SUMMARY
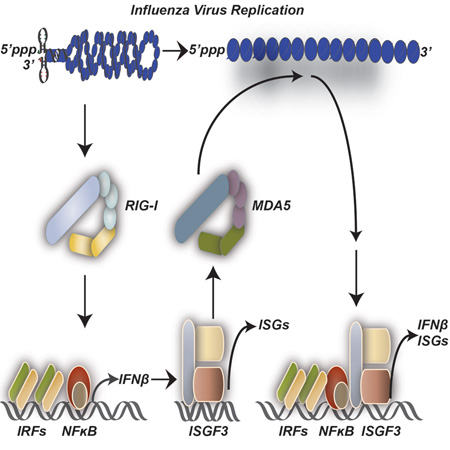
Responding to an influenza A virus (IAV) infection demands an effective intrinsic cellular defense strategy to slow replication. To identify contributing host factors to this defense, we exploited the host microRNA pathway to perform an in vivo RNAi screen. To this end, IAV, lacking a functional NS1 antagonist, was engineered to encode individual siRNAs against antiviral host genes in an effort to rescue attenuation. This screening platform resulted in the enrichment of strains targeting virus-activated transcription factors, specific antiviral effectors, and intracellular pattern recognition receptors (PRRs). Interestingly, in addition to RIG-I, the PRR for IAV, a virus with the capacity to silence MDA5 also emerged as a dominant strain in wild type, but not in MDA5-deficient mice. Transcriptional profiling of infected knockout cells confirmed RIG-I to be the primary PRR for IAV but implicated MDA5 as a significant contributor to the cellular defense against influenza A virus.
Graphical Abstract
INTRODUCTION
Virus infection and subsequent replication poses a significant threat to the host cell. As a result, every living species has evolved mechanisms to both recognize infection and counteract it through a variety of means. In chordates, this response is mediated by the detection of pathogen-associated molecular patterns (PAMPs) through specialized PAMP recognition receptors (PRRs) - ultimately resulting in the secretion of Type I and III interferons (IFN-I and IFN-III, respectively) (Loo and Gale, 2011). In the vast majority of these cells, virus replication generates aberrant nucleic acid structures such as an exposed 5’ triphosphate (5ppp), di-phosphate (5pp) or double stranded RNA (dsRNA) (Loo and Gale, 2011). Sensing of viral nucleic acid is achieved by RIG-I (Retinoic-acid-inducible protein I, encoded by Ddx58) and/or MDA5 (melanoma-differentiation-associated gene 5, encoded by Ifih1), which are often referred to as RIG-I-like receptors (RLRs) to distinguish them from Toll like receptors (TLRs) (Loo and Gale, 2011). The RLR family consists of RIG-I, MDA5, and a third truncated member, LGP2 (laboratory of genetics and physiology 2), which consist of N-terminal tandem caspase activation and recruitment domains (CARDs), adenosine triphosphatase (ATPase) activity, and a C-terminal RNA binding domain (Loo and Gale, 2011). While LGP2 lacks CARD domains, engagement of RIG-I or MDA5 is thought to result in multimerization and assembly of a large complex at the mitochondria membrane that is mediated by K63 ubiquitin chains and polymerization of the mitochondrial antiviral-signaling protein MAVS (also called IPS-I, Cardif, and VISA) (Gack et al., 2007; Hou et al., 2011; Jiang et al., 2012). Complex assembly is responsible for the activation of the IKK and IKK-like kinases, which activate NF-κB and IRF3/7, respectively, resulting in the formation of the interferon beta (IFNB) enhanceosome (Loo and Gale, 2011). Transcriptional induction of IFNB results in both autocrine and paracrine activation of the JAK/STAT pathway and the coordinated upregulation of ~500 interferon stimulated genes (ISGs) (Loo and Gale, 2011). In addition, many ISGs can also be directly induced in response to virus infection as a result of PRR detection and IRF3/7 activation (Honda et al., 2005; Schmid et al., 2010).
Understanding the function of individual ISGs as they pertain to the cellular antiviral defense strategy is important. This information can be used to understand viral pathology, it can provide potential therapeutic drug targets, and it can inform individuals of their susceptibility to infection in this era of personalized genomics (Everitt et al., 2012; Zhang et al., 2014). For these reasons, ISG function has been studied in a variety of ways including large-scale overexpression screens where loss of virus replication is used to identify genes critical for coordinating this response (Li et al., 2013; Schoggins et al., 2011). While these screens successfully identified many of the primary orchestrators of the antiviral response, this strategy is unable to implicate multi-subunit effector molecules – where expression of any single transcript is incapable of eliciting an antiviral activity. In contrast to this approach, many high-throughput siRNA screens have also been utilized to identify host/pathogen interactions as a result of loss of function – an approach that will most certainly be further explored using CRISPR methodology (Brass et al., 2009; König et al., 2010; Krishnan et al., 2008; Li et al., 2009; Sessions et al., 2009; Shalem et al., 2014). While loss of function studies such as these have implicated a vast array of host transcripts that can influence virus replication, they are limited to indirect measurements of virus output and are not performed in vivo. While these studies have many valuable attributes and have resulted in significant findings, there is a need to adapt this approach to viruses replicating in the context of a physiological infection. Therefore, in an effort to expand upon the findings of traditional in vitro siRNA screens, we developed a platform to enable screening in vivo, thereby eliminating the need for a transformed cell line and allowing the direct measure of viral output (tenOever, 2013). To this end, we combined the silencing potential of siRNA libraries with the capacity of viruses to produce them in the form of artificial microRNAs (amiRNAs) (Varble et al., 2013).
While endogenous miRNAs are not believed to play a significant role in the direct response to most RNA viruses, this RNA-based regulatory system can be exploited to generate customized small interfering RNAs (siRNAs) (tenOever, 2013). In brief, host miRNAs derive as primary transcripts (pri-miRNA) and are recognized by a nuclear microprocessor composed of the RNAse III nuclease Drosha and its dsRNA binding partner DGCR8 (Bartel, 2009). The microprocessor cleaves the pri-miRNA into a precursor (pre-miRNA) hairpin that contains a signature 2nt 3’ overhang that ensures its cytoplasmic export and subsequent processing by the only other mammalian RNAse III nuclease, Dicer (Bartel, 2009). The resulting duplex RNA (~19–21nts) is then loaded into the RNA induced silencing complex (RISC) where it mediates post-transcriptional gene silencing on host mRNAs through partial complementarity. While miRNA-mediated repression rarely exceeds two fold, silencing can be potentiated if complementarity between miRNA and target is free of mismatches (tenOever, 2013). As both miRNA structure and targets can be engineered, this dynamic allows one to artificially interface miRNA biology with the field of mammalian virology (tenOever, 2013).
Here we apply an artificial miRNA-based screening approach to IAV in an effort to identify host restriction factors that significantly impair replication in vivo. We achieve this by enabling siRNA delivery through a replication-competent mouse-adapted IAV (A/Puerto Rico/8/34 (H1N1), herein referred to as PR8). In brief, we expanded on our findings that IAV can be engineered to synthesize a functional miRNA (Chua et al., 2013; Schmid et al., 2014; Varble et al., 2010) with the fact that the miR-124 scaffold was amenable to sequence changes (Varble et al., 2010). These findings permitted us to engineer IAV strains with the capacity to eliminate a desired host transcript in a sequence-dependent manner. We thus attenuated the parental PR8 strain by disrupting NS1 function in an attempt to restore replication with the addition of the siRNA. As reverse genetics of IAV demands individual rescues, we limited our library of siRNAs to one hundred host genes that are induced in response to IAV infection or have been previously implicated in the antiviral response. To this end, we generated an IAV-based library of viruses (two viruses/target gene) that are identical at a protein level, but with each virion encoding a unique mouse-specific siRNA capable of silencing a single virus-induced host transcript. This library was then administered to mice and selective pressure was used to parse out those host factors that restore replication of the attenuated virus. In vivo screening implicated a myriad of genes previously reported as mediators of the host response to IAV as well as MDA5, a PRR not predicted to be involved. This work reveals an unappreciated roll for MDA5 in the in vivo response of IAV and demonstrates the unique value in this screening platform.
RESULTS
Attenuating IAV to generate a platform for siRNA-mediated phenotypic rescue
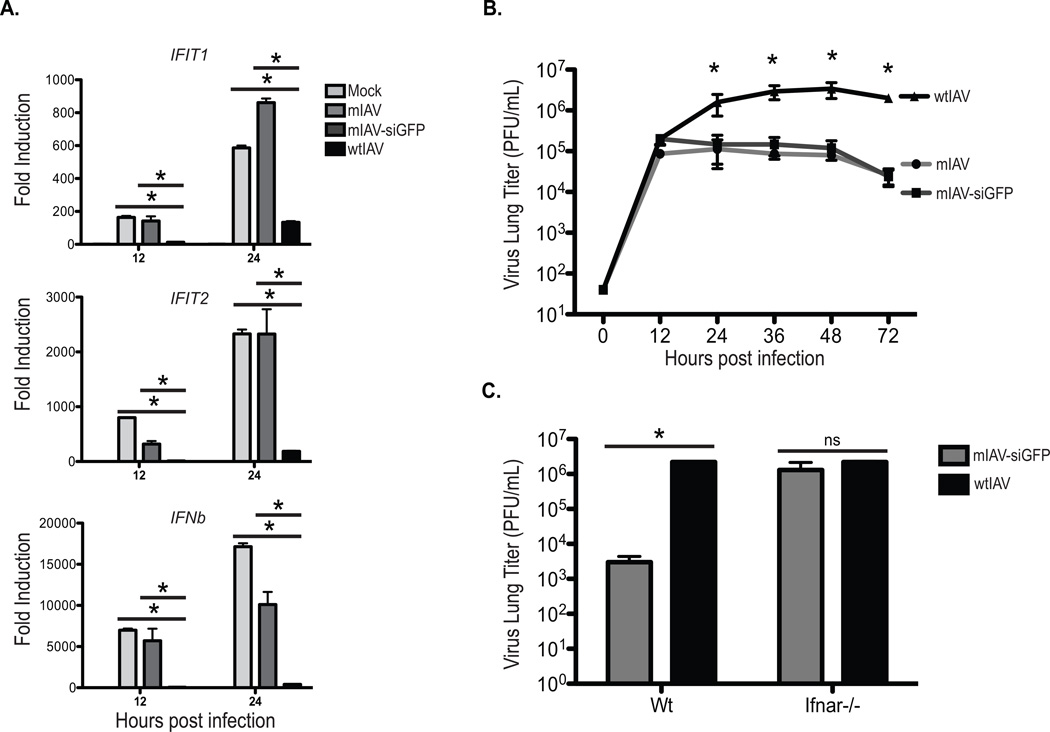
In an effort to identify the components of the IFN-mediated host antiviral defense system that effectively inhibit IAV replication, we chose to first disrupt the capacity of the virus from blocking PAMP detection. To this end, we mutated NS1, the major antagonist of the host antiviral defenses (Garcia-Sastre et al., 1998). Disruption of NS1 activity was achieved by a three amino acid substitution that impairs dsRNA binding (Donelan et al., 2003). These mutations render the virus incapable of blocking PAMP recognition and result in a virus that is attenuated by ~3logs (Donelan et al., 2003). In addition to this, we split the NS1 and NEP (also called NS2) open reading frames (ORFs) to permit a miRNA insertion point while also being mindful not to change the levels of svRNA or NEP on segment eight as these elements are critical to the IAV life cycle (Chua et al., 2013; Donelan et al., 2003; Perez et al., 2010; Varble et al., 2010). In agreement with published literature, virus rescue of the split NS1 mutant virus (mIAV) resulted in a strain that demonstrated significant transcriptional induction of IFNB (Ifnb) and ISGs (Ifit1 and Ifit2) as compared to wild type virus (Donelan et al., 2003; Varble et al., 2010) (Figure 1A). This increased IFN-based response was equally evident when the mIAV was engineered to encode an artificial siRNA against GFP (mIAV-siGFP) (Figure 1A). This cellular response could be further corroborated at a protein level with other ISGs, such as STAT1 (Figure S1). Furthermore, increased induction of these antiviral defenses in response to IAV infection was found to be inversely proportional to the level of virus replication (Figure 1B), a phenotype that was not present in mice lacking a functional IFN-I system (Figure 1C). Taken together, these results demonstrate the effectiveness of the host IFN-I response in the absence of NS1 and irrespective of whether the virus encodes a miRNA.
Figure 1. Virus-encoded siRNAs do not impose further attenuation on mIAV.
(A) Quantitative RT-PCR (qPCR) of IFIT1, IFIT2, and IFNb in A549 cells either mock treated or infected (MOI=0.1) with mIAV, mIAV-siGFP, or wtIAV. Error bars, SD, *p<0.05.
(B) Multi-cycle growth curves in A549s infected (MOI=0.01) with mIAV, mIAV-siGFP, or wtIAV. Error bars, SD, *p<0.05.
(C) Virus lung titers of wild type or Ifnar1−/− B6 mice infected with either the mIAV-siGFP, or wtIAV viruses. Mice were infected with 1 × 106 pfu of each virus and lungs were harvested 24 hpi. Error bars, SD, *p<0.05.
mIAV-derived siRNAs are capable of silencing during productive infection
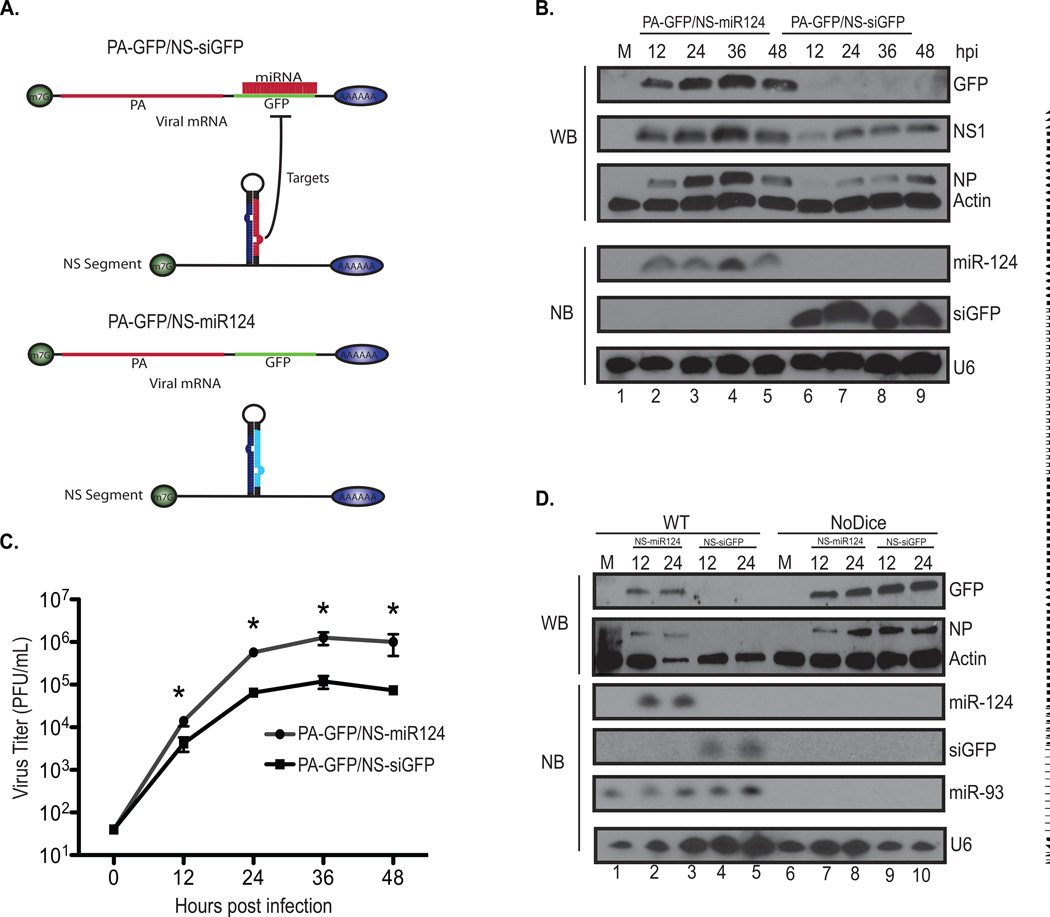
As our screen was intended to implicate important antiviral host genes in a physiological setting, we wanted to determine whether IAV infection provided adequate time to silence a transcript in the context of a single cell infection. This was especially important given that IAV, lacking a functional NS1, has been reported to induce the ribosylation of RISC (Seo et al., 2013). Therefore, to test our targeting strategy, we first determined whether we could engineer a miRNA-dependent, self-inactivating virus. To this end, we generated a virus encoding enhanced Green Fluorescent Protein (herein referred to simply as GFP) downstream of the polymerase PA component on segment three and combined this bicistronic segment with a recombinant NS segment encoding either miR-124 or a miR-124-based amiRNA targeting GFP (Figure 2A). We reasoned that should the virus with the capacity to silence GFP (and PA indirectly) achieve self-targeting, the biology of screening in this manner would be successful. Upon virus rescue, we observed that the virus encoding the siRNA against GFP resulted in significant attenuation and a corresponding loss of nucleoprotein (NP) and NS1 in contrast to the control virus expressing miR-124 (Figure 2B, 2C and S2A). To confirm that the attenuation observed by the self-targeted virus was siRNA-dependent, we infected human 293 cells deficient in Dicer (herein referred to as NoDice cells (Bogerd et al., 2014)) and observed no attenuation at the protein level or viral output confirming we had achieved siRNA-mediated self-targeting (Figure 2D and S2B). This result confirmed that we had achieved siRNA-mediated self-targeting (Figure 2D and S2B) and demonstrated that IAV can be engineered to generate a functional siRNA for which silencing of the target transcript can occur during the course of the viral life cycle and prior to ribosylation of RISC.
Figure 2. The kinetics of virus-derived siRNAs are sufficient to knockdown virus or IFN-induced genes.
(A) Schematic depicting a self-targeted virus (PA-GFP/ NS-siGFP) and the control virus (PA-GFP/NS-miR124). Shown is the PA segment encoding GFP and the NS segment encoding an siRNA targeting GFP (siGFP) or a non-targeting small RNA (miR-124).
(B) Multi-cycle growth curves in A549s infected (MOI=0.01) with viruses in (A). Error bars, SD, *p<0.05.
(C) Top three panels depict western blots of whole cell extract derived from A549 cells either mock treated or infected (MOI=0.01) with PA-GFP/NS-siGFP or PA-GFP/NS-miR124. Immunoblots were probed for GFP, IAV NP, IAV NS1, and Actin, which was used as a loading control. Bottom three panels depict northern blots of total RNA derived from A549s. Northern blots were probed for small RNA expression of microRNA-124 (miR-124) the GFP targeting siRNA (siGFP) and the host splicing RNA, U6, which was used as a loading control.
(D) Top two panels depict western blots of whole cell extract derived from wild type 293T cells or NoDice cells (293T cells lacking Dicer). Cells were either mock treated or infected (MOI=0.01) with PA-GFP/NS-siGFP or PA-GFP/NS-miR124 at the time points indicated. Immunoblots were probed for GFP, IAV NP, and Actin as in (C). Bottom four panels depict northern blots of total RNA derived from the conditions above. Northern blots were probed for small RNA expression of miR-124, siGFP, miR-93, and U6.
Enhancing replication of an attenuated IAV strain through siRNA incorporation
Given that the dynamics of virus-based silencing is best suited for genes that are transcriptionally induced in response to infection, we first performed RNAseq on mIAV-infected cells to detail the complete repertoire of virus-induced genes that demonstrated low mRNA levels at baseline (Figure S3A and Table S1). Using genes that demonstrated less than 50 reads at baseline but a 5-fold induction in response to virus, we collated a list of ISGs for siRNA design to which we also included a number of known virus-activated factors (Table S2). While previous siRNA libraries have traditionally been modeled to mimic hsa-miR-30a (Silva et al., 2005; Zeng et al., 2002), we wanted to determine whether modeling the library after another scaffold would improve processing and silencing. To this end, we tested the ability of a hairpin targeting EGFP at position 122 either in the background of hsa-miR-30a (pEGFP122-30) or mmu-miR-124-2 (pEGFP122-124) (Figure S3B). We determined that processing and silencing of this hairpin was more efficient utilizing mmu-miR-124-2 as a scaffold (Figure S3C, and 3D). In this regard, we applied the published attributes of effective siRNAs to target our antiviral gene list and ensured the 5’ ends of the desired stem where A/U rich and less thermodynamically stable than the corresponding 3’ ends of the resulting duplex RNA (Fellmann et al., 2011). These conditions were incorporated to ensure that the 3’ (3p) side of the hairpin would be loaded into the RISC – opposed to the 5p side (Figure 3A). Furthermore, in addition to using an attenuated, mouse-adapted strain of IAV, we also designed our siRNAs to be murine-specific to ensure there would be no biosafety concerns (Office of Science Policy, 2012). We then designed two artificial miRNAs per virus-induced gene and generated a library comprised of more than 200 unique hairpins that were individually synthesized and cloned into segment eight of our mIAV platform (Table S2). Recombinant mIAV strains were subsequently rescued individually, sequence verified, tittered, and used to generate a variety of stoichiometrically balanced libraries.
Figure 3. Constructing a library of siRNAs targeting virus or IFN induced genes.
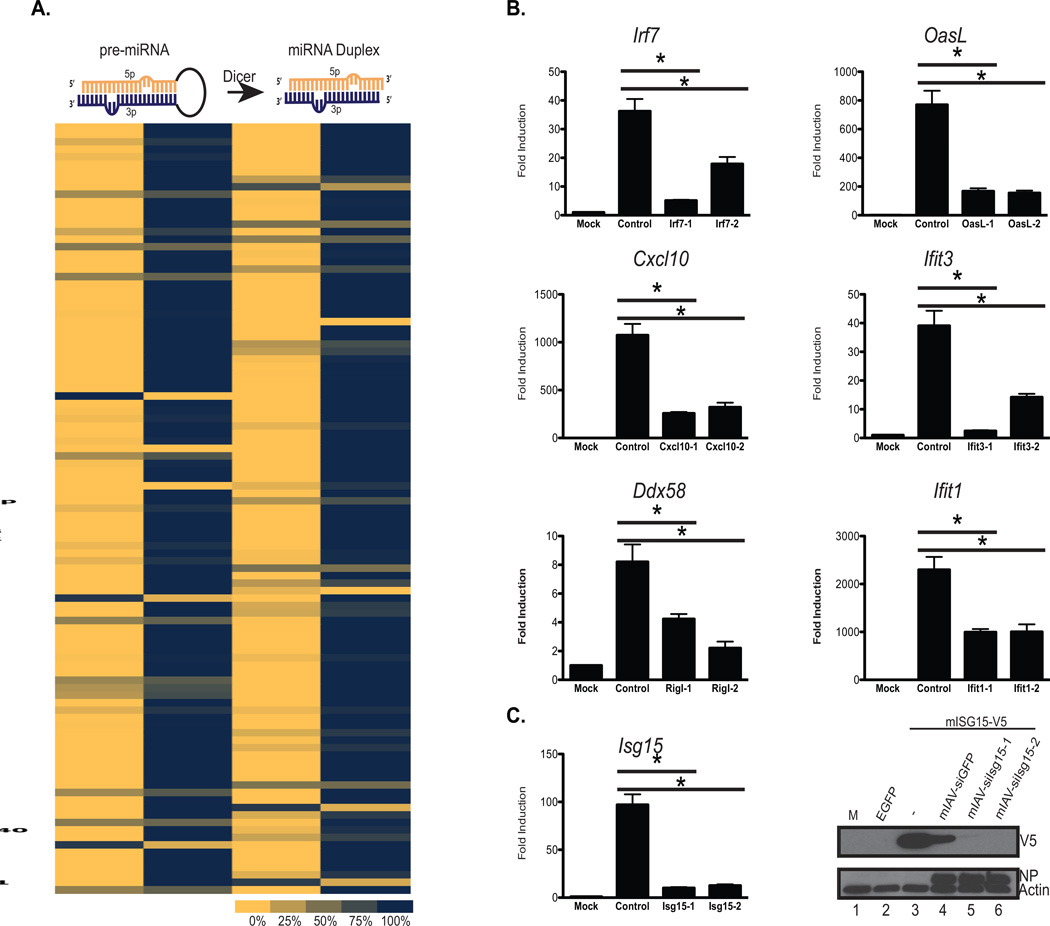
(A) Schematic depicting the structure of pre- and mature miRNA duplex with the 5p (orange) and 3p (blue) strands highlighted. Beneath this is a heat map of relative read counts derived from small RNAseq analysis from small RNAs derived from A549s infected with the IAV library. Results are shown as the proportion of the 5p and 3p produced for each siRNA.
(B) Quantitative RT-PCR (qPCR) of mRNA derived from mouse embryonic fibroblasts mock treated or infected (MOI=3) with viruses targeting IRF7, CXCL10, RIG-I, OASL, IFIT3, and IFIT1. Each qPCR was generated to determine the expression profile of the cognate target. Each virus set included two unique siRNA hairpins (−1 or −2). Error bars, SD, *p<0.05.
(C) RNA and protein analysis of ISG15 from MEFs mock treated or infected (MOI=3) with the corresponding siISG15 viruses (Isg15-1 or Isg15-2). Top panel depict qPCR analysis for ISG15 as described in (B), bottom panel depicts a western blot of whole cell extract derived from 293 cells transfected with epitope tagged (V5) murine ISG15 and infected with the aforementioned viruses. Immunoblots were probed form V5, IAV NP, and Actin.
In an effort to propagate the mIAV-based siRNA libraries, we administered them to transformed human alveolar cells (A549), where specific targeting should not occur, and the small RNA population was monitored 12hrs post infection to ascertain the accuracy of processing. In agreement with previous studies, we found mIAV infection did not significantly alter the miRNA profiles of the host (Figure S3E), whereas the individual profiles of miR-124- based small RNAs demonstrated that 83% of the hairpins processed only the 3p strand, 13% processed both 3p and 5p, 2% processed only 5p, and 2% failed to produce an siRNA (Figure 3A). Intracellular levels of artificial mIAV-based siRNAs were found to be comparable to the most abundant read numbers for endogenous miRNAs, supporting earlier findings that RNA virus-mediated production can generate up to 60000 copies of the siRNA/cell within 8hrs of infection (Varble et al., 2010). Furthermore, individual verification of >10% of these hairpins found them to have potent silencing potential at the mRNA level for transcripts encoding PRRs, transcription factors, and components of the antiviral response (Figure 3B). This activity could also be corroborated at a protein level (Figure 3C). Collectively, these data show we can successfully generate IAV with the capacity to silence a host factor in the context of a productive infection.
in vivo RNAi screen identifies host factors that restrict IAV replication
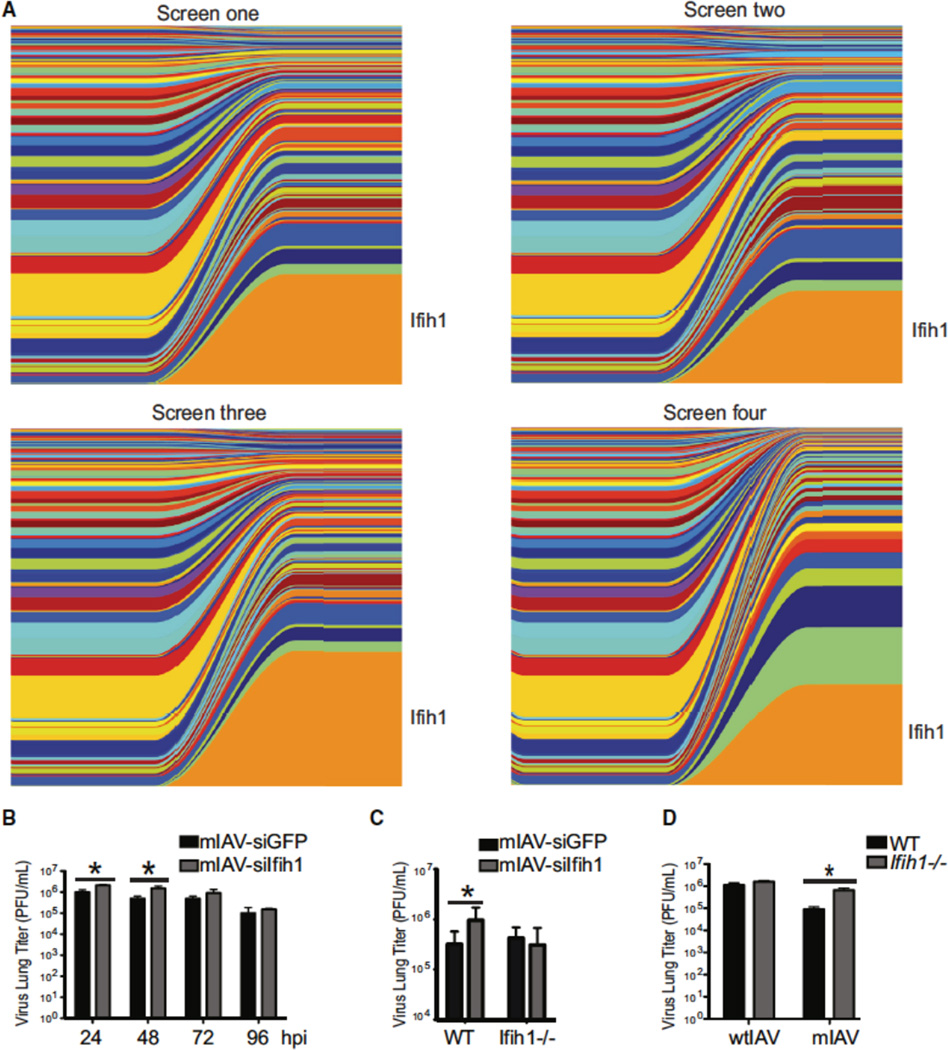
Following assembly and characterization of the virus-induced siRNA gene library (Figure 3), we administered the complete library to individual animals at a dose of 1 × 106 plaque forming units (pfu) – the estimated lethal dose for 50% of the animals (LD50) (Donelan et al., 2003). At this dose we were confident that all of the hairpins represented in the library would be present during infection. Following 96hrs of infection, ~12 generations for the virus, animals were sacrificed and the lungs were removed and homogenized. The supernatant from lung homogenates demonstrated an average titer of 1 × 104 pfu from each of the animals screened - reflecting the poor replication capacity of mIAV (Figure S4A). Furthermore, the cellular fraction from for each lung was used to isolate total RNA from which the small RNA library could be profiled using standard RNAseq. Individual reads mapping to the miR-124-based library were aligned and consolidated using Bowtie and custom made shell scripts and subsequently graphed using MatLab where individual hairpins were designated unique colors (Figure 4A). Input levels were determined by RNA derived from the virus library prior to in vivo administration. Based on the width of the band, one can ascertain the overall percentage of a given hairpin in the population (Varble et al., 2014). As such, the over-representation of a common color across all four animals depicts a hairpin whose relative increase was consistent in each screen. Interestingly, this line of experimentation demonstrated that both viruses designed to silence MDA5 (Ifih1-1 and -2) were enriched greater than 50-fold in every screen; of which one dominated every virus population (from 0.26% of input to 26%–31% of output) (Table S3). In addition to MDA5, gene enrichment of greater than 25 fold was also observed across all four screens for more than 30 host factors, six of which were represented by both gene-specific hairpins. While some evidence of off target effects was present, the predominant enrichment of host factors known to be implicated in the cellular response to IAV (i.e. PRRs, NFκB, and IRFs), suggested that the screen was successful (Table S3).
Figure 4. Infection with the library in mice selects for a virus targeting Ifih1.
(A) Graphical depiction of siRNA composition derived from deep sequencing data of mice infected with 1 × 104 pfu of the pooled library. Left side of each graph represents the proportion of unique viral strains, as defined by their siRNA. Right side of each graph depicts in vivo selection dynamics following four days of infection.
(B) Levels of virus replication from mice infected with 1 × 106 pfu of mIAV-siGFP or -siIfih1 and assessed at the indicated time points. Error bars, SD, *p<0.05.
(C) Lung titers from wild-type or Ifih1−/− mice infected with 1 × 106 pfu of mIAV-siGFP or -siIfih1 as measured 24 hours post infection. Error bars, SD, *p<0.05.
(D) Lung titers from wild-type mice or Ifih1−/− mice infected with 1 × 104 pfu of wtIAV or mIAV measured 48 hours post infection. Error bars, SD, *p<0.05.
To ensure that virus selection was not determined by the stoichiometic composition of the starting library, we performed three additional screens with unique sub-libraries composed of ~80 hairpins each for which we excluded both Ifit1-targeting viruses. Each sub-library (A, B or C) was administered to a small cohort of mice and lungs were harvested four days post infection. In agreement with our primary screen, we again observed a strong selection for viruses targeting PRRs (Ddx58), NFκB (p50 and p65), and members of the IRF family (Irf5 and Irf7) (Table S4). Taken together, these results demonstrate the robustness of this screening platform and illustrate that selection can be used to identify host factors that play a significant role in inhibiting virus replication.
Given that the PRR responsible for IAV has been demonstrated to be RIG-I (Gitlin et al., 2006; Kato et al., 2006), we decided to further investigate the enrichment of the dominant MDA5-targeting virus. To first ensure that this virus did not encode a mutation that conferred a replication advantage independent of the siRNA, we plaque-purified and sequenced the virus using the RNAseq platform and determined that it did not undergo any mutations from the parental strain beyond the two custom stems of the recombinant hairpin (Table S5). Next, to ensure that the increased representation was due to a murine-specific silencing event, we compared virus replication rates in mIAV strains encoding hairpins to GFP (mIAV-siGFP) or the selected hairpin designed to target MDA5 (mIAV-siIfih1) (Figure S4A). To this end, we infected Madin-Darby canine kidney (MDCK) cells with the aforementioned viruses and monitored replication at 0, 12, 24, 36, and 48hrs post infection. Virus titers for both mIAV-siGFP and – siIfih1 were found to rapidly increase in the first 12hrs and subsequently plateau at a concentration of 1 × 107 pfu/mL with no significant strain differences. Together these data suggest the virus-encoded hairpin does not confer a replication advantage in a non-murine system. To determine if mIAV-siIfih1 maintained an elevated level of replication independent of the original in vivo competition experiment where it was selected, we infected wild type mice with mIAV-siGFP or –Ifih1 and determined lung titers at 24, 48, 72, and 96hpi (Figure 4B). Titer data found a modest, but significant increase in mIAV-siIfih1 titers as compared to mIAV-siGFP at 24 and 48hpi – thereafter showing comparable titers as the animals began clearing virus.
Next, to ascertain why mIAV-siIfih1 demonstrated elevated titers, we examined the targeting potential of the artificial miRNA selected. The mIAV-siIfih1 hairpin was designed to target Ifih1, the gene encoding MDA5, and was found to be abundantly expressed by the virus (Figure S4B). However, our earlier analysis of hairpin processing demonstrated that this hairpin was also one of those identified for generating a 5p strand (Figure 3A). Interestingly, the 5p strand of the miRNA was a near perfect match for Setd7, a methyltransferase implicated in controlling the transcriptional output of p53 (Kurash et al., 2008) (Figure S4C). To first determine which of these targets, Setd7 and/or Ifih1, may be responsible for the increased representation of mIAV-siIfih1, we ascertained silencing capacity of this hairpin for both transcripts. To this end, viruses designed to target GFP or Ifih1 were used to infect murine fibroblasts and the endogenous levels of Ifih1 and Setd7 were determined by qPCR and Western blot (Figure S4D and S4E). This approach found that the mIAV-siIfih1 virus was a potent silencer of Ifih1 but failed to suppress Setd7, presumably owing to the fact that the conditions for optimal silencing were not fulfilled by this siRNA for this unintentional target. The capacity of mIAV-siIfih1 to silence Ifih1 directly could be further confirmed in cells lacking Dicer, where no loss of transcript was observed (Figure S4F). These data demonstrate that the increase in mIAV-siIfih1 virus replication cannot derive from the silencing of SETD7 and suggest this activity is the result of targeting MDA5.
Next, to directly test whether increased replication of this NS1 mutant virus encoding the Ifih1 siRNA was the result of decreased MDA5, we infected wild type and Ifih1−/− mice to determine whether this would result in a loss of phenotype (Figure 4C). To this end, mIAV-siGFP or -siIfih1 viruses were administered intranasally and lung titers were determined at 48hpi. In contrast to wild type mice, where the mIAV-siIfih1 strain again demonstrated a modest but significant elevation in viral titer, no significant replication differences were observed in the absence of MDA5 (Figure 4C). Finally, to confirm that these results were not the indirect by-product of our engineered viruses, we additionally infected wild type and Ifih1−/− mice with strains lacking miRNAs. To this end, we administered wtIAV or mIAV to WT and Ifih1−/− mice and found that, in the absence of a functional NS1, the loss of MDA5 resulted in a significant elevation in titer (Figure 4D).
Defining the contribution to individual PRRs in response to IAV
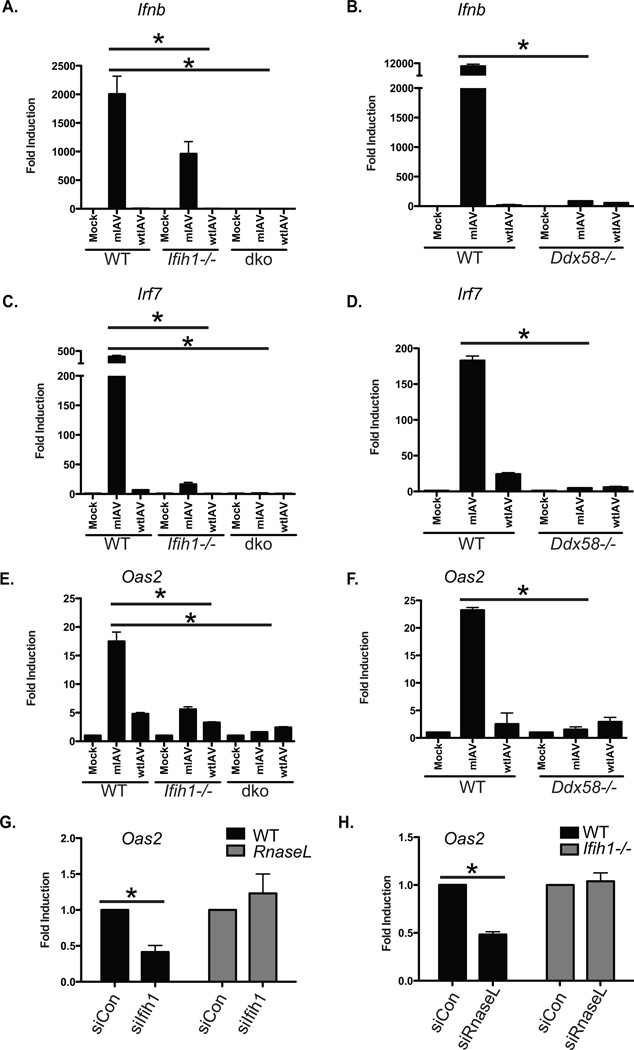
Cellular recognition of IAV infection has been demonstrated to be the result of recognition of the 5’ triphosphate PAMP found on the end of each viral genomic segment (Schlee et al., 2009). It was therefore unexpected to select an IAV strain with the capacity to silence MDA5, the PRR implicated in recognition of complex dsRNAs, which had been reported not to be involved in IAV-mediated induction of IFNB (Gitlin et al., 2006; Kato et al., 2006; Pichlmair et al., 2009). To address this apparent contradiction, we determined the induction of IFNB (Ifnb) in primary murine embryonic fibroblasts lacking MDA5 (Ifih1−/−) or both MDA5 and RIG-I (Mda5−/−/Ddx58−/−, herein referred to simply as double knockout out; dko) (Figure 5A). In agreement with Gitlin et al., (Gitlin et al., 2006) we found that loss of MDA5 results in only a moderate decrease of Ifnb message which was completely ablated in the absence of both PRRs (Figure 5A). To determine the relative contributions of MDA5 and RIG-I, we next performed the same experiment in RIG-I deficient fibroblasts (Ddx58−/−) and, again in agreement with past studies (Gitlin et al., 2006; Kato et al., 2006), found RIG-I expression was essential for the transcriptional induction of Ifnb (Figure 5B).
Figure 5. MDA5 is a non-redundant pattern recognition receptor for IAV.
(A) qPCR of Ifnb derived from wild-type, Ifih1−/−, or Ddx58/Ifih1−/− (dko) murine fibroblast cells either mock treated or infected with mIAV or wtIAV (MOI=2). Error bars, SD, *p<0.05.
(B) qPCR of Ifnb derived from wild-type, or Ddx58−/− murine fibroblast cells either mock treated or infected with mIAV or wtIAV (MOI=2). Error bars, SD, *p<0.05.
(C) qPCR as described in (A) for Irf7.
(D) qPCR as described in (B) for Irf7.
(C) qPCR as described in (A) for Oas2.
(D) qPCR as described in (B) for Oas2.
F) qPCR of Oas2 derived from wild-type, or RnaseL−/− murine fibroblast cells either mock treated or infected with mIAV (MOI=2). Error bars, SD, *p<0.05.
(G) qPCR of Oas2 derived from wild-type, or Ifih1−/− murine fibroblast cells either mock treated or infected with mIAV (MOI=2). Error bars, SD, *p<0.05.
To ascertain the underlying cause for the selection of the MDA5 siRNA-containing virus, we next examined whether loss of this PRR impacted the cellular response to virus – independent of the transcriptional induction of Ifnb. To this end, we analyzed Ifih1−/−, Ddx58−/−, and dko cells for Irf7 transcript levels, an interferon inducible transcription factor that serves as a master regulator for both IFN-dependent and –independent gene induction (Honda et al., 2005; Schmid et al., 2010) (Figure 5C and D). Interestingly, despite transcriptional induction of Ifnb, the absence of MDA5 was found to reduce Irf7 induction by more than 20 fold (Figure 5C). Loss of Irf7 was also observed in Ddx58−/− and the dko fibroblasts (Figure 5C and D). Comparable to Irf7, transcriptional induction patterns of Oas2, Ifit1, Stat1, and Isg15 also demonstrated dependence on both RIG-I and MDA5 (Figure 5E, 5F and S5A–F). These results suggest that MDA5 contributes to the amplification of the antiviral state but in a manner that is downstream of RIG-I-mediated viral recognition, a process that is normally inhibited by NS1 (Mibayashi et al., 2007).
As loss of MDA5 was impacting the amplification of the host response, opposed to the induction of IFN-I, we next sought to determine the molecular mechanism for virus selection. In this regard, we found it noteworthy that screening in the absence of the Ifih1-targeting viruses resulted in the selection of clones targeting OAS isoforms, the virus-induced activators of RNAseL (Table S4). Given the selection for OAS isoforms and the recent implication of the OAS/RNAseL system in MDA5 activation (Luthra et al., 2011; Malathi et al., 2007), we next investigated whether this intrinsic cellular defense against IAV was responsible for amplifying the host antiviral response. Indeed, this same pathway has been suggested to be an effective antiviral activity against IAV and one of the primary targets of the NS1 protein (Cooper et al., 2015; Min and Krug, 2006). Moreover, the recent finding that MDA5 aids in exposing viral dsRNA to the host machinery would suggest that MDA5 could enhance the activity of the OAS/RNAseL system (Yao et al., 2015). Given this, we determined whether MDA5-dependent induction of OAS2 was impacted by the expression of RNaseL. Interestingly, while depletion of MDA5 resulted in a 50% loss of the OAS2 transcript in wild type cells, this phenotype was absent in fibroblasts deficient for RNase L expression (Figure 5G and S5G). Moreover, if we silenced RNaseL, we also reduced OAS2 expression, but only in the presence of MDA5 (Figure 5H and S5H). Taken together, these data suggest that MDA5 contributes to an enhancement in the antiviral response through the recognition of the OAS/RNAseL system.
Determining the biological role for MDA5 in IAV infection
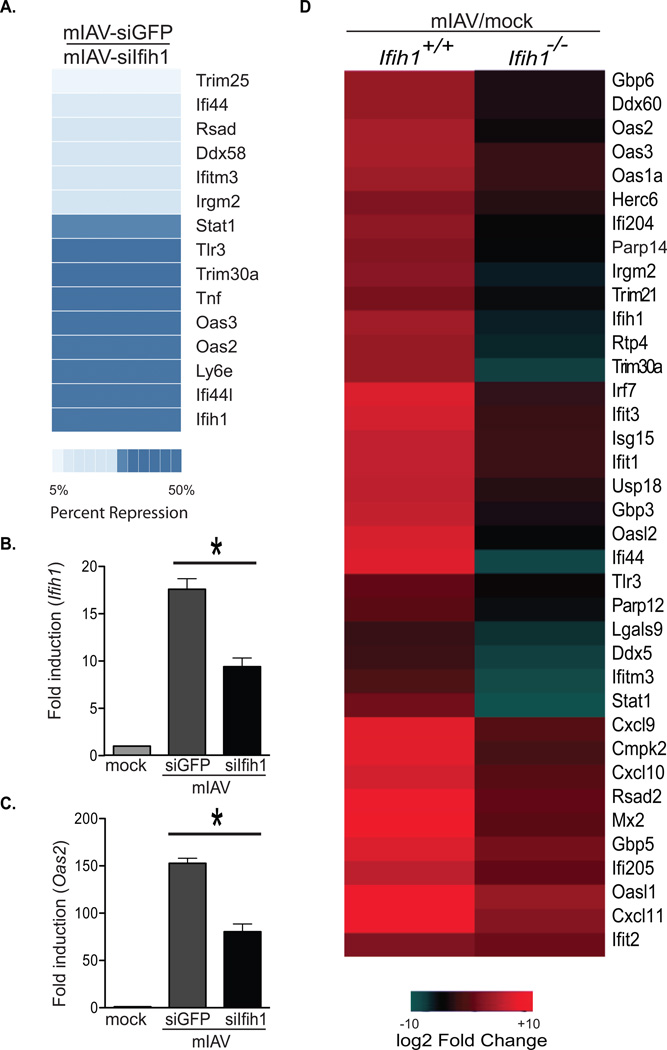
Next, to better define the biological role of MDA5 in the cellular response to IAV, we repeated the infections of mIAV-siGFP and -siIfih1 in murine fibroblasts and monitored the transcriptomes using biological replicates of mRNAseq data (Figure 6A). In agreement with earlier results, mRNAseq confirmed Ifih1 as being significantly reduced with no change observed in Setd7 levels between experimental conditions (Table S6). Interestingly, comparing mIAV-siGFP to –siIfih1 transcriptomes, we found silencing of a small subset of known virus-induced host genes including: Ifih1, Stat1, Oas2 and Oas3, results that could be independently corroborated by qPCR (Figure 6B–C and S6).
Figure 6. Infection with mIAV-siIfih1 results in an altered ISG profile.
(A) Transcriptional profiling of murine fibroblasts infected (MOI=2) with mIAV-siGFP or -siIfih1 as determined by mRNAseq. The heat map depicts fold repression of mIAV-siGFP-induced genes in response to mIAV-siIfih1.
(B) qPCR of Ifih1 derived from samples described in (A). Error bars, SD, *p<0.05.
(C) qPCR as described in (B) for Oas2
(D) Heat map depicting expression changes in a subset of antiviral genes in wild-type or Ifih1−/− murine fibroblasts infected with mIAV (MOI=2) for 12 hours. Values represent log2 fold induction of infected over uninfected cells.
To further ensure that the MDA5 contribution to the IAV screen selection was independent of the engineered hairpin, we infected MDA5 knockout cells with mIAV, lacking any hairpin, and repeated the mRNAseq profiling methodology. Transcriptome mapping of these cells, like mIAV-siIfih1, showed no defect in Ifnb induction but demonstrated a significantly muted induction of ISGs (Figure 6D). Taken together, these results suggest that MDA5, while not directly responsible for IFN-I induction upon IAV infection, participates in a process which amplifies the host antiviral response.
Conservation of MDA5 function as a restriction factor for IAV
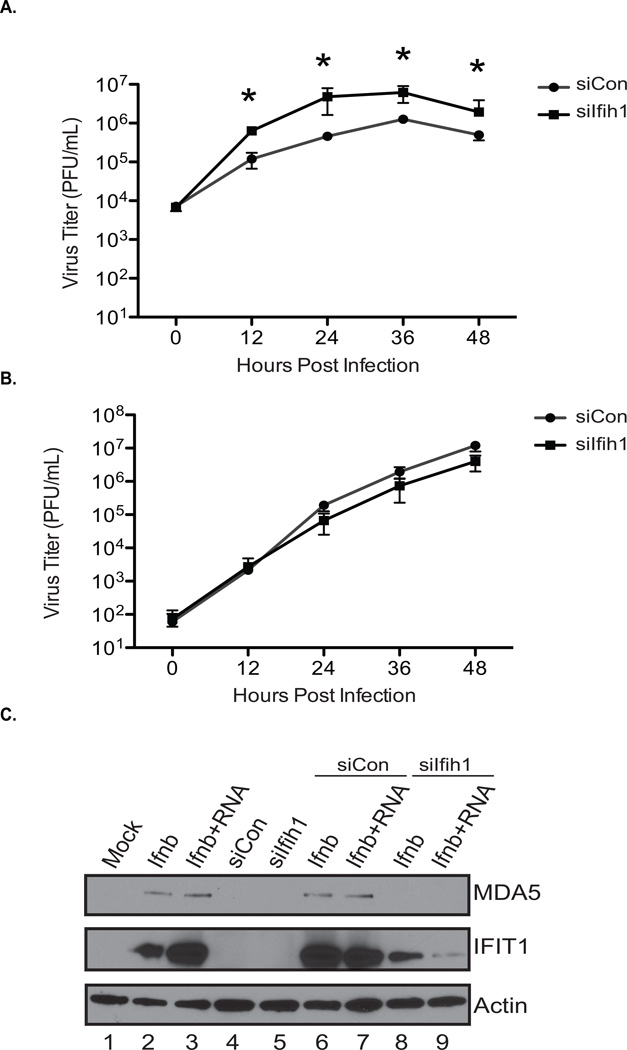
Lastly, to verify the general involvement of MDA5 in the response to IAV, we examined whether loss of this transcript in human cells also resulted in increased virus replication and a muting of the antiviral response as observed in mice. As our artificial miRNAs were designed to be murine-specific, we used standard siRNA transfection to silence MDA5 in the human respiratory cell line A549. Following complete silencing of MDA5 (Figure S7A), we infected cells with mIAV, lacking a hairpin, and measured titers at 0, 12, 24, 36, and 48hpi (Figure 7A). In agreement with our murine data, these studies found that loss of MDA5 significantly enhanced virus replication by ~1 log at each time point measured. This phenotype was lost when wild type IAV was administered – further supporting the hypothesis that wild type NS1 can interfere with MDA5 function (Figure 7B). Lastly, to determine whether the increased replication in the absence of MDA5 correlated to a loss of antiviral gene induction, we again treated cells with control or IFIH1-specific siRNAs and subsequently treated cells with recombinant IAV PAMP RNA and/or recombinant IFN-I to mimic the conditions of infection in the absence of virus replication or expression of viral antagonists (Figure 7C). These data demonstrated that, in agreement with mRNAseq analyses from murine infections, loss of MDA5 results in decreased expression of virus-induced genes including IFIT1 for which levels were reduced by more than 1000 fold (Figure 7C and S7B).
Figure 7. MDA5 function of IAV detection is conserved between mouse and human cells.
(A) Multi-cycle growth curves performed in A549 cells treated with a scrambled (scbl) or IFIH1- specific siRNAs. Cells were infected (MOI=0.01) with mIAV and supernatants collected and plaqued at the indicated time points. Error bars, SD, *p<0.05.
(B) Multi-cycle growth curves as described in (A) using wild type IAV in place of mIAV.
(C) Western Blot of whole cell extract derived from A549 cells treated as in (A) and subsequently transfected with IAV RNA and/or administered IFNβ. Immunoblots were probed for MDA5, IFIT1, and Actin.
Given that virus-mediated induction of Ifit1 was also found to require RIG-I (Figure S5), we next aimed to address why our screen did not result in the dominant selection of a mIAV-siDdx58 virus. To this end, we generated a recombinant virus that targeted human RIG-I and performed a multi-cycle growth curve in the A549 cell line (Figure S7C). As expected, we observe a significant decrease in RIG-I levels which correlated to an increase in the IAV nucleoprotein (NP). In addition, titers from these experiments demonstrated an ~1 log increase in viral growth when RIG-I expression was reduced, a phenotype which was lost in cells lacking RIG-I (Figure S7D and S7E). Given these results, we next sought to determine what the baseline levels of RIG-I are in vivo - as high baseline levels of RIG-I would explain why targeting the mRNA would not offer the selective advantage of other host antiviral genes. In agreement with the Human Proteome Map data, western blot of whole lung demonstrated robust levels of RIG-I, opposed to a complete lack of expression of MDA5 (Figure S7F and S7G). Taken together, these data suggest that the lack of a dominant RIG-I selection is the result of high basal levels of this PRR in vivo and implicate MDA5 in the amplification of a RIG-I-dependent response.
DISCUSSION
The cellular response to virus infection can determine the overall outcome for the host. For this reason, elucidating how such a defense system is orchestrated is critical to understanding viral pathogenesis. In this study, we utilize an in vivo siRNA screening technique that allows us to define host factors with significant inhibitory potential in the context of a bona fide infection. This method involves generating siRNAs modeled after endogenous miRNAs to increase their silencing potential. It should be noted that we do not observe any dramatic difference in host miRNA levels in the context of an IAV infection, regardless of whether it encodes its own siRNA (Langlois et al., 2012; Varble et al., 2010), suggesting this approach would not have indirectly impaired the antiviral response. Given this, we provided an attenuated strain of IAV with the individual capacity to silence antiviral response genes. These efforts identified MDA5, a pattern recognition receptor that is generally believed to recognize complex dsRNA, a PAMP not associated with IAV infection (Pichlmair et al., 2009; Weber et al., 2006). In fact, two separate studies directly investigated whether IAV, lacking a functional NS1, were detected by MDA5 (Gitlin et al., 2006; Kato et al., 2006). In both reports, the authors concluded that detection of IAV was based exclusively on RIG-I, however, these conclusions were based on the transcriptional induction of Ifnb – results which are in agreement with those presented here (Gitlin et al., 2006; Kato et al., 2006). In fact, it was the lack of the IFN-I phenotype that prompted us to ascertain how the transcriptome might have been influenced by the loss of MDA5 to result in the in vivo selection process. Given that MDA5 had previously been implicated in the induction of a subset of chemokines in response to IAV lacking NS1 (Kim et al., 2014), we hypothesized that this PRR may contribute to the antiviral defenses of the cell in an IFN-independent manner. Interestingly, RNAseq data from IAV encoding an siRNA to MDA5 or from IAV-infected MDA5 deficient cells both demonstrated a common subset of antiviral genes that were lost without a defect in IFN-I. While it is interesting that loss of MDA5 (Ifih1) also resulted in decreased RIG-I expression (Ddx58), selection of the Ifih1 virus presumably resulted from the direct loss of an antiviral genes, notably OAS2 and OAS3, especially given the implication of the OAS/RNaseL antiviral system inhibiting IAV replication in the absence of wild type NS1 (Min and Krug, 2006). In this regard, it is interesting to note that this same discovery was made with paramyxovirus, in which the V accessory protein has also been found to be a potent antagonist of MDA5 leading many to conclude early on that only RIG-I was involved in the host response to these negative stranded RNA viruses (Andrejeva et al., 2004; Childs et al., 2007; Motz et al., 2013).
The antiviral function of MDA5 as it relates to both orthomyxoviruses and paramyxoviruses is noteworthy. As these viruses do not produce a significant amount of dsRNA in the course of their life cycle, with the exception of defective interfering particle formation (Tapia et al., 2013; Yount et al., 2008), it suggests that MDA5 may have additional recognition capacities. This is certainly true for the recent implication of MDA5 in its ability to detect 5’ caps lacking 2’O-methyl groups (Zust et al., 2011). While aberrant caps are unlikely to be involved in the detection of either orthomyxoviruses or paramyxoviruses, the recent discovery that MDA5 can serve to displace viral proteins from masking dsRNA would certainly explain why silencing MDA5 provides a selective advantage to IAV in our in vivo screen (Yao et al., 2015). In this regard, the fact that MDA5 is both virus- and IFN-inducible hints at the possibility that MDA5 may also detect aberrant RNAs formed in the context of the host IFN-I response – including the by-products of RNaseL cleavage. Such cross talk would thus explain why silencing of MDA5 has a modest impact on IFIT1 even in the absence of virus-derived PAMP (Figure 7C).
It is also of note that the dynamics of this screening method seem to select for only the most advantageous siRNAs. While quadruplicate screens independently selected for both viruses designed to target MDA5, only one emerged to dominate the overall population. Similarly, while well-characterized antiviral host factors were also enriched in each screen, these viruses did not amplify to the extent of mIAV-Ifih1. It is our hypothesis that this phenotype is a result of the complex dynamics that result in the context of this in vivo screen. For example, aside from differences in silencing efficiency for a given siRNA, the baseline levels of the target protein, as well as its inherent stability, would have significant implications on whether silencing the cognate mRNA would provide a replication advantage; this is perhaps best exemplified by the data on RIG-I (Figure S7).
Finally, it should be noted that the knowledge obtained from this screening platform could in no way be used to enhance the pathogenesis of IAV. Given the antagonistic potency of NS1 (Chua et al., 2013), the additional silencing capacity is unlikely to provide any replication advantage to an evolutionarily optimized wild type strain. However, generating a library of IAV strains lacking a functional NS1 allows for selection of increased fitness only as it relates to that population. To this end, this technology can be safely adopted to identify factors responsible for virus tropism, transmission, or replication and will undoubtedly continue to result in a greater understanding of the interactions between IAV and its host.
EXPERIMENTAL PROCEDURES
Artificial microRNA Design
For constructing the artificial miRNA targeting library, we selected 100 IAV-induced genes from mIAV-GFP infection in A549 cells (Table S1). Genes were chosen based on their known antiviral function or overall induction in response to infection and prioritized to favor those with low base line expression (Table S2). The online tool DSIR (http://biodev.cea.fr/DSIR/DSIR.html) was used to design multiple siRNAs for each transcript. The highest scoring siRNAs for each gene, that also conformed to more recent “rules” for artificial miRNAs (Fellmann et al., 2011), were modeled after mmu-miR-124-2 in which the loop, and the first 50 nucleotides upstream and downstream of the stem were left intact (Table S3). The resulting hairpins were then chemically synthesized individually (DNA 2.0) and introduced by InFusion cloning (Clonetech) into a BstEII site of the modified segment 8 of IAV as previously described (Varble et al., 2010).
Small RNA and mRNA deep sequencing
Small RNA sequencing from the artificial miRNA library was performed from total RNA harvested 24 hrs post IAV infection and processed as previously described (Pfeffer et al., 2005). The monitoring of virus populations was accomplished by generating cDNA from viral RNA using Superscript II and random hexamers. Primers containing Illumina linkers were used to amplify the amiRNA hairpins and deep sequencing analysis was done on a MiSeq instrument (Illumina). Custom Shell and Python scripts were used to analyze the data using Star as the primary aligner. MatLab and R were used to visualize virus populations and transcriptome data, respectively. For mRNAseq, isolated mRNA was processed using the Truseq Illumina kit (Illumina) following the manufacturer’s instructions and run on a HiSeq 2500 instrument (Illumina).
Supplementary Material
Acknowledgements
We wish to thank Drs. B. Cullen (Duke University) for NoDice 293 cells, A. Tarakhovsky (Rockefeller University) for murine cells lacking Dicer, M. Gale (Washington University) for Ddx58−/−, Ifih1−/−, and Ddx58/Ifih1 knockckout murine fibroblasts, and R.H. Silverman for the RnaseL knockout murine fibroblasts. This work was supported by the Burroughs Wellcome Fund. BRT is supported by the National Institute of Allergy and Infectious Diseases grants R01AI110575 and R01AI093571. AAB is supported by the National Institute of Health grant 1F31AI110130-01A1. CBL is supported by the National Institute of Allergy and Infectious Diseases grant R01AI083284.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci U S A. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Whisnant AW, Kennedy EM, Flores O, Cullen BR. Derivation and characterization of Dicer- and microRNA-deficient human cells. RNA. 2014;20:923–937. doi: 10.1261/rna.044545.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs K, Stock N, Ross C, Andrejeva J, Hilton L, Skinner M, Randall R, Goodbourn S. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology. 2007;359:190–200. doi: 10.1016/j.virol.2006.09.023. [DOI] [PubMed] [Google Scholar]
- Chua MA, Schmid S, Perez JT, Langlois RA, Tenoever BR. Influenza A virus utilizes suboptimal splicing to coordinate the timing of infection. Cell Rep. 2013;3:23–29. doi: 10.1016/j.celrep.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DA, Banerjee S, Chakrabarti A, Garcia-Sastre A, Hesselberth JR, Silverman RH, Barton DJ. RNase L targets distinct sites in influenza A virus RNAs. J Virol. 2015;89:2764–2776. doi: 10.1128/JVI.02953-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donelan NR, Basler CF, Garcia-Sastre A. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J Virol. 2003;77:13257–13266. doi: 10.1128/JVI.77.24.13257-13266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, Chin CR, Feeley EM, Sims JS, Adams DJ, et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellmann C, Zuber J, McJunkin K, Chang K, Malone CD, Dickins RA, Xu Q, Hengartner MO, Elledge SJ, Hannon GJ, et al. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Mol Cell. 2011;41:733–746. doi: 10.1016/j.molcel.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Kinch LN, Brautigam CA, Chen X, Du F, Grishin NV, Chen ZJ. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity. 2012;36:959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kim WK, Jain D, Sanchez MD, Koziol-White CJ, Matthews K, Ge MQ, Haczku A, Panettieri RA, Jr, Frieman MB, Lopez CB. Deficiency of melanoma differentiation-associated protein 5 results in exacerbated chronic postviral lung inflammation. Am J Respir Crit Care Med. 2014;189:437–448. doi: 10.1164/rccm.201307-1338OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, et al. Human host factors required for influenza virus replication. Nature. 2010;463:813–817. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F, et al. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurash JK, Lei H, Shen Q, Marston WL, Granda BW, Fan H, Wall D, Li E, Gaudet F. Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Mol Cell. 2008;29:392–400. doi: 10.1016/j.molcel.2007.12.025. [DOI] [PubMed] [Google Scholar]
- Langlois RA, Varble A, Chua MA, Garcia-Sastre A, tenOever BR. Hematopoietic-specific targeting of influenza A virus reveals replication requirements for induction of antiviral immune responses. Proc Natl Acad Sci U S A. 2012;109:12117–12122. doi: 10.1073/pnas.1206039109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ding SC, Cho H, Chung BC, Gale M, Jr, Chanda SK, Diamond MS. A short hairpin RNA screen of interferon-stimulated genes identifies a novel negative regulator of the cellular antiviral response. MBio. 2013;4:e00385–e00313. doi: 10.1128/mBio.00385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci U S A. 2009;106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra P, Sun D, Silverman RH, He He. Activation of IFN-β expression by a viral mRNA through RNase L and MDA5. Proc Natl Acad Sci U S A. 2011;108:2118–2123. doi: 10.1073/pnas.1012409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malathi K, Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mibayashi M, Martinez-Sobrido L, Loo YM, Cardenas WB, Gale M, Jr, Garcia-Sastre A. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol. 2007;81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min JY, Krug RM. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2'–5' oligo (A) synthetase/RNase L pathway. Proc Natl Acad Sci U S A. 2006;103:7100–7105. doi: 10.1073/pnas.0602184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motz C, Schuhmann KM, Kirchhofer A, Moldt M, Witte G, Conzelmann KK, Hopfner KP. Paramyxovirus V proteins disrupt the fold of the RNA sensor MDA5 to inhibit antiviral signaling. Science. 2013;339:690–693. doi: 10.1126/science.1230949. [DOI] [PubMed] [Google Scholar]
- Office of Science Policy, N.I.o.H. United States Goverment policy for oversight of life sciences dual use research of concern. 2012 [Google Scholar]
- Perez JT, Varble A, Sachidanandam R, Zlatev I, Manoharan M, Garcia-Sastre A, tenOever BR. Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc Natl Acad Sci U S A. 2010;107:11525–11530. doi: 10.1073/pnas.1001984107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Rehwinkel J, Kato H, Takeuchi O, Akira S, Way M, Schiavo G, Reis e Sousa C. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, et al. Recognition of 5' triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid S, Mordstein M, Kochs G, Garcia-Sastre A, Tenoever BR. Transcription factor redundancy ensures induction of the antiviral state. J Biol Chem. 2010;285:42013–42022. doi: 10.1074/jbc.M110.165936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid S, Zony LC, tenOever BR. A versatile RNA vector for delivery of coding and noncoding RNAs. J Virol. 2014;88:2333–2336. doi: 10.1128/JVI.03267-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo GJ, Kincaid RP, Phanaksri T, Burke JM, Pare JM, Cox JE, Hsiang TY, Krug RM, Sullivan CS. Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe. 2013;14:435–445. doi: 10.1016/j.chom.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, Rodgers MA, Ramirez JL, Dimopoulos G, Yang PL, Pearson JL, et al. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- Tapia K, Kim WK, Sun Y, Mercado-Lopez X, Dunay E, Wise M, Adu M, Lopez CB. Defective viral genomes arising in vivo provide critical danger signals for the triggering of lung antiviral immunity. PLoS Pathog. 2013;9:e1003703. doi: 10.1371/journal.ppat.1003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- tenOever BR. RNA viruses and the host microRNA machinery. Nat Rev Microbiol. 2013;11:169–180. doi: 10.1038/nrmicro2971. [DOI] [PubMed] [Google Scholar]
- Varble A, Benitez AA, Schmid S, Sachs D, Shim JV, Rodriguez-Barrueco R, Panis M, Crumiller M, Silva JM, Sachidanandam R, et al. An in vivo RNAi screening approach to identify host determinants of virus replication. Cell Host Microbe. 2013;14:346–356. doi: 10.1016/j.chom.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Varble A, Chua MA, Perez JT, Manicassamy B, Garcia-Sastre A, tenOever BR. Engineered RNA viral synthesis of microRNAs. Proc Natl Acad Sci U S A. 2010;107:11519–11524. doi: 10.1073/pnas.1003115107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Dittmann M, Peisley A, Hoffmann HH, Gilmore RH, Schmidt T, Schmid-Burgk JL, Hornung V, Rice CM, Hur S. ATP-Dependent Effector-like Functions of RIG-I-like Receptors. Mol Cell. 2015 doi: 10.1016/j.molcel.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount JS, Gitlin L, Moran TM, Lopez CB. MDA5 participates in the detection of paramyxovirus infection and is essential for the early activation of dendritic cells in response to Sendai Virus defective interfering particles. J Immunol. 2008;180:4910–4918. doi: 10.4049/jimmunol.180.7.4910. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- Zhang X, Bogunovic D, Payelle-Brogard B, Francois-Newton V, Speer SD, Yuan C, Volpi S, Li Z, Sanal O, Mansouri D, et al. Human intracellular ISG15 prevents interferon-alpha/beta over-amplification and auto-inflammation. Nature. 2014 doi: 10.1038/nature13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, et al. Ribose 2'-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.