence on expression levels of UGT1A1 [8,9]. However, their minor allele frequencies are variable across populations and therefore lead to varying effects of Irinotecan application depending on ethnicity [10,11]. For example, rs4148323 (G71R) which is almost non-polymorphic in European and African population has a minor allele frequency of about 20% in Asians while TA7 repeat is less frequent in this population (~10%) [12].
Homozygosity for rare pathogenic coding variants on the other hand are responsible for severe deficiency of enzymatic activity that range from lethal hyperbilirubinemia (Crigler-Najjar type 1) with zero enzyme activity to Crigler-Najjar type 2 (CN-2) with very low enzyme activity [13, 14]. These pathogenic variants are extremely rare in the general population.
Founded in 2007, the eMERGE Network is a consortium of multiple adult and pediatric institutions using biorepositories linked to electronic medical record (EMR) systems for large-scale genomic research [15]. As part of this network, we conducted this study in order to test the validity of phenotypic data derived from EMR and the capability of this network for genetic research.
Genome-wide association studies (GWAS) for total serum bilirubin or other liver function tests previously have been reported in different ancestries of well characterized adult populations [16–24]; however, the genetic contribution in a pediatrics population has not been specifically evaluated. There is a tendency toward more extreme phenotypes in pediatrics and some genomic effects may be age dependent or stronger in earlier stage of development than later on. In addition, understanding genomic loci that operate in pediatric population are especially important for a better health care since there is an opportunity for early preventive measures as well as long term implementation of a treatment plan in order to avoid further hepatotoxicity and hyperbilirubinemia. Finally the joint effect model of pediatric and adults will provide a better estimate of percentage of variance explained. In this study, we investigate whether susceptibility loci identified by GWAS in adults are associated in children and adolescents. For this purpose, we conducted quantitative GWAS analyses for liver function tests in a primarily European-derived population and compared the observed size effects between pediatric and adult participants from the eMERGE Network.
Materials and Methods
Ethics statement
The eMERGE Network is a collaboration of institutions with biobanks linked to EMRs. The detail of recruitment and biological sample collection of eMERGE cohorts has been described previously [15]. De-identified samples linked to EMR were supplied from multiple investigators from different institutions with approval from their respective institutional review boards (IRBs) including Cincinnati Children’s Hospital Medical Center, Boston Children’s Hospital, Geisinger Health System, Group Health Research Institute and Marshfield Clinic [15]. All study participants provided written consent prior to study enrollment as part of the DNA biobank in each site. Written informed consent was obtained from the next of kin, caretakers, or guardians on behalf of the minors/children enrolled in the study and have been approved by the IRB.
Study participants
De-identified samples from five different eMERGE sites were evaluated (Table 1). Children and teens, aged through 19 years old composed the pediatric population. For GWAS and PheWAS analyses only those self-reported to have European ancestry and with normal ranges of liver function tests were selected (Table 1), (details described under “Phenotyping”).
Table 1. The demographic distribution of EMR-linked eMERGE cohorts.
| Cohorts | eMERGE-Sites | #Europeans | M/F | Platforms | Mean age (±SD) |
|---|---|---|---|---|---|
| Pediatrics | BCH | 148 | 92/56 | Affymetrix-Axiom | 13.30 (5.47) |
| CCHMC | 419 | 272/147 | Illumina-610 | ||
| 217 | 128/89 | Illumina-Omni-1 | |||
| 184 | 112/72 | Illumina-Omni-5 | |||
| 99 | 12/87 | Affymetrix-6 | |||
| CCHMC-Total | 919 | 524/395 | 11.52 (5.35) | ||
| Adults | Marshfield | 728 | 339/389 | Illumina660W | |
| 50 | 7/43 | Affymetrix-6 | |||
| Marshfield-Total | 778 | 346/432 | 64.9 (11.46) | ||
| Geisinger | 691 | 459/232 | Illumina-Omni-Express | 70.7 (13.81) | |
| GroupHealth | 657 | 288/369 | Illumina660W | ||
| 101 | 63/38 | Illumina-Omni-Express | |||
| GroupHealth-Total | 758 | 351/407 | 67.4 (14.92) | ||
| Total | 3294 | 1772/1522 |
Genotyping and imputation
High throughput SNP genotyping was carried out previously in each respective facility as shown in Table 1. Quality control (QC) of the data was performed before imputation. In each genotyped cohort, standard quality control criteria were met and single nucleotide polymorphisms (SNPs) were removed if (a) >5% of the genotyping data was missing, (b) out of Hardy-Weinberg equilibrium (HWE, p < 0.0001) in controls, or a minor allele frequency (MAF) <1%. Samples with call rates <98% were excluded. Recently all eMERGE cohorts have undergone whole genome imputation as described [25]. The imputation pipeline includes SHAPEIT2/IMPUTE2 program and the publicly available 1000-Genomes Project as the reference haplotype panel composed of 1092 samples (release version 2 from March 2012 of the 1000 Genomes Project Phase I, ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/release/20110521) [26].
The basic quality controls for eMERGE imputed data provided to us included a threshold of 0.90 for the genotype posterior probability and info score > 0.7 [25]. Info score is the information metric that IMPUTE2 reports [26]. This metric typically takes values between 0 and 1, where values near 1 indicate that a SNP has been imputed with high certainty. This is often used to remove poorly imputed SNPs. Principal component analysis (PCA) was performed to identify outliers and hidden population structure using EIGENSTRAT [27]. The first three principal components explained most of the variance and were retained and used as covariates during the association analysis in order to adjust for population stratification. In addition, UGT1A1 whole exome sequencing data were available to use from the eMERGE-PGx project (a Network collaboration with Pharmacogenomics Research Network (PGRN)) dataset derived from 5163 independent samples from different ancestries [28]. To increase the resolution of the association signal at UGT1A1, this collection was then phased and used as a reference panel for secondary imputation using the SHAPEIT2/IMPUTE2 pipeline described above [26].
Phenotyping
Levels of serum bilirubin are significantly elevated in a number of diseases. Therefore, we first excluded conditions that either cause overproduction (e.g hemolytic diseases) or decreased in excretion of bilirubin (e.g hepatobiliary diseases). These include ICD-9 codes of 570 to 579.99 with 101 distinct diseases (Chronic liver disease and cirrhosis, Alcoholic fatty liver, drug induced liver disease, gallbladder disease), diagnostic codes of 280 to 289.99 for blood related diseases with 133 categories including hemolytic anemia or Sickle cell disease as well as blood related malignancy with ICD-9 codes of 200 to 209.99 with 391 categories. We also excluded those undergoing chemotherapy or transplant. The same exclusion rules were applied to other liver function enzymes.
Since transient physiologic hyperbilirubinemia is common in the neonatal period, we also excluded children less than one year old. Due to the nature of electronic medical records and since ICD-9 codes, per se, may not be specific and due to underdiagnosed conditions or errors, an additional filtration step was applied in which we excluded samples that still had any out of normal range values (>2 mg/dl) for total serum bilirubin [29,30]. After these filtration steps, the latest lab measures were used for analyses. For other liver enzymes (ALT, ASP, GGT and ALP) and because of wider normal data ranges, natural log-transformations were generated and used for Linear-regression analyses to preserve normal distribution.
A phenome-wide association analysis (PheWAS) was performed in which presence or absence of each PheWAS code that mapped from translated ICD-9 codes were considered as a binary phenotype as described previously [31]. These PheWAS codes were used to define case and control group. Control groups for Crohn’s Disease (CD), for instance, excluded CD, ulcerative colitis and several other related gastrointestinal complaints. The current PheWAS map is available at (http://phewascatalog.org).
Genome-wide Association Statistics
After performing quality control measures mentioned above, we tested 3,301,391 autosomal SNPs for quantitative association study. Linear-regression analyses assuming an additive genetic model was used on latest total serum bilirubin, ALT, ALP, and AST and GGT using PLINK software package and adjusted by age, sex, principal components, sites and genotyping platforms [32]. In addition, previously known variants associated with serum bilirubin level from the NHGRI catalog were selected as a priori list of 9 autosomal candidate genes and evaluated separately in order to confirm these effects, in which false discovery rate (FDR) methods were used to correct for multiple testing using the Benjamini–Hochberg procedure implemented in PLINK [32]. PLINK was also used for conditional analyses and pairwise SNP-SNP interactions (epistasis) [32]. The ‘‘epistasis” option in PLINK provides a logistic regression test for interaction that assumes an allelic model for interactions and their principal effects in which PLINK makes a model based on allele dosage for each SNP [32]. To graphically display results, LocusZoom and Golden Helix programs were used [33, Golden Helix GenomeBrowse® visualization tool (Version 8.3.0). Bozeman, MT: Golden Helix, Inc. Available from http://www.goldenhelix.com).
Results
Characteristics of the study participants are presented in Table 1. After exclusions of series of disease conditions described in the Methods, the latest serum bilirubin level and other liver function tests were used for analyses. As shown in Table 1, the mean ± SD age was 67.7±13.40 for adult participants and 12.4±5.43 for pediatrics participants. The mean ± SD of total bilirubin level was 0.56±0.28. All studies showed significantly higher bilirubin levels in males than females, as expected. The mean of total serum bilirubin in males was 0.59±0.31 mg/dl and 0.52±0.26 mg/dl in females. Similarly, it was higher in adults than children (mean± SD of 0.61±0.28 in adults, 0.44 ±0.28 in children). Therefore for all analyses, age- and sex-adjusted analyses (covariates described in Materials and Methods) were conducted.
Consistent with previous reports, we identified strong genetic signals at UGT1A1 locus at 2q37. Fig 1 shows the Manhattan plot of this GWAS effect. The overall low inflation rate of (λ = 1.004) was observed. Indeed, the best SNP, rs887829, produced p = 1.30x10-118, beta = 0.15. This is equal to an OR = 6.35 when we consider 10% of tail distribution as cases and controls (Table 2). As shown in Table 2, the effect was also strong in the pediatric-only cohort with 1067 participants (p = 8.26x10-47, beta = 0.18).
Fig 1. (A and B) Manhattan plot and Q–Q plot of genome-wide markers for total serum bilirubin in 3294 European samples respectively.
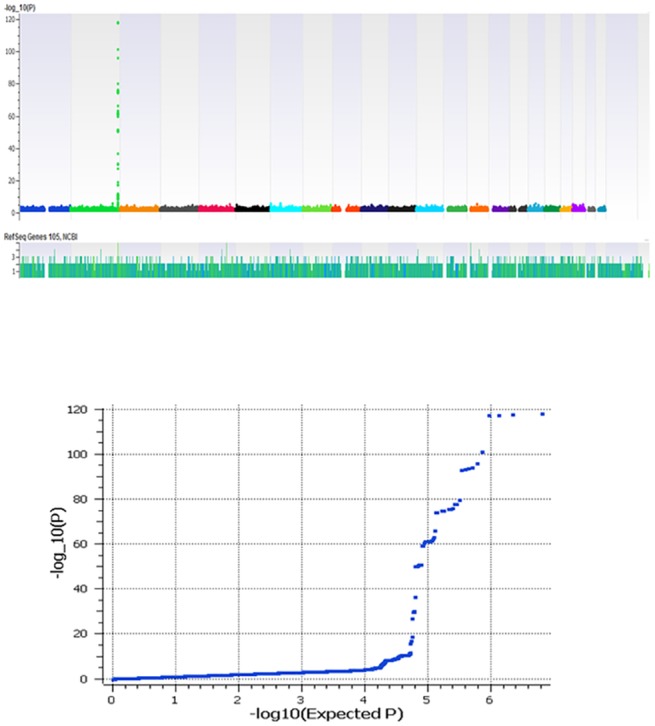
Table 2. Quantitative and case-control association result of top SNPs in UGT1A1 for total serum bilirubin levels in pediatric and adult subgroups.
| Sample size | CHR | SNP | Position * | Beta | p value † | |
| Pediatrics | 1067 | 2 | rs887829-T | 234668570 | 0.177 | 8.26E-47 |
| Adult | 2227 | 2 | rs887829-T | 234668570 | 0.147 | 2.00E-63 |
| Total | 3294 | 2 | rs887829-T | 234668570 | 0.157 | 1.30E-118 |
| tail-pheno | Case/Control | OR | p value ‡ | |||
| Decile | 354/370 | 2 | rs887829-T | 234668570 | 6.347 (5.02–8.01) | 8.76E-59 |
*Map Position: NCBI build 37.
†linear regression association test.
‡Chi-square test.
Next, in order to increase the resolution of this association signal we imputed additional variants using the PGRN sequencing dataset as a reference panel [28]. As a result additional association signals for a number of imputed variants were detected that pass the quality threshold (info score >0.7). In particular, the TA7 indel repeat (rs8175347) that was in linkage disequilibrium with the top marker (rs887829, r2 = 0.99) was imputed and with similar magnitude of effect (p = 9.78x10-117, beta = 0.15). Another imputed intronic marker, rs111741722, produced the highest effect in this study (p = 5.41x10-119, beta = 0.15). Fig 2 shows the haplotypic LD structure of the TA repeat with nearby markers that all generate similar magnitude of results. Indeed, in this region more than 176 SNPs (or indels) had p<10–8 spanning 150Kb on the long arm of chromosome 2q37.1 (Fig 3). As shown in Fig 2, SNP rs4124874, which also influences gene expression [9] was an incomplete proxy for TA repeat (r2 = 0.60 and produced p = 6.66x10-75, beta = 0.12). After conditional analysis controlling for this marker, the TA repeat locus still remained significant (p = 3.58x10-42); on the other hand conditioning on the TA repeat diminished significantly all association signal for rs4124874, p = 0.90 suggesting that the TA repeat mainly explains global variations in this region. As shown in Fig 2, TA repeat was in perfect LD with 5 SNP markers (rs887829, rs111741722, rs6742078, rs4148324, rs4148325) spanning less than 10KB, therefore, further conditional analyses were not possible.
Fig 2. The LD structure between markers in peak association signals.
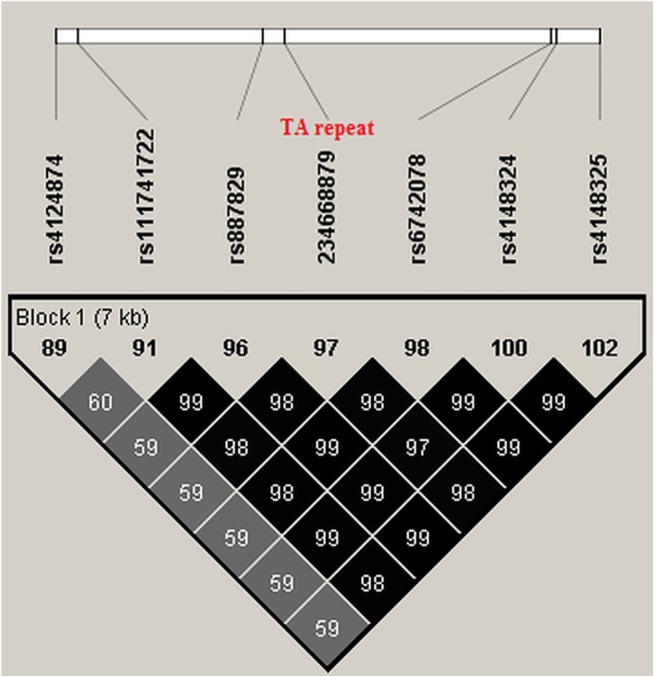
r2 = correlation coefficient as a measure of linkage disequilibrium.
Fig 3. A schematic representation of association map at UGT1A region for total serum bilirubin.

The TA repeat location is shown with vertical line.
Other effects
Apart from UGT1A1 effect, no other effects reach a genome significance level (P<10–8) in our cohorts (Fig 1A). There is one other locus at 12p12 (SLCO1B1-SLCO1B3) that is consistently reported to be associated with total serum bilirubin at less magnitude [16,17]. In addition, several other genes suggestively have been reported to be associated. We, therefore, more thoroughly evaluated these variants and obtained 9 distinct lead SNPs from the NHGRI catalog and confirmed several previously reported associations for bilirubin at the level of FDR<0.05. These include SLCO1B1-SLCO1B3, TDRP and ZMYND8 (Table 3). SLCO1B1 is an organic anion transporter (OATP1B1) important for uptake of bilirubin in liver. The nonsynonymous SNP, rs4149056 V174A, which alters uptake activity of this transporter produced p = 0.0004, beta = 0.03 in our combined cohorts (Table 3). In this region SNPs located at SLCO1B3 was also associated including rs10841661 (Table 3). There was a 350Kb distance between these two family members of B1 and B3 with r2 = 0.002. Therefore, conditional analysis controlling for rs4149056 still shows association effects at rs10841661 (p = 0.0008) suggesting an independent effect in SLCO1B3.
Table 3. Other effects associated with total serum bilirubin at FDR<0.05.
| CHR | Gene | SNP | BP | Minor allele* | BETA | P value | P value- Adjusted for rs887829 | P value- Interaction with rs887829 | REF |
|---|---|---|---|---|---|---|---|---|---|
| 8 | TDRP | rs17665859 | 445601 | C | 0.037 | 0.003 | 0.009 | 0.28 | 17 |
| 12 | SLCO1B3 | rs10841661 | 20984832 | T | 0.025 | 0.0003 | 0.0006 | 0.24 | 20 |
| 12 | SLCO1B3 | rs17680137 | 21015906 | G | 0.029 | 0.001 | 0.0013 | 0.75 | 20 |
| 12 | SLCO1B1 | rs4149056 | 21331549 | C | 0.032 | 0.0004 | 0.00009 | 0.21 | 16 |
| 20 | ZMYND8 | rs7262634 | 45834279 | C | 0.042 | 0.005 | 0.0192 | 0.2 | 24 |
Conditional P values adjusted for lead SNP in UGT1A1 (rs887829) as well as pairwise SNPxSNP interactions with (rs887829) are shown.
*All effects are for minor alleles.
We next tested for any effect of the UGT1A1 gene on the association of bilirubin levels with these genes and SNP rs887829, the top marker on the UGT1A1 locus in the model, was used for conditional analyses. As shown in Table 2, no association disappeared after controlling for rs887829, suggesting that part of the variability of total serum bilirubin can be explained by these other genes. Furthermore, although no significant gene-gene interaction was observed, some association improved (Table 3).
Rare effects
In the UGT1A1 gene, there are more than 50 known stop codon, gain of function or frame shift mutations reported in families with CN-type 1 or 2 [34]. These mutations are extremely rare and homozygosity is link to autosomal recessive early lethality especially in CN type I. In PGRN sequencing data we identified 16 of these variants in 44 participants but all were carriers (heterozygous); Serum total bilirubin were available for 9 of them that all were within normal range consistent with a recessive effect. No homozygote recessive or compound heterozygote participants were detected.
As mentioned above the SNP rs4148323 (G71R) mutation is more common in the Asian population. This variant is associated with a milder form of hyperbilirubinemia and Gilbert’s syndrome. PGRN sequencing data revealed two Asian participants that were homozygous for coding SNP rs4148323 (G71R). The means of total serum bilirubin for these participants were 1.52 (95% CI = 1.032–2.018) and 0.85 (95%CI 0.68–1.0) respectively. They have not been diagnosed clinically with jaundice according to EMR, nor did they have any risk alleles in TA repeat, rs4124874 or any other detected rare variants.
Other Liver function test
Except for serum total alkaline phosphatase, no other liver enzymes produced a genome wide significant effect (P<1x10-8). The effect of the ABO blood group locus and total alkaline phosphatase that has been replicated in different studies was confirmed in our study (best p ≤1x10-15 for SNP rs600038, Table 4, Fig 4A). The effect was consistent across pediatric and adult cohorts. ABO blood type is known to be correlated with serum alkaline phosphatase, therefore we next identified perfect surrogate SNPs tagging ABO alleles in our dataset which include: rs8176704 for the A allele, rs505922 (or rs612169) for the O allele and rs8176746 (or rs8176672) for the B allele [35,36]. As shown in Fig 4B, after controlling for all three ABO SNP determinants, the association signal dramatically reduced for the lead SNP rs600038 with p-conditional = 0.02, consistent with previous studies. Of note, the linkage disequilibrium between lead SNP rs600038 and three ABO SNP determinants (rs505922 and rs8176746, rs8176704) were r2 = 0.39, 0.03 and 0.01 respectively.
Table 4.
| Enzyme | Gene | CHR | SNP | BP | Minor allele | BETA | P value | REF |
|---|---|---|---|---|---|---|---|---|
| ALP | ABO | 9 | rs651007 | 136153875 | T | -0.034 | 1.06E-14 | 22,23 |
| ABO | 9 | rs600038 | 136151806 | C | -0.034 | 8.608e-15 | 22,23 | |
| ABO | 9 | rs579459 | 136154168 | C | -0.034 | 9.436e-15 | 22,23 |
Fig 4. (A) The association signal between total serum ALP and the ABO blood locus. (B): The association of lead SNPs at the ABO locus with serum ALP and their relation with ABO tag SNPs (rs8176746, rs8176704, rs505922); A significant drop in association effect in lead SNPs after controlling for ABO-tag SNPs are shown (red and black dots represent before and after conditional analyses respectively).

PheWAS approach
To explore pleiotropy of the TA-repeat in UGT1A1 for other clinical diagnoses, we performed a PheWAS study in adult and pediatric cohorts separately using available ICD-9 codes as previously described [31]. As expected, since we excluded all participants with hyperbilirubinemia and related conditions, we did not find any associations for these conditions. There were 377 distinct diagnostic codes for adults with minimum of 100 participants in each category. After correcting for multiple comparisons, no results remained significant. The best overall effect was a trend toward associations with cerebrovascular disease and ischemic stroke in adult-only cohorts adjusting for age, gender, site, platforms and PCs (unadjusted p = 0.0008, adjusted p = 0.056, OR = 0.75, 95%CI 0.63–0.88). There were 464 cases with this diagnosis and 1433 controls. Of note this was a protective effect for the TA7 repeat. In addition, results for other atherosclerosis-related conditions such as ischemic cardiovascular disease were in the same direction but only suggestive (data not shown). The number of samples for these conditions were small in pediatrics cohorts (<10).
Discussion
In this GWAS analysis from five eMERGE sites we confirmed the substantial contribution of UGT1A1 on human serum bilirubin levels and a number of other transporter genes, including SLCO1B1 and B3 that influence variation in bilirubin levels. This is the largest GWAS study for serum bilirubin in a pediatric cohort. We found that this effect is strong in pediatric as well as adult populations (Table 2). Further imputation using PGRN sequencing data revealed that the TA indel repeat (UGT1A1*28, rs8175347) is in strong LD with 5 additional SNPs (r2>0.98) and mainly responsible for the GWAS signal at this location. Indeed, this locus by itself explained 14% of total variations of serum bilirubin in our cohorts comparable with previous studies [16]. In comparison only 0.2% of total variation of serum bilirubin can be explained by rs4149056 SNP in SLCO1B1 in our cohort. Similar to previous studies, we did not detect a significant gene gene interaction between SLCO1B1/B3 and UGT1A1 (p = 0.21, Table 2) [16]. We confirmed a number of other effects as shown in Table 3 including TDRP and ZMYND8. We had 90% power to detect such association in our studies (average MAF = 0.10, beta 0.03–0.05, OR = 1.1–1.2).
We applied a successful strategy using electronic medical records to identify individuals with normal variations in serum bilirubin and other liver function tests by excluding more than 300 blood and liver-related diseases and conditions (see method). Electronic health records have enormous phenotypic heterogeneity. There are many reasons that a patient may have hyperbilirubinemia that range from a simple viral hepatitis to hemolytic crises due to medications. Including serum bilirubin for these non-genetic conditions may produce noise and skew the normal distribution of the quantitative phenotype under study. Obviously additional studies are necessary to address any disease specific condition. Similarly we applied stringent quality criteria and thresholds for genotyping data and adjusted all results by age, sex, site of study, platform and principal components. We evaluated the eMERGE PGRN dataset for rare variants at UGT1A1 and as a result we identified two Asian participants with homozygous for the rs4148323 minor allele resulting in a (p.Gly71Arg) coding change. Interestingly they did not have additional risk alleles in the TA repeat or rs4124874 variants suggesting an independent effect for (p.Gly71Arg). One of these participants shows a subclinical hyperbilirubinemia (latest total bilirubin = 2.1 mg/dl) possibly associated with the effect of this missense variant [37]. As mentioned above, this variant is rare in other ancestries. Other available liver function tests were normal in these two participants. In addition, we identified 44 heterozygote (or carrier) participants with other potential deleterious mutations such as (rs34993780, p.Tyr486Asp). Since homozygous variants are extremely rare in population (1:1000000) with severe early hyperbilirubinema (CN typ1 or 2), we did not expect to find these homozygotes in our population panel cohorts. Interestingly no compound heterozygote was detected.
Among other liver function tests, we confirmed the effect of the ABO locus on serum alkaline phosphatase. This effect has been consistently replicated in other studies [22,23]. Approximately 90% of serum alkaline phosphatase (ALP) originates from liver, bone and kidney, while 10% from intestine and 1% from placenta [38]. Most of the intestinal ALP is attached to ABO antigens on the surface of erythrocytes by a glycosyl-phosphatidylinositol anchor, however, with different binding capacity depending on different blood groups. Erythrocytes of blood type A bind to almost all intestinal ALP, while erythrocytes of blood type B/O bind to a much lesser degree. This results in an increase of intestinal ALP in serum of individuals with blood type B/O [39]. The observation that association between rs600038 and serum ALP dramatically reduced after adjusting for ABO-related variants support this physiologic process; although a residual effect exists (p = 0.02, Fig 4B) that may indicate other regulatory roles of this locus for serum alkaline phosphatase or underlying confounding effects since the ABO locus is associated with a variety of metabolic related phenotypes including serum levels of intercellular adhesion molecule-1 (ICAM-1), E-selectin, and P-selectin that participate in inflammatory processes by promoting adhesion of leukocytes to vascular wall endothelium, effects on low-density lipoprotein and total serum cholesterol levels as well as coronary artery disease and stroke [35,40,41].
The major strengths of our study include careful quality control and minimal population stratification. Electronic medical records, however, have several study limitations including human errors in extraction of lab measurements or errors in billing code and clinical diagnosis. In addition the population under study may have wide spectrum of different diseases and therefore cannot represent a random sample from the normal general population. We have controlled all relevant medical diagnoses in the analyses to avoid potential sampling bias. The associations with serum bilirubin levels and UGT1A1 were in fact highly consistent with previous publications that often recruit well-characterized participants.
Total serum bilirubin is associated with several clinical outcomes, including cardiovascular disease, diabetes and drug metabolism that warrant additional study. In fact, bilirubin is a potent antioxidant and higher levels of serum bilirubin may offer a therapeutic advantage in oxidative stress-mediated conditions [42]. In this context, we detected a suggestive protective effect of TA7 repeat against chronic cerebrovascular disease that point to this direction (OR = 0.75). Our adult cohort, consist of elderly participants with the mean age of 67.7 (95% CI 63.00–72.33) that could well-represent a long term sequela of bilirubin on vascular system. Obviously excluding potential liver disease patients in this study was a key step to clarify the inverse associations between bilirubin and stroke as discussed in other published works [43]. Finally, susceptibility to severe neutropenia following the administration of Irinotecan in TA7 homozygotes with colon cancer is particularly interesting and this data will be used for future pharmacogenetic studies across the eMERGE network.
Acknowledgments
We are grateful to the individuals who participated in this study. This work was supported by collaborative sites in the eMERGE Network and funded by NHGRI through the following grants: U01HG006828 (Cincinnati Children’s Hospital Medical Center/Boston Children’s Hospital); U01HG006389 (Essentia Institute of Rural Health, Marshfield Clinic Research Foundation and Pennsylvania State University); U01HG006382 (Geisinger Clinic); U01HG006375 (Group Health Cooperative/University of Washington); U01HG006385 (Vanderbilt University Medical Center serving as the Coordinating Center) and the PGRNSeq dataset (eMERGE PGx); U01HG004438 (CIDR) serving as a Sequencing Center. We would also like to thank other members of the eMERGE consortium for their support and feedback.
Data Availability
Data files are available from dbGaP under eMERGE accession numbers: phs000494.v1.p1, phs000495.v1. phs000360.v2.p1, phs000387.v1.p1.
Funding Statement
This study was supported by the National Institutes of Health (http://www.nih.gov/) grants: U01HG006828-NHGRI-Cincinnati Children’s Hospital Medical Center/Boston Children’s Hospital (to JBH, IAH, BN, BLC, KM, TL,CP); U01HG006389-NHGRI-Essentia Institute of Rural Health, Marshfield Clinic Research Foundation and Pennsylvania State University (to PLP, MHB, TEK, MDR, SSV); U01HG006382–NHGRI-Geisinger Clinic (to MSW, KMB); U01HG006375-NHGRI-Group Health Cooperative/University of Washington (to GPJ, JG); U01HG006385-NHGRI-Vanderbilt University Medical Center serving as the Coordinating Center and the PGRNSeq dataset (eMERGE PGx); U01HG004438-NHGRI- The Center for Inherited Disease Research (CIDR) serving as a Sequencing Center for eMERGE. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bathum L, Petersen HC, Rosholm JU, Hyltoft Petersen P, Vaupel J, Christensen K. Evidence for a substantial genetic influence on biochemical liver function tests: results from a population-based Danish twin study. Clin Chem. 2001;47:81–87. [PubMed] [Google Scholar]
- 2. Bosma PJ, Chowdhury JR, Bakker C, Gantla S, de Boer A, Oostra BA, et al. The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert's syndrome.N Engl J Med. 1995;333:1171–5. [DOI] [PubMed] [Google Scholar]
- 3. Lampe JW, Bigler J, Horner N K, Potter J D. UDPglucuronosyltransferase (UGT1A1*28 andUGT1A6*2) polymorphisms in Caucasians and Asians: Relationships to serum bilirubin concentrations. Pharmacogenetics.1999; 9, 341–349. [DOI] [PubMed] [Google Scholar]
- 4. Premawardhena A, Fisher CA, Liu YT, Verma I C, De Silva S, Arambepola M, et al. The global distribution of length polymorphisms of the promoters of the glucuronosyltransferase 1 gene (UGT1A1): Hematologic and evolutionary implications. Blood Cells Mol Dis,2003; 31, 98–101. [DOI] [PubMed] [Google Scholar]
- 5. Lin JP, O’Donnell CJ, Schwaiger JP, Cupples LA, Lingenhel A, Hunt SC, et al. Association between the UGT1A1*28 allele, bilirubin levels, and coronary heart disease in the Framingham Heart Study. Circulation.2006; 114, 1476–1481. [DOI] [PubMed] [Google Scholar]
- 6. Zucker SD, Qin X, Rouster SD, Yu F, Green RM, Keshavan P, et al. Mechanism of indinavir-induced hyperbilirubinemia. Proc Natl Acad Sci USA.2001; 98: 12671–12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Innocenti F, Vokes EE, Ratain MJ. Irinogenetics: what is the right star? J Clin Oncol. 2006;24:2221–4. [DOI] [PubMed] [Google Scholar]
- 8. Aono S, Yamada Y, Keino H, Hanada N, Nakagawa T, Sasaoka Y, et al. Identification of defect in the genes for bilirubin UDP-glucuronosyl-transferase in a patient with Crigler-Najjar syndrome type II. Biochem Biophys Res Commun 1993;197:1239–44. [DOI] [PubMed] [Google Scholar]
- 9. Sugatani J, Yamakawa K, Yoshinari K, Machida T, Takagi H, Mori M, et al. Identification of a defect in the UGT1A1 gene promoter and its association with hyperbilirubinemia. Biochem Bio¬phys Res Commun 2002;292:492–7. [DOI] [PubMed] [Google Scholar]
- 10. Jinno H, Tanaka-Kagawa T, Hanioka N, Saeki M, Ishida S, Nishimura T, et al. Glucuronidation of 7-ethyl-10-hydroxycamp¬tothecin (SN-38), an active metabolite of irinotecan (CPT-11), by human UGT1A1 variants, G71R, P229Q, and Y486D. Drug Metab Dispos 2003;31:108–13. [DOI] [PubMed] [Google Scholar]
- 11. Gagné JF, Montminy V, Belanger P, Journault K, Gaucher G, Guillemette C. Common human UGT1A polymorphisms and the altered metabolism of irinotecan active metabolite 7-ethyl-10-hy¬droxycamptothecin (SN-38). Mol Pharmacol 2002;62:608–17. [DOI] [PubMed] [Google Scholar]
- 12. Kim JY, Cheong HS, Park BL, Kim LH, Namgoong S, Kim JO, et al. Comprehensive variant screening of the UGT gene family.Yonsei Med J. 2014;55:232–9. 10.3349/ymj.2014.55.1.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sneitz N, Bakker CT, de Knegt RJ, Halley DJ, Finel M, Bosma PJ. Crigler–Najjar syndrome in The Netherlands: identification of four novel UGT1A1 alleles, genotype-phenotype correlation, and functional analysis of 10 missense mutants. Hum Mutat. 2010;31:52–59. 10.1002/humu.21133 [DOI] [PubMed] [Google Scholar]
- 14. Sampietro M, Iolascon A. Molecular pathology of Crigler-Najjar type I and II and Gilbert’s syndromes. Haematologica.1999; 84, 150–157. [PubMed] [Google Scholar]
- 15. McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med. Genomics 2011;4:13 10.1186/1755-8794-4-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson AD, Kavousi M, Smith AV, Chen MH, Dehghan A, Aspelund T, et al. Genome-wide association meta-analysis for total serum bilirubin levels.Hum Mol Genet. 2009;18:2700–10. 10.1093/hmg/ddp202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bielinski SJ, Chai HS, Pathak J, Talwalkar JA, Limburg PJ, Gullerud RE, et al. Mayo Genome Consortia: a genotype-phenotype resource for genome-wide association studies with an application to the analysis of circulating bilirubin levels. Mayo Clin Proc. 2011;86:606–14. 10.4065/mcp.2011.0178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dai X, Wu C, He Y, Gui L, Zhou L, Guo H, et al. A genome-wide association study for serum bilirubin levels and gene-environment interaction in a Chinese population. Genet Epidemiol. 2013;37:293–300. 10.1002/gepi.21711 [DOI] [PubMed] [Google Scholar]
- 19. Kang TW, Kim HJ, Ju H, Kim JH, Jeon YJ, Lee HC, et al. Genome-wide association of serum bilirubin levels in Korean population. Hum Mol Genet. 2010;19:3672–8. 10.1093/hmg/ddq281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sanna S, Busonero F, Maschio A, McArdle PF, Usala G, Dei M, et al. common variants in the SLCO1B3 locus are associated with bilirubin levels and unconjugated hyperbilirubinemia. Hum Mol Genet. 2009;18:2711–8. 10.1093/hmg/ddp203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen G, Ramos E, Adeyemo A, Shriner D, Zhou J, Doumatey AP, et al. UGT1A1 is a major locus influencing bilirubin levels in African Americans. Eur J Hum Genet. 2012;20:463–8. 10.1038/ejhg.2011.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yuan X, Waterworth D, Perry JR, Lim N, Song K, Chambers JC, et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet. 2008;83:520–8. 10.1016/j.ajhg.2008.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chambers JC, Zhang W, Sehmi J, Li X, Wass MN, Van der Harst P, et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011;43:1131–8. 10.1038/ng.970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nelson D, Yoshida EM, Paulson MS, Hengen PN, Ge D, Kanwar B, et al. Genome-wide association study to characterize serum bilirubin elevations in patients with HCV treated with GS-9256, an HCV NS3 serine protease inhibitor.Antivir Ther. 2014;19:679–86. 10.3851/IMP2747 [DOI] [PubMed] [Google Scholar]
- 25. Verma SS, de Andrade M, Tromp G, Kuivaniemi H, Pugh E, Namjou-Khales B, et al. Imputation and quality control steps for combining multiple genome-wide datasets.Front Genet. 2014;5:370 10.3389/fgene.2014.00370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda). 2011;1, 457–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38, 904–9. [DOI] [PubMed] [Google Scholar]
- 28. Rasmussen-Torvik LJ, Stallings SC, Gordon AS, Almoguera B, Basford MA, Bielinski SJ, et al. Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems.Clin Pharmacol Ther. 2014;96:482–9. 10.1038/clpt.2014.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Berk PD, Korenblat KM. Approach to the patient with jaundice or abnormal liver test results In: Goldman L, Ausiello D, eds. Cecil Medicine. 24th ed. Philadelphia, Pa: Saunders Elsevier; 2011:chap 149. [Google Scholar]
- 30. Pratt DS. Liver chemistry and function tests In: Feldman M, Friedman LS, Brandt LJ, eds. Sleisenger and Fordtran's Gastrointestinal and Liver Disease. 9th ed. Philadelphia, Pa: Saunders Elsevier;2010:chap 73. [Google Scholar]
- 31. Namjou B, Marsolo K, Caroll RJ, Denny JC, Ritchie MD, Verma SS, et al. Phenome-wide association study (PheWAS) in EMR-linked pediatric cohorts, genetically links PLCL1 to speech language development and IL5-IL13 to Eosinophilic Esophagitis.Front Genet. 2014. November 18;5:401 10.3389/fgene.2014.00401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81, 559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010;26: 2336–2337. 10.1093/bioinformatics/btq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rahim F, Galehdari H, Mohammadi-As J, Saki N. Regression Modeling and Meta-Analysis of Diagnostic Accuracy of SNP-Based Pathogenicity Detection Tools for UGT1A1 Gene Mutation.Genet Res Int. 2013;2013:546909 10.1155/2013/546909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barbalic M, Dupuis J, Dehghan A, Bis JC, Hoogeveen RC, Schnabel RB, et al. Large-scale genomic studies reveal central role of ABO in sP-selectin and sICAM-1 levels.Hum Mol Genet. 2010;19:1863–72. 10.1093/hmg/ddq061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Qi L, Cornelis MC, Kraft P, Jensen M, van Dam RM, Sun Q, et al. Genetic variants in ABO blood group region, plasma soluble E-selectin levels and risk of type 2 diabetes.Hum Mol Genet. 2010;19:1856–62. 10.1093/hmg/ddq057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yamamoto K, Sato H, Fujiyama Y, Doida Y, Bamba T. Contribution of two missense mutations (G71R and Y486D) of the bilirubin UDP glycosyltransferase (UGT1A1) gene to phenotypes of Gilbert’s syndrome and Crigler-Najjar syndrome type II. Biochim Biophys Acta 1998; 1406: 267–273 [DOI] [PubMed] [Google Scholar]
- 38. Hirano K, Matsumoto H, Tanaka T, Hayashi Y, Iino S, Domar U, et al. Specific assays for human alkaline phosphatase isozymes. Clin Chim Acta1987;166:265–273. [DOI] [PubMed] [Google Scholar]
- 39. Bayer PM, Hotschek H, Knoth E: Intestinal alkaline phosphatase and the ABO blood group system–a new aspect. Clin Chim Acta 1980;108:81–87. [DOI] [PubMed] [Google Scholar]
- 40. Paterson AD, Lopes-Virella MF, Waggott D, Boright AP, Hosseini SM, Carter RE, et al. Genome-wide association identifies the ABO blood group as a major locus associated with serum levels of soluble E-selectin.Arterioscler Thromb Vasc Biol. 2009;29:1958–67. 10.1161/ATVBAHA.109.192971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schunkert H, König IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease.Nat Genet. 2011;43:333–8. 10.1038/ng.784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perlstein TS, Pande RL, Beckman JA, Creager MA. Serum total bilirubin level and prevalent lower extremity peripheral arterial disease: National Health and Nutrition Examination Survey (NHANES) 1999 to 2004. Arterioscler Thromb Vasc Biol 2008;28:166–172. [DOI] [PubMed] [Google Scholar]
- 43. Vítek L. Does hyperbilirubinemia protect from coronary heart disease? Am J Cardiol. 2001;88:1218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data files are available from dbGaP under eMERGE accession numbers: phs000494.v1.p1, phs000495.v1. phs000360.v2.p1, phs000387.v1.p1.


