Abstract
Identifying predictors of mild cognitive impairment (MCI) and Alzheimer's disease (AD) can lead to more accurate diagnosis and facilitate clinical trial participation. We identified 320 participants (93 cognitively normal or CN, 162 MCI, 65 AD) with baseline magnetic resonance imaging (MRI) data, cerebrospinal fluid (CSD) biomarkers, and cognition data in the Alzheimer's Disease Neuroimaging Initiative database. We used independent component analysis (ICA) on structural MR images to derive 30 gray matter covariance patterns (ICs) across all participants. These ICs were used in iterative and stepwise discriminant classifier analyses to predict diagnostic classification at 24 months for CN vs. MCI, CN vs. AD, MCI vs. AD, and stable MCI (MCI-S) vs. MCI progression to AD (MCI-P). Models were cross-validated with a “leave-10-out” procedure. For CN vs. MCI, 84.7% accuracy was achieved based on cognitive performance measures, ICs, p-tau181p, and ApoE ε4 status. For CN vs. AD, 94.8% accuracy was achieved based on cognitive performance measures, ICs, and p-tau181p. For MCI vs. AD and MCI-S vs. MCI-P, models achieved 83.1% and 80.3% accuracy, respectively, based on cognitive performance measures, ICs, and p-tau181p. ICA-derived MRI biomarkers achieve excellent diagnostic accuracy for MCI conversion, which is little improved by CSF biomarkers and ApoE ε4 status.
Keywords: Data reduction, MCI, AD, Alzheimer's Disease Neuroimaging Initiative
1. Introduction
Identifying predictors of mild cognitive impairment (MCI) and Alzheimer's disease (AD) can lead to earlier, more accurate diagnosis and facilitate participant selection for clinical trials (Sperling et al., 2011; Reiman et al., 2012). The typical diagnostic progression from cognitively normal (CN), to MCI (Petersen et al., 1999; Gauthier et al., 2006), and finally AD reflects a systematic pattern of progressive atrophy (Whitwell et al., 2007; McDonald et al., 2009; Risacher et al., 2010), which corresponds to the spread of amyloid and tau neuropathology (Arnold et al., 1991; Braak and Braak, 1991; Jack et al., 2010). Measures of cognitive function (Gomar et al., 2011), regional atrophy (Bakkour et al., 2009; Davatzikos et al., 2011), and cerebrospinal fluid (CSF) biomarkers have been used to predict MCI or AD diagnosis. Prognostic classification accuracy based solely on radiologically detected atrophy (Cuingnet et al., 2011) or CSF biomarkers, such as amyloid-β 1-42 (Aβ1-42), tau, and phosphorylated tau at Thr181 (p-tau181p), varies widely, but is generally sub-optimal (Arnold et al., 1991; Braak and Braak, 1991; Shaw et al., 2007; Visser et al., 2009; Liu et al., 2013a).
The limited accuracy of individual biomarkers in predicting MCI and AD has motivated researchers to examine the combined prognostic value of structural magnetic resonance imaging (MRI), cognitive performance scores, CSF biomarkers, and genetic polymorphisms (Mueller et al., 2005; Davatzikos et al., 2011; Shaffer et al., 2013). A variety of approaches have been used to derive MRI biomarkers from whole brain images for use in diagnostic classifiers (Cuingnet et al., 2011). We chose to use independent component analysis, or ICA, to analyze structural MRI data (Xu et al., 2009) collected in the Alzheimer's Disease Neuroimaging Initiative (ADNI) (Mueller et al., 2005).
ICA is a data-driven, multivariate approach for reducing data dimensionality of images with minimal bias. ICA can be used to decompose whole brain gray matter (GM) maps into maximally independent spatial GM covariance patterns called independent components, or ICs. We applied ICA to MRI GM maps across participants diagnosed as CN, MCI, or AD. We examined how generated ICs can be used for prognostic diagnostic classification. Shaffer and colleagues (2013) recently used ICA as a data-reduction technique on a cohort of MCI participants from ADNI, followed by an additive logistic regression analysis to examine to what degree structural MRI ICs, CSF biomarkers, fluorodeoxyglucose positron emission tomography (FDG-PET), and other factors could distinguish between participants with MCI that remained stable (MCI-S) or that progressed to AD (MCI-P). We modified and extended this approach to a wider range of diagnostic classifications. While FDG-PET scans may also be useful in diagnostic classification (Shaffer et al., 2013), we focused on MRI-derived measures since MRI is far more commonly used in clinical practice.
Critically, we emphasize that ICA was used across all subjects only as a data-reduction technique. No ICs were extracted on the basis of any diagnostic classification, which would bias classifiers and otherwise require using training and validation datasets. Rather, ICs were used to account for variance across the AD spectrum and to reduce the dimensionality of the brain. Shaffer and colleagues (2013) notably used ICA in the same way.
We used baseline MRI, cognitive data, and CSF biomarkers to predict clinical diagnosis of CN, MCI, or AD at 24 months. Specifically, for each diagnostic pair, we performed a series of discriminant classification analyses iteratively to assess the additive prognostic value of 30 structural ICs derived from all subjects, cognitive data, CSF biomarkers (Shaw et al., 2007), and apolipoprotein ε4 genotyping (ApoE ε4 status) (Liu et al., 2013a). We also performed stepwise discriminant classification analyses to see if relatively sparse models achieved comparable diagnostic accuracy. Finally, to determine the clinical and biological significance of derived ICs, we assessed their relationships with cognitive performance measures and CSF biomarkers.
2. Methods
2.1. Ethics statement
Institutional review boards approved the ADNI protocol (“ADNI 1 Procedures Manual” at http://adni.loni.ucla.edu/methods/documents/) at their respective institutions. Participants at their respective ADNI sites gave written informed consent.
2.2. Subjects
All retrospective, de-identified data were obtained from the ADNI database (www.loni.ucla.edu/ADNI). The National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies, and non-profit organizations launched ADNI in 2003. ADNI is a $60 million, 5-year public-private partnership. Details about the initiative are described elsewhere by the principal investigator (Weiner et al., 2012).
For our purposes, between October to November 2012, we gathered data from 93 CN, 162 MCI, and 65 AD participants enrolled in ADNI that had the following characteristics: (1) baseline CSF biomarkers; (2) baseline 1.5-Tesla T1-weighted structural scans; (3) ApoE ε4 status; (4) baseline neuropsychological performance; and (5) clinical diagnosis at baseline and at the month 24 visit. Participants with MCI at baseline either remained stable by 24 months (i.e., MCI-S; n = 86) or progressed to AD (i.e., MCI-P; n = 76). We chose to examine the ability of baseline measures to predict diagnosis after 24 months because predictions within this timeframe are clinically meaningful at the first evaluation of a patient. In addition, there is already evidence that structural MRI of patients with MCI may contain sufficient information to predict conversion to AD 2 years in advance (Whitwell et al.,2007).
2.3. Cognition and CSF measures
Table 1 presents baseline demographic and cognitive data, including the 70-point Alzheimer's Disease Assessment Scale-cognitive subscale, or ADAS-cog (Rosen et al., 1984); the Clinical Dementia Rating-sum of boxes (Morris, 1993); and the Mini-Mental State Examination, or MMSE (Folstein et al., 1975). For discriminant classification analyses, we used the composite executive function (Gibbons et al., 2012) and memory factor scores (Crane et al., 2012) available on the ADNI website. These scores are the product of a factor analysis of ADNI memory and executive function tests. Clinical diagnosis, MMSE, and CDR scores for participants at baseline and the month 24 visit are listed in Supplementary Table 1.
Table 1.
Baseline Demographics, Diagnostic and Cognitive Data
| Diagnosis | N | Sex | Age | Education | CDR - Sum of Boxes | MMSE | ADAS-COG 70 |
|---|---|---|---|---|---|---|---|
| CN | 93 | 48F, 45M | 75.40 ± 5.15 | 15.71 ± 0.50 | 0.11 ± 0.50 | 29.01 ± 1.28 | 6.70 ± 3.60 |
| MCI | 162 | 107F, 55M | 74.2 ± 7.43 | 15.99 ± 2.93 | 1.52 ± 0.91 | 27.07 ± 1.79 | 11.07 ± 4.55 |
| AD | 65 | 36F, 29F | 75.43 ± 7.62 | 15.06 ± 3.21 | 4.22 ± 1.59 | 23.52 ± 1.95 | 18.08 ± 6.01 |
Mean ± SD where applicable. CDR = Clinical Dementia Rating.
MMSE = Mini-Mental State Examination. ADAS-COG = Alzheimer's
Disease Assessment Scale-Cognitive 11 item.
Standardized ADNI criteria were used to diagnose CN based on (1) a CDR of 0, (2) MMSE inclusively between 24 and 30, (3) no evidence of depression, and (4) no memory complaints. MCI was diagnosed based on (1) a CDR of 0.5, (2) an MMSE inclusive of 24 to 30, (3) preserved activities of daily living, and (4) worse performance on the WMS-Revised Logical Memory II test using education-adjusted scores. Probable AD was diagnosed based on (1) a CDR of 0.5 or 1.0, 2) an MMSE inclusive of 20 to 26, and (3) having met other criteria defined by NINCD-ADRDA (McKhann et al., 1984).
CSF was collected by lumbar puncture and assayed for tau, p-tau181p, and Aβ1-42 as previously described (Shaw et al., 2007). For ApoE ε4 status, participants were dichotomously classified as having zero ApoE ε4 alleles (“Non-ApoE ε4”) versus one or two ApoE ε4 alleles (“ApoE ε4”).
2.4. MRI acquisition, pre-processing, and ICA
T1-weighted MPRAGE images of typically 1.25 × 1.25 × 1.2 mm were acquired on 1.5 -Tesla MR imaging instruments using a standardized protocol (Jack et al., 2008). Images underwent standardized quality control inspection and system-specific correction for imaging artifacts at the Mayo Clinic. Adjustments included gradient non-linearity correction and intensity non-uniformity correction.
As summarized in Supplementary Fig. 1A, scans were preprocessed using the toolbox VBM8 (Eggert et al., 2012) (http://dbm.neuro.uni-jena.de/vbm/download/), an add-on to SPM8 (http://www.fil.ion.ucl.ac.uk/spm/software/spm8/). Briefly, this technique is an extension of the unified segmentation algorithm (Ashburner and Friston, 2005), a technique whereby bias correction, registration, and segmentation are done in one unified generative model. VBM8 utilizes a maximum a posteriori approach to remove noise and correct for partial volume estimation. Image registration is optimized by nonlinearly conforming a given ADNI brain image to a set of probabilistic tissue class maps in Montreal Neurological Institute (MNI) space using Diffeomorphic Anatomical Registration Through Exponentiated Lie Algebra, or DARTEL (Ashburner, 2007). Images were modulated to compensate for normalization to MNI space and were segmented into GM, white matter, and CSF. Images were inspected to ensure proper segmentation and removal of meninges and other non-brain structures. GM maps were smoothed with a 6-mm Gaussian kernel (Xu et al., 2009).
Next, we applied spatial ICA to all 320 smoothed GM maps (Calhoun et al., 2001) using the Infomax algorithm as applied in the SBM toolbox (http://www.nitrc.org/projects/gift), as summarized in Supplementary Fig. 1B (Xu et al., 2009). Based on previous component estimation analyses (Xu et al., 2009; Kapogiannis et al., 2013), 30 ICs were chosen as the number of GM covariance patterns that balanced reduced dimensionality through eigen decomposition while maximally retaining original source image variance. Exploratory analyses with 20 ICs resulted in poor spatial specificity for smaller, important structures such as the hippocampus, while 40 or 50 IC solutions produced fragmentary ICs that did not encompass larger parietal or prefrontal areas. Infomax ICA was conducted three times to reliably estimate eigen decomposition. SBM ICA decomposes structural MRI data into subject loading coefficients (or loading scores) and component maps. Specifically, each GM map image was converted into a vector and was arrayed in a subject-by-image matrix of all GM maps. This matrix was then decomposed into a mixing matrix (expressing the relationship between the subjects and the 30 ICs) and a source matrix (expressing the relationship between the 30 ICs and the image voxels). Each row of the mixing matrix contained the loading coefficients or loading scores for one IC and expresses how this IC contributed to each of the subjects. Each column of the mixing matrix expressed how different subjects contributed to an individual IC. Conversely, the rows of the source matrix expressed how each IC contributed to each of the voxels, and its columns indicate how each voxel contributed to each of the ICs. Additional information on these matrices can be found in previous reports that used SBM ICA (Calhoun et al., 2006; Xu et al., 2009) and Supplementary Fig. 1B. In effect, a higher loading weight for a given IC strongly reflects higher GM volume (Calhoun et al., 2006; Xu et al., 2009). Source matrices were reconstructed into 3D z-scored images of ICs in the MNI coordinate system.
2.5. Statistics
All statistical analyses were conducted in SPSS 20.0 (Chicago, IL). Discriminant classification analysis was used to predict month-24 diagnostic classification for the following diagnostic pairs: (1) CN vs. MCI, (2) CN vs. AD, (3) MCI vs. AD, and (4) MCI-S vs. MCI-P (Supplementary Fig. 1C). We did not use logistic regression because it requires larger sample sizes per independent variable entered to maintain model reliability (Spicer, 2005). The accuracy (i.e., percentage of subject pairs that were correctly classified), sensitivity, and specificity with respective confidence intervals were calculated for each classification model. Receiver operating characteristic (ROC) curves plotting sensitivity against 1-specificity were created for each classifier. The area under the curve (AUC) was used as a measure of classification accuracy. The goal was to identify the model that had the best accuracy for a given diagnostic classification. To examine if a more complex model had significantly higher accuracy than a less complex one, an F-test change statistic was computed by comparing derived F-values. This process was iteratively repeated from the most complex to least complex models.
We performed classifier analyses in an iterative manner, examining increasingly complex models. At each one of these iterations, all variables were entered in a single step, so that all canonical discriminant functions were included in the composite classifier. The least complex model included three nuisance variables (age, sex, and education), followed by successively adding the 30 ICs, then adding the two cognitive factors, then adding CSF biomarkers, and finally adding ApoE ε4 status for the most complex model. Age, sex, and education were always included because they are standard nuisance variables in several classifier articles (e.g., Shaffer et al., 2013). CSF biomarkers were not entered simultaneously to avoid multicollinearity. The CSF biomarker that best improved model accuracy was kept in the model.
In addition, we performed stepwise analyses to determine if smaller sets of variables produce classification accuracies comparable to more complex models. For stepwise classifier analysis, independent variables (the three nuisance variables, ICs, cognitive factors, etc.) were entered in a stepwise fashion, based on Wilks’ lamda and using F value of 3.84 as criterion for entry and F value of 2.71 as criterion for removal in subsequent steps.
All models were cross-validated using a “leave-10-out” approach. To determine which of the 30 ICs contributed more to a given diagnostic classification, a multivariate analysis of covariance (MANCOVA) was conducted for each diagnostic pair (e.g., CN vs. AD), in which diagnosis at 24 months was the independent variable, the loading factor weights of the 30 ICs across participants were the dependent variables, and age, sex, and years of education were covariates. Then, to assess the biological plausibility and external validity of these ICs against established measures reflecting AD pathogenesis, we computed their two-tailed Pearson's correlations with CSF biomarkers and cognitive factors. Again, the 30 ICs were derived from all subjects regardless of diagnostic outcome, in order to minimize bias.
3. Results
3.1. ICA results
The eigenvalues of the 30 ICs, derived from 320 brains and agnostic to baseline or 24-month diagnosis, accounted for 95.4% of total source variance. The variance inflation factor (VIF) for all ICs within all diagnostic classification models did not exceed 3.4, confirming that ICs are not multicollinear. Supplementary Fig. 2 shows and describes all 30 ICs.
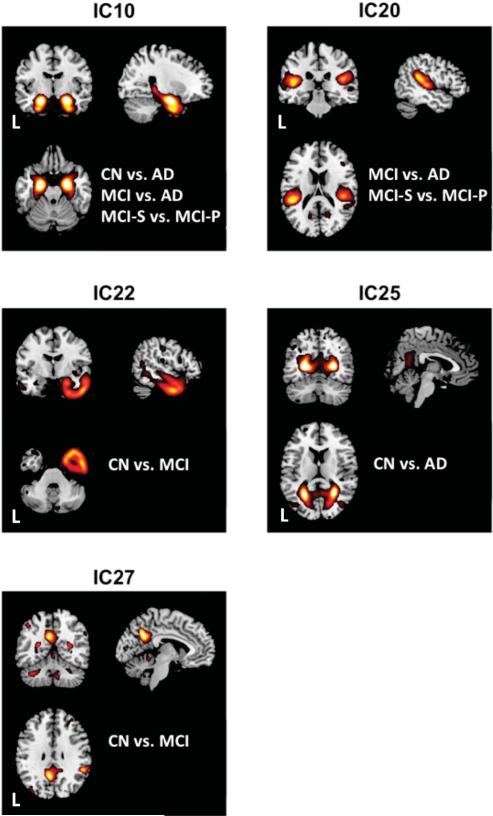
Fig. 1 and Supplementary Fig. 3, respectively, illustrate the ICs that consistently had the largest F-values in the MANCOVAs, and therefore contributed the most to the various classification analyses. We named these ICs conventionally after the regions primarily included, as follows: (1) IC 10 (“Medial temporal lobe, or MTL IC”) comprised bilateral hippocampus, medial temporal cortex, and immediately proximal inferior temporal cortex; (2) IC 14 (“middle frontal gyrus, or MFG IC”) comprised right middle frontal gyrus and superior temporal sulcus; (3) IC 20 (“temporoparietal junction, or TPJ IC”) comprised bilateral TPJ; (4) IC 25 (“calcarine IC”) comprised bilateral calcarine sulcus, lingual gyrus, and precuneus; (5) IC 26 (“inferior temporal-occipital IC”) comprised parts of inferior temporal, fusiform, posterior occipital, and inferior parietal gyri; (6) IC 30 (“anterior cerebellar IC”) comprised anterior cerebellum, but also superior occipital and parietal gyri; (7) IC 22 (“Right anterior temporal lobe” IC) comprised most of the right anterior temporal lobe; and (8) IC 27 (“posteromedial cortex IC”) comprised precuneus and posterior cingulate gyrus.
Fig. 1. Stepwise-selected representative ICs.
Stepwise discriminant analysis consistently selected 5 independent components (ICs) as the most significant GM covariance patterns among the different group classifications (e.g., CN vs. AD). The ICs were IC10 (“medial temporal lobe”), IC20 (“tempoparietal junction”), IC22 (“right anterior temporal lobe”), IC25 (“calcarine”), and IC27 (“posteromedial cortex”). Diagnostic labels for each group classifier are listed with a given IC for orientation purposes. AD = Alzheimer's Disease; CN = cognitively normal; IC = Independent Component; L = Left; MCI = Mild Cognitive Impairment. Brains are oriented in neurological space.
3.2. CN vs. MCI
Table 2 and Supplementary Fig. 4 depict classifier model performance and ROC curves for the CN vs. MCI discriminant classification. Based on the MANCOVA for CN vs. MCI, the three most significant ICs in all models were the MTL IC [F(1,172) = 7.595, P = 0.006], medial parietal-occipital IC [F(1,172) = 11.538, P < 0.001], and anterior cerebellar IC [F(1,172) = 10.907, P < 0.001] (Supplementary Fig. 3). The best accuracy achieved was 84.7% for a model that included demographic covariates, the two cognitive factors, the 30 ICs, p-tau181p, and ApoE ε4 status. Based on the F-test change statistic, however, the Covariates + Cognition + MRI + CSF + ApoE model did not perform better than the Covariates + Cognition + MRI + CSF model [F(1,171) = .804, P = .371]. The Covariates + Cognition + MRI + CSF model also did not achieve better accuracy compared with the Covariates + Cognition + MRI model [F(1,172) = 1.298, P = 0.256]. The Covariates + Cognition + MRI model was a more accurate classifier than the Covariates + Cognition or Covariates + MRI models [F(1,173) = 12.365, P < 0.001]. The Covariates + MRI model performed worse than the Covariates + Cognition model [F(1,174) = 33.771, P < 0.001]. The model selected by the stepwise method had 8.1% less accuracy relative to the Covariates + Cognition + MRI + CSF + ApoE iterative model. The stepwise model included (1) the memory factor [F(1,167) = 59.095, P < 0.001]; (2) the right anterior temporal lobe IC [F(2,166) = 35.709, P < 0.001]; (3) the posteromedial cortex IC [F(3,165) = 26.993, P < 0.001]; (4) CSF tau [F(4,164) = 21.789, P < 0.001]; (5) IC13, an IC primarily encompassing inferior frontal operculum and lateral temporal regions [F(5,163) = 18.835, P < 0.001]; and (6) IC9, an IC encompassing dorsal prefrontal cortex and middle cingulate cortex [F(6, 162) = 16.888, P < 0.001].
Table 2.
CN and MCI Classification Results for Iterative Models and the Stepwise Model
| Model | AUC + CI | ACC (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|
| Covariates | .604 (.521-.688) | 60.1 | 68.1 | 53.0 |
| Covariates + Cognition | .832 (.771-.893) | 78.4 | 73.1 | 83.1 |
| Covariates + MRI | .819 (.758-.880) | 74.3 | 68.8 | 79.3 |
| Covariates + Cognition + MRI | .896 (.851-.941) | 83.3 | 76.7 | 89.1 |
| Covariates + Cognition + MRI + CSF | .901 (.857-.945) | 83.0 | 76.7 | 88.5 |
| Covariates + Cognition + MRI + CSF + ApoE | .908 (.867-.950) | 84.7 | 78.0 | 90.7 |
| Stepwise Model | .868 (.817-.918) | 76.6 | 67.6 | 84.7 |
ACC = accuracy; ApoE = Apolipoprotein ε4 status; AUC = area under the curve; CI = confidence interval (95%). Covariates included age, sex, and education. Iterative models used the p-tau181p CSF biomarker. The bold in the upper part of the table denotes the model that had the highest classification accuracy and a significant increase in the F-value relative to less complex models.
3.3. CN vs. AD
Classifier model performance and ROCs for CN vs. AD are depicted in Table 3 and Supplementary Fig. 5. Based on the MANCOVA for CN vs. AD, the three most significant ICs in all models were the MTL IC [F(1,230) = 65.643, P < 0.001], MFG IC (F(1,230) = 51,712, P < 0.001), and inferior temporal-occipital IC [F(1,230) = 34.564, P < 0.001] (Supplementary Fig. 3). The highest accuracy iterative model achieved 94.8% accuracy and included demographic covariates, the two cognitive factors, the 30 ICs, and p-tau181p. Based on the F-value change statistic, this Covariates + Cognition + MRI + CSF model did not have a better fit than the Covariates + Cognition + MRI model [F(1,230) = 3.333, P = 0.069]. The Covariates + Cognition + MRI model achieved better accuracy than the Covariates + Cognition and Covariates + MRI models [F(1,231) = 17.531, P < 0.001]. The Covariates + MRI model was a less accurate classifier than the Covariates + Cognition model [F(1,233) = 35.883, P < 0.001]. The model selected by the stepwise method had 3.1% lower accuracy relative to the highest accuracy iterative model and included (1) the memory factor [F(1,228) = 319.479, P < 0.001]; (2) ApoE ε4 status [F(1,227) = 82.019, P < 0.001]; (3) the MTL IC [F(3,226) = 124.117, P < 0.001]; (4) the calcarine IC [F(4,225) = 96.279, P < 0.001]; and IC17, an IC primarily comprising the striatum, posterior cerebellum, and to a lesser extent ventral PFC [F(5,224) = 78.893, P < 0.001].
Table 3.
CN and AD Classification Results for Iterative Models and the Stepwise Model
| Model | AUC + CI | ACC (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|
| Covariates | .556 (.481-.632) | 60.2 | 100 | 0 |
| Covariates + Cognition | .958 (.930-.987) | 93.0 | 94.5 | 91.8 |
| Covariates + MRI | .935 (.904-.966) | 87.4 | 91.7 | 82.5 |
| Covariates + Cognition + MRI | .980 (.965-.995) | 94.3 | 94.9 | 94.0 |
| Covariates + Cognition + MRI + CSF | .981 (.967-.995) | 94.8 | 94.9 | 94.6 |
| Covariates + Cognition + MRI + CSF + ApoE | .983 (.971-.995) | 93.5 | 95.7 | 90.2 |
| Stepwise Model | .972 (.953-.992) | 91.7 | 92.0 | 90.7 |
ACC = accuracy; ApoE = apolipoprotein ε4 status; AUC = area under the curve; CI = confidence interval (95%). Covariates included age, sex, and education. Iterative models used the p-tau181p CSF biomarker. The bold font in the upper part of the table denotes the model that had the highest classification accuracy and a significant increase in the F-value relative to less complex models.
3.4. MCI vs. AD
Classifier model performance for MCI vs. AD and ROC curves are depicted in Table 4 and Supplementary Fig. 6. Based on the MANCOVA for MCI vs. AD, the three most predictive ICs in all models were the MTL IC [F(1,220) = 18.227, P < 0.001], MFG IC [F(1,220) = 22.955, P < 0.001], and inferior temporal-occipital IC [F(1,220) = 25.898, P < 0.001] (Supplementary Fig. 3). The highest accuracy iterative model achieved a classification accuracy of 83.1% and included demographic covariates, the two cognitive factors, 30 ICs, and tau. Based on F-test change statistics, however, this Covariates + Cognition + MRI + CSF model did not have a better fit than the Covariates + Cognition + MRI model [F(1,220) = 1.381, P = 0.241]. The Covariates + Cognition + MRI model performed better than the Covariates + MRI and Covariates + Cognition models [F(1,221) = 6.492, P = 0.012]. The Covariates + MRI model achieved lower accuracy than the Covariates + Cognition model [F(1,222) = 38.642, P < 0.001]. The model selected by the stepwise method had 5.3% lower accuracy relative to the highest accuracy iterative model and included (1) the memory factor [F(1,217) = 75.689, P < 0.001]; 2) the TPJ IC [F(2,216) = 47.163, P < 0.001]; (3) the MTL IC [F(3,215) = 34.576, P < 0.001]; and (4) the executive function factor [F(4,214) = 27.268, P < 0.001].
Table 4.
MCI and AD Classification Results for Iterative Models and the Stepwise Model
| Model | AUC + CI | ACC (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|
| Covariates | .471 (.392-.549) | 62.7 | 100.0 | 0 |
| Covariates + Cognition | .833 (.781-.886) | 78.3 | 86.5 | 64.5 |
| Covariates + MRI | .843 (.792-.894) | 77.6 | 84.7 | 65.7 |
| Covariates + Cognition + MRI | .886 (.843-.929) | 81.4 | 86.1 | 73.0 |
| Covariates + Cognition + MRI + CSF | .885 (.842-.928) | 83.1 | 89.1 | 73.0 |
| Covariates + Cognition + MRI + CSF + ApoE | .867 (.820-.913) | 82.2 | 87.6 | 73.0 |
| Stepwise Model | .863 (.817-.910) | 77.8 | 84.3 | 66.9 |
ACC = accuracy; ApoE = apolipoprotein ε4 status; AUC = area under the curve; CI = confidence interval (95%). Covariates included age, sex, and education. Iterative models used the p-tau181p CSF biomarker. The bold font in the upper part of the table denotes the model that had the highest classification accuracy and a significant increase in the F-value relative to less complex models.
3.5. MCI-S vs. MCI-P
Classifier model performances for MCI-S vs. MCI-P are depicted in Table 5, with ROCs in Supplementary Fig. 7. Based on the MANCOVA for MCI-S vs. MCI-P, the three most predictive ICs in all models were the MTL IC [F(1,156) = 22.703, P < 0.001], TPJ IC [F(1,156) = 11.621, P < 0001], and inferior temporal-occipital IC [F(1,156) = 16.137, P < 0.001] (Supplementary Fig. 3). The highest accuracy model achieved 80.3% accuracy and included demographic covariates, the two cognitive factors, the 30 ICs, and p-tau181p. Based on the F-change statistic, this Covariates + Cognition + MRI + CSF model did not perform better than the Covariate + Cognition + MRI model [F(1,156) = 2.275, P = 0.133]. The Covariates + Cognition + MRI model was a more accurate classifier than the Covariates + Cognition or Covariates + MRI models [F(1,157) = 4.670, P = 0.032]. The Covariate + MRI model performed better than the Covariate + Cognition model [F(1,158) = 43.658, P < 0.001]. The model selected by the stepwise method had 4.8% less accuracy than the most accurate iterative model. It included (1) the memory factor [F(1,154) = 35.632, P < 0.001]; (2) the TPJ IC [F(2,153) = 24.210, P < 0.001] and the MTL IC [F(3,152) = 19.070, P < 0.001].
Table 5.
MCI-S and MCI-P Classification Results for Iterative Models and a Stepwise Model
| Model | AUC + CI | ACC (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|
| Covariates | .479 (.390-.569) | 53.5 | 25.2 | 78.4 |
| Covariates + Cognition | .769 (.697-.841) | 73.3 | 74.2 | 72.5 |
| Covariates + MRI | .853 (.794-.912) | 79.6 | 75.6 | 83.2 |
| Covariates + Cognition + MRI | .874 (.820-.928) | 80.0 | 78.3 | 81.5 |
| Covariates + Cognition + MRI + CSF | .878 (.825-.931) | 80.3 | 81.7 | 79.0 |
| Covariates + Cognition + MRI + CSF + ApoE | .855 (.798-.913) | 79.9 | 80.3 | 79.6 |
| Stepwise Model | .828 (.764-.892) | 75.5 | 74.8 | 76.0 |
ACC = accuracy; ApoE = apolipoprotein ε4 status; AUC = area under the curve; CI = confidence interval (95%). Covariates included age, sex, and education. Iterative models used the p-tau181p CSF biomarker. The bold font in the upper part of the table denotes the model that had the highest classification accuracy and a significant increase in the F-value relative to less complex models.
3.6. ICs, CSF biomarkers and cognitive factors
We computed correlations between ICs 10, 20, 22, 26, and 27, the ICs that were most consistently featured in the various classification analyses (Fig. 1 and Supplementary Fig. 3) with the cognitive factors and CSF biomarkers.
Supplementary Table 2 shows that for all ICs except the posteromedial parietal IC, higher factor loadings representing more gray matter predicted, to varying degrees, higher memory and executive function scores, higher Aβ1-42, and lower tau and p-tau181p.
4. Discussion
In this report, comparable to Shaffer et al. (2013), ICA was used solely as a data-reduction technique for baseline structural MR images from ADNI. ICs were generated and combined with cognitive performance measures and other indices in classifier models to predict diagnosis in the CN-MCI-AD spectrum after 24 months. One key conclusion is how robustly cognition predicted diagnostic classifications. Gomar and colleagues (2011) recently showed that cognitive measures were better predictors of MCI conversion to AD than most biomarkers. However, we advise caution because the cognitive performance measures we used were derived, in part, from the same ADNI participants to whom we applied diagnostic classification. While other reports have also used these factors in classifiers (Crane et al., 2012; Habeck et al., 2012; Nir et al., 2013), we would like to caution readers to this source of potential bias and the possibility of inflated classification accuracy based on using these cognitive factors.
Nonetheless, our findings suggest that there is added accuracy gained by incorporating MRI metrics to predict diagnostic classification. Specifically, models that incorporated covariates, ICs, and cognition had significantly higher accuracies for each diagnostic classification than models based solely on cognition or MRI. This increased accuracy was highest in distinguishing CN vs. MCI (4.9%) and CN vs. AD (6.9%). Interestingly, for each dichotomous diagnostic classification, the ICs that contributed most to the classification significantly reflected patterns of atrophy recognized by prior research (McDonald et al., 2009; Risacher et al., 2010). In addition, these ICs were correlated with cognitive factors and AD biomarkers, supporting their biological plausibility and external validity as useful indices of AD atrophy. CSF biomarkers or ApoE ε4 status appeared to offer no significant additive value for all diagnostic classifications. Results will be discussed separately for each diagnostic dichotomy.
4.1. CN vs. MCI
A variety of techniques have been used to derive MRI biomarkers for classification between CN and MCI or CN and MCI-P (Chincarini et al., 2011; Cuingnet et al., 2011; Hinrichs et al., 2011; Wolz et al., 2011; Chu et al., 2012; Liu et al., 2012). Liu and colleagues (Liu et al., 2012) have to date achieved the best CN vs. MCI prognostic accuracy (87.85%) using a random patch-based ensemble classifier. However, it is unclear if they made diagnostic predictions for baseline diagnoses or a subsequent time point, where baseline prediction would yield higher accuracy. In comparison, we achieved a poorer accuracy of 74.3% based on ICs alone and an accuracy of 83.3% by combining cognitive scores and ICs. CSF biomarkers and ApoE ε4 status appear to have little to no added statistical utility in improving the CN vs. MCI classifier, validating previous reports (Vemuri et al., 2009; Walhovd et al., 2010).
In our analysis, the ICs that contributed most to this classification were the MTL IC, right anterior temporal lobe IC, posteromedial cortex IC, and calcarine IC. Recent studies using ADNI data (McDonald et al., 2009; Risacher et al., 2010) have similarly indicated that atrophy in MCI is found in temporal and parietal-occipital regions. It is important to note that these ICs correspond to the distribution of tau pathology and neurodegeneration in early AD, which first target allocortical MTL regions, followed by multimodal temporal and posteromedial parietal and occipital cortical areas (Arnold et al., 1991; Braak and Braak, 1991).
4.2. CN vs. AD
A variety of methodologies have been applied to distinguish between participants who appear to be normally aging versus those with AD (Chincarini et al., 2011; Cuingnet et al., 2011; Wolz et al., 2011; Chu et al., 2012; Liu et al., 2012, 2014; Yuan et al., 2012). Thus far, using MRI, Chincarini and colleagues (2011) have achieved the best results (AUC = 0.97) using a random forest classifier and machine learning approach among a priori designated areas in MTL. Wee and colleagues (2013) achieve a comparable result integrating cortical volume, thickness, and correlative matrices of morphological features. Our AUC was similar to both studies (AUC = 0.935). Our ICA method achieved better accuracy than many ROI or dimensionality-reducing techniques, as surveyed elsewhere (Cuingnet et al., 2011). The iterative model including MRI and cognitive factors performed comparably (AUC = 0.980) to the Chincarini MRI model and similar multimodal feature sets (Zhang and Shen, 2012). It should be noted that while the most accurate CN vs. AD model included CSF biomarkers, the gain in accuracy (0.5%) was non-significant. Including ApoE ε4 status in a classifier model similarly produces a non-significant increase of 1.4%.
The ICs that contributed most to this classification were the MTL, calcarine, MFG, and inferior temporal-occipital ICs. The overall pattern is highly similar to one identified based on the cortical thickness of select ROIs (Wolz et al., 2011) and reflects the diffuse atrophy typical of AD (McDonald et al., 2009; Risacher et al., 2010) and the progression of tau neuropathology (Arnold et al., 1991; Braak and Braak, 1991).
4.3. MCI vs. AD and MCI-S vs. MCI-P
In distinguishing MCI-S versus MCI-P, the ICA model achieved better accuracy (79.6%) than other techniques (Aksu et al., 2011; Chincarini et al., 2011; Cuingnet et al., 2011; Gomar et al., 2011; Hinrichs et al., 2011; Chu et al., 2012; Liu et al., 2013b; Shaffer et al., 2013). Shaffer and colleagues (2013) also used baseline structural ICA as a data-reduction tool to perform diagnostic classification for MCI-S vs. MCI-P. They derived four ICs and achieved an AUC of 0.741, which was not significantly higher than the AUC of just the covariates, whereas models incorporating ICs from FDG-PET images achieved an AUC of 0.874. By contrast, the AUC we derive for 30 ICs was 0.853 and did outperform models including covariates and covariates plus cognitive factors. Furthermore, the addition of CSF and ApoE ε4 status did not significantly increase accuracy. Critically, the AUCs from our study and in Shaffer et al. are comparable, suggesting that our classifier models using ICs derived from all subjects regardless of diagnosis were not biased.
To the best of our knowledge, for MCI-S vs. MCI-P, the accuracy we achieved with MRI ICA is excellent and on par with the accuracy (81.5%) reported by Misra and co-workers (Misra et al., 2009), while our AUC (0.853) is higher. Wee and colleagues (Wee et al., 2013) also achieved a high degree of accuracy when predicting MCI conversion 3 years after baseline (75.05%). Thus, presently, ICA represents an optimal MR data-reduction technique for classifying MCI-S and MCI-P, but not other diagnostic classifications. The Covariates + Cognition + MRI model was again the model with the highest significant F-value. While including CSF or ApoE ε4 status, respectively, increased accuracy by 0.6% or 2.4%, these increases were non-significant. For MCI vs. AD, including CSF biomarkers and ApoE ε4 status again led to non-significant increases in accuracy, suggesting that CSF biomarkers and ApoE genotyping may have little additive prognostic value.
In our analysis, the ICs that contributed most to the MCI vs. AD classification were the MTL, MFG, and inferior temporal-occipital ICs. For MCI-S vs. MCI-P, the most significant ICs included MTL, TPJ, and inferior temporal-occipital ICs. These findings are consistent with the fact that with transition to clinical AD, tangle pathology (Arnold et al., 1991; Braak and Braak, 1991) and atrophy spread to frontal regions and the superior temporal sulcus (Whitwell et al., 2007; McDonald et al., 2009; Risacher et al., 2010).
4.4. Limitations and conclusions
Patterns of GM covariance predicted future classification in the CN-MCI-AD diagnostic spectrum, presumably by reflecting patterns of atrophy already established 2 years in advance of diagnosis. Different ICs emerged along the AD spectrum. Specifically, atrophy in the MTL was predictive across the spectrum. Preserved anterior temporal and posteromedial areas were predictive of future CN status, while TPJ atrophy was predictive for MCI participants who convert to clinical AD. Importantly, these temporal and parietal ICs were associated with neuropsychological performance and CSF biomarkers, confirming similar associations with regional GM volumetric measures seen in the literature. Our MRI ICA data-reduction approach achieved excellent accuracy in distinguishing MCI-S and MCI-P. When combined with neuropsychological performance, our other diagnostic classifiers utilizing ICA perform less well or comparably to other ADNI multimodal classifiers. One limitation is that the accuracy of some of the discriminant classifiers should be interpreted with caution. Factors such as the number of ICs selected, the sample size of groups, and clinical heterogeneity can lead to variations in model fit. Future ICA studies in larger datasets should confirm our findings, in particular regarding MCI conversion. It is also important to again highlight that the ADNI memory and executive function factors were derived from some, but not all, of the same participants to whose data we applied discriminant classification. Thus, classifier model accuracies may be inflated. Another limitation is that ICA cannot currently be translated into a clinical setting for diagnostic classification. Nonetheless, advances in clinical grade software could automate this process and classify participants based on an exemplar dataset. While the diagnostic utility of CSF biomarkers and ApoE ε4 status should not be discounted, they appear to offer little additive benefit compared with MRI biomarkers (Vemuri et al., 2009; Walhovd et al., 2010).
Supplementary Material
Acknowledgements
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI). ADNI data are disseminated by the Laboratory for NeuroImaging at the University of California, Los Angeles. The authors thank Jeffrey Metter and Eleanor Simonsick for their constructive comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aksu Y, Miller DJ, Kesidis G, Bigler DC, Yang QX. An MRI-derived definition of MCI-to-AD conversion for long-term, automatic prognosis of MCI patients. PloS One. 2011;6:e25074. doi: 10.1371/journal.pone.0025074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold SE, Hyman BT, Flory J, Damasio AR, Van Hoesen GW. The topographical and neuroanatomical distribution of neurofibrillary tangles and neuritic plaques in the cerebral cortex of patients with Alzheimer's disease. Cerebral Cortex. 1991;1:103–116. doi: 10.1093/cercor/1.1.103. [DOI] [PubMed] [Google Scholar]
- Ashburner J. A fast diffeomorphic image registration algorithm. Neurolmage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Unified segmentation. Neurolmage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Bakkour A, Morris JC, Dickerson BC. The cortical signature of prodromal AD: regional thinning predicts mild AD dementia. Neurology. 2009;72:1048–1055. doi: 10.1212/01.wnl.0000340981.97664.2f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing ofAlzheimer-related changes. Acta Neuropathologica. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Giuliani NR, Pekar JJ, Kiehl KA, Pearlson GD. Method for multimodal analysis of independent source differences in schizophrenia: combining gray matter structural and auditory oddball functional data. Human Brain Mapping. 2006;27:47–62. doi: 10.1002/hbm.20166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ. A method for making group inferences from functional MRI data using independent component analysis. Human Brain Mapping. 2001;14:140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chincarini A, Bosco P, Calvini P, Gemme G, Esposito M, Olivieri C, Rei L, Squarcia S, Rodriguez G, Bellotti R, Cerello P, De Mitri I, Retico A, Nobili F. Local MRI analysis approach in the diagnosis of early and prodromal Alzheimer's disease. NeuroImage. 2011;58:469–480. doi: 10.1016/j.neuroimage.2011.05.083. [DOI] [PubMed] [Google Scholar]
- Chu C, Hsu AL, Chou KH, Bandettini P, Lin C. Does feature selection improve classification accuracy? Impact of sample size and feature selection on classification using anatomical magnetic resonance images. NeuroImage. 2012;60:59–70. doi: 10.1016/j.neuroimage.2011.11.066. [DOI] [PubMed] [Google Scholar]
- Crane PK, Carle A, Gibbons LE, Insel P, Mackin RS, Gross A, Jones RN, Mukherjee S, Curtis SM, Harvey D, Weiner M, Mungas D. Development and assessment of a composite score for memory in the Alzheimer's Disease Neuroimaging Initiative (ADNI). Brain Imaging and Behavior. 2012;6:502–516. doi: 10.1007/s11682-012-9186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuingnet R, Gerardin E, Tessieras J, Auzias G, Lehericy S, Habert MO, Chupin M, Benali H, Colliot O. Automatic classification of patients with Alzheimer's disease from structural MRI: a comparison of ten methods using the ADNI database. NeuroImage. 2011;56:766–781. doi: 10.1016/j.neuroimage.2010.06.013. [DOI] [PubMed] [Google Scholar]
- Davatzikos C, Bhatt P, Shaw LM, Batmanghelich KN, Trojanowski JQ. Prediction ofMCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiology ofAging. 2011;32:2322, e2319–2327. doi: 10.1016/j.neurobiolaging.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert LD, Sommer J, Jansen A, Kircher T, Konrad C. Accuracy and reliability of automated gray matter segmentation pathways on real and simulated structural magnetic resonance images of the human brain. PloS One. 2012;7:e45081. doi: 10.1371/journal.pone.0045081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. Journal of Psychiatric Research. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Gauthier S, Reisberg B, Zaudig M, Petersen RC, Ritchie K, Broich K, Belleville S, Brodaty H, Bennett D, Chertkow H, Cummings JL, de Leon M, Feldman H, Ganguli M, Hampel H, Scheltens P, Tierney MC, Whitehouse P, Winblad B. Mild cognitive impairment. Lancet. 2006;367:1262–1270. doi: 10.1016/S0140-6736(06)68542-5. [DOI] [PubMed] [Google Scholar]
- Gibbons LE, Carle AC, Mackin RS, Harvey D, Mukherjee S, Insel P, Curtis SM, Mungas D, Crane PK. A composite score for executive functioning, validated in Alzheimer's Disease Neuroimaging Initiative (ADNI) participants with baseline mild cognitive impairment. Brain Imaging and Behavior. 2012;6:517–527. doi: 10.1007/s11682-012-9176-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomar JJ, Bobes-Bascaran MT, Conejero-Goldberg C, Davies P, Goldberg TE. Utility of combinations ofbiomarkers, cognitive markers, and risk factors to predict conversion from mild cognitive impairment to Alzheimer disease in patients in the Alzheimer's disease neuroimaging initiative. Archives of General Psychiatry. 2011;68:961–969. doi: 10.1001/archgenpsychiatry.2011.96. [DOI] [PubMed] [Google Scholar]
- Habeck C, Risacher S, Lee GJ, Glymour MM, Mormino E, Mukherjee S, Kim S, Nho K, DeCarli C, Saykin AJ, Crane PK. Relationship between baseline brain metabolism measured using [(1)(8)F]FDG PET and memory and executive function in prodromal and early Alzheimer's disease. Brain Imaging and Behavior. 2012;6:568–583. doi: 10.1007/s11682-012-9208-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs C, Singh V, Xu G, Johnson SC. Predictive markers forAD in a multi-modality framework: an analysis of MCI progression in the ADNI population. NeuroImage. 2011;55:574–589. doi: 10.1016/j.neuroimage.2010.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr., Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, J LW, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DL, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. Journal ofMagnetic Resonance Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr., Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW, Petersen RC, Trojanowski JQ. Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. Lancet Neurology. 2010;9:119–128. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapogiannis D, Reiter DA, Willette AA, Mattson MP. Posteromedial cortex glutamate and GABA predict intrinsic functional connectivity of the default mode network. NeuroImage. 2013;64:112–119. doi: 10.1016/j.neuroimage.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nature Reviews. Neurology. 2013a;9(2):106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang D, Shen D. Ensemble sparse classification of Alzheimer's disease. NeuroImage. 2012;60:1106–1116. doi: 10.1016/j.neuroimage.2012.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang D, Shen D. Hierarchical fusion of features and classifier decisions forAlzheimer's disease diagnosis. Human Brain Mapping. 2014;35(4):13051319. doi: 10.1002/hbm.22254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Mattila J, Ruiz MA, Paajanen T, Koikkalainen J, van Gils M, Herukka SK, Waldemar G, Lotjonen J, Soininen H. Predicting AD conversion: comparison between prodromal AD guidelines and ComputerAssisted PredictAD Tool. PloS One. 2013b;8:e55246. doi: 10.1371/journal.pone.0055246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald CR, McEvoy LK, Gharapetian L, Fennema-Notestine C, Hagler DJ, Jr., Holland D, Koyama A, Brewer JB, Dale AM. Regional rates of neocortical atrophy from normal aging to earlyAlzheimer disease. Neurology. 2009;73:457–465. doi: 10.1212/WNL.0b013e3181b16431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- Misra C, Fan Y, Davatzikos C. Baseline and longitudinal patterns ofbrain atrophy in MCI patients, and their use in prediction of short-term conversion to AD: results from ADNI. Neurolmage. 2009;44:1415–1422. doi: 10.1016/j.neuroimage.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JC. The Clinical Dementia Rating (CDR): currentversion and scoring rules. Neurology. 1993;43:2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L. Ways toward an early diagnosis in Alzheimer's disease: the Alzheimer's Disease Neuroimaging Initiative (ADNI). Alzheimer's & dementia : the journal of the Alzheimer's Association. 2005;1:55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nir TM, Jahanshad N, Villalon-Reina JE, Toga AW, Jack CR, Weiner MW, Thompson PM. Effectiveness of regional DTI measures in distinguishing Alzheimer's disease, MCI, and normal aging. NeuroImage. Clinical. 2013;3:180–195. doi: 10.1016/j.nicl.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Archives of Neurology. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- Reiman EM, Brinton RD, Katz R, Petersen RC, Negash S, Mungas D, Aisen PS. Considerations in the design of clinical trials for cognitive aging. The Journals of Gerontology. Series A. Biol Sciences and Medical Sciences. 2012;67:766–772. doi: 10.1093/gerona/gls124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risacher SL, Shen L, West JD, Kim S, McDonald BC, Beckett LA, Harvey DJ, Jack CR, Jr., Weiner MW, Saykin AJ. Longitudinal MRI atrophy biomarkers: relationship to conversion in the ADNI cohort. Neurobiology of Aging. 2010;31:1401–1418. doi: 10.1016/j.neurobiolaging.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen WG, Mohs RC, Davis KL. A new rating scale for Alzheimer's disease. The American Journal of Psychiatry. 1984;141:1356–1364. doi: 10.1176/ajp.141.11.1356. [DOI] [PubMed] [Google Scholar]
- Shaffer JL, Petrella JR, Sheldon FC, Choudhury KR, Calhoun VD, Coleman RE, Doraiswamy PM. Predicting cognitive decline in subjects at risk for Alzheimer disease by using combined cerebrospinal fluid, MR imaging, and PET biomarkers. Radiology. 2013;266:583–591. doi: 10.1148/radiol.12120010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw LM, Korecka M, Clark CM, Lee VM, Trojanowski JQ. Biomarkers of neurodegeneration for diagnosis and monitoring therapeutics. Nature reviews. Drug Discovery. 2007;6:295–303. doi: 10.1038/nrd2176. [DOI] [PubMed] [Google Scholar]
- Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, Iwatsubo T, Jack CR, Jr., Kaye J, Montine TJ, Park DC, Reiman EM, Rowe CC, Siemers E, Stern Y, Yaffe K, Carrillo MC, Thies B, Morrison-Bogorad M, Wagster MV, Phelps CH. Toward definingthe preclinical stages ofAlzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimer's Dementia. 2011;7:280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer J. Making Sense of Multivariate Data Analysis. Sage Publications; Los Angeles: 2005. [Google Scholar]
- Vemuri P, Wiste HJ, Weigand SD, Shaw LM, Trojanowski JQ, Weiner MW, Knopman DS, Petersen RC, Jack CR., Jr. MRI and CSF biomarkers in normal, MCI, and AD subjects: predicting future clinical change. Neurology. 2009;73:294–301. doi: 10.1212/WNL.0b013e3181af79fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser PJ, Verhey F, Knol DL, Scheltens P, Wahlund LO, Freund-Levi Y, Tsolaki M, Minthon L, Wallin AK, Hampel H, Burger K, Pirttila T, Soininen H, Rikkert MO, Verbeek MM, Spiru L, Blennow K. Prevalence and prognosticvalue of CSF markers ofAlzheimer's disease pathology in patients with subjective cognitive impairment or mild cognitive impairment in the DESCRIPA study: a prospective cohort study. Lancet Neurology. 2009;8:619–627. doi: 10.1016/S1474-4422(09)70139-5. [DOI] [PubMed] [Google Scholar]
- Walhovd KB, Fjell AM, Brewer J, McEvoy LK, Fennema-Notestine C, Hagler DJ, Jr., Jennings RG, Karow D, Dale AM. Combining MR imaging, positron-emission tomography, and CSF biomarkers in the diagnosis and prognosis of Alzheimer disease. AJNR. American Journal of Neuroradiology. 2010;31:347–354. doi: 10.3174/ajnr.A1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee CY, Yap PT, Shen D. Prediction of Alzheimer's disease and mild cognitive impairment using cortical morphological patterns. Human Brain Mapping. 2013;34:3411–3425. doi: 10.1002/hbm.22156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, Harvey D, Jack CR, Jagust W, Liu E, Morris JC, Petersen RC, Saykin AJ, Schmidt ME, Shaw L, Siuciak JA, Soares H, Toga AW, Trojanowski JQ. The Alzheimer's Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimer's & Dementia. 2012;8:S1–68. doi: 10.1016/j.jalz.2011.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Przybelski SA, Weigand SD, Knopman DS, Boeve BF, Petersen RC, Jack CR., Jr. 3D maps from multiple MRI illustrate changing atrophy patterns as subjects progress from mild cognitive impairment to Alzheimer's disease. Brain. 2007;130:1777–1786. doi: 10.1093/brain/awm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolz R, Julkunen V, Koikkalainen J, Niskanen E, Zhang DP, Rueckert D, Soininen H, Lotjonen J. Multi-method analysis ofMRI images in early diagnostics ofAlzheimer's disease. PloS One. 2011;6:e25446. doi: 10.1371/journal.pone.0025446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Groth KM, Pearlson G, Schretlen DJ, Calhoun VD. Source-based morphometry: the use of independent component analysis to identify gray matter differences with application to schizophrenia. Human Brain Mapping. 2009;30:711–724. doi: 10.1002/hbm.20540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Wang Y, Thompson PM, Narayan VA, Ye J. Multi-source feature learning for joint analysis of incomplete multiple heterogeneous neuroimaging data. NeuroImage. 2012;61:622–632. doi: 10.1016/j.neuroimage.2012.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Shen D. Predicting future clinical changes ofMCI patients using longitudinal and multimodal biomarkers. PloS One. 2012;7:e33182. doi: 10.1371/journal.pone.0033182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.