Abstract
New approaches to optimizing cancer drug development in both the laboratory and the clinic will be required to fully achieve the goal of individualized, precision cancer therapy. Improved preclinical models that more closely reflect the now recognized genomic complexity of human cancers are needed. Here we describe a collaborative research project integrating core resources of The Jackson Laboratory (JAX) Basic Science Cancer Center with genomics and clinical research facilities at the UC Davis Comprehensive Cancer Center (UCD) to establish a clinically and genomically-annotated patient-derived xenograft (PDX) platform designed to enhance new drug development and strategies for targeted therapies. Advanced stage non-small cell lung cancer (NSCLC) was selected for initial studies due to emergence of a number of “druggable” molecular targets, and recent recognition of substantial inter- and intra-patient tumor heterogeneity. Additionally, clonal evolution after targeted therapy interventions makes this tumor type ideal for investigation of this platform. Employing the immunodeficient NSG mouse, over 200 NSCLC tumor biopsies have been xenotransplanted. During the annotation process, patient tumors and subsequent PDXs are compared at multiple levels, including histomorphology, clinically applicable molecular biomarkers, global gene expression patterns, gene copy number variations and DNA/chromosomal alterations. NSCLC PDXs are grouped into panels of interest by oncogene subtype and/or histologic subtype. Multi-regimen drug testing, paired with pre- and post-therapy next generation sequencing (NGS) and timed tumor pharmacodynamics, enables determination of efficacy, signaling pathway alterations and mechanisms of sensitivity-resistance in individual models. This approach should facilitate derivation of new therapeutic strategies and the transition to individualized therapy.
Introduction
Substantial advances have been made in understanding the molecular biology driving carcinogenesis as well as cancer-associated proliferative and anti-apoptotic signaling pathways. A wide variety of potentially “druggable” molecular targets for cancer therapy have emerged from these studies. While a large number of molecular-targeted agents have subsequently been tested, most of which showed substantial activity in available preclinical models, relatively few have been successful in the clinic (1, 2, 3, 4, 5). Thus, there remains an unmet need in drug development for new approaches toward testing patient-relevant treatment models in the laboratory, prior to or concurrent with initiation of clinical trials. The present preclinical evaluation process for new anti-cancer agents, based predominantly on human cancer cell lines or cell line-based xenograft models, has proven largely ineffective at predicting therapeutic potential in patients themselves (6, 7). Despite promising preclinical results, only five percent of cancer drugs under development are eventually approved for use. The majority fail due to lack of efficacy in Phase III clinical trials, as exemplified by recent trials for non-small cell lung cancer (NSCLC) (8). Chief among the many obstacles to development of effective anti-cancer regimens is the complexity of tumor signal transduction networks, with parallel pathways, crosstalk, compensatory feedback mechanisms and extensive interactions between tumor cells and their microenvironment, which cannot be replicated in conventional preclinical models taken in isolation. Adding to these complexities are the inter- and intra-patient tumor heterogeneity characteristic of human cancers and the innate adaptability of tumor mutator phenotypes, resulting in rapid development of resistance mechanisms. Taken together, these challenges necessitate new ways of thinking regarding the role of preclinical models in cancer drug development. (9)
Addressing an unmet need for improving drug development strategies in NSCLC
This need for improved preclinical platforms is particularly relevant to NSCLC, a worldwide health care epidemic for which most systemic therapies offer only modest benefit. For most patients with advanced NSCLC, the therapeutic decision-making process has remained largely empiric, based on factors such as historical treatment precedent, individual patient characteristics, and physician or patient preferences. Although the feasibility of selecting treatment for individual cancer patients based on tumor molecular profiles (personalized therapy or precision medicine) is already being explored in NSCLC, these efforts have been hampered considerably by tumor heterogeneity and the complexity of the underlying biologic pathways, as well as suboptimal preclinical models in which individualized therapeutic strategies can be tested [1, 3, 7]. Further, improved strategies to identify and overcome mechanisms of de novo and acquired resistance to treatment are essential to increasing survival and cure rates (10, 11, 12, 13).
Already, transgenic or knock-in preclinical systems such as genetically engineered mouse models (GEMMs) have proven to be invaluable tools for understanding carcinogenesis, tumor biology and target validation for therapeutics in ways not addressable with other modeling approaches, (14, 15, 16, 17, 18) Nevertheless, GEMMs represent non-human cancers lacking the range of tumor heterogeneity and complexity of biologic pathways inherent to human cancers and present within PDXs. While preliminary studies have already demonstrated that both chemotherapeutic agents and biologic therapies can be administered and assessed in a systematic fashion using in vivo xenotransplantation PDX models, systematic development, clinical-genomic annotation and drug testing paradigms have yet to be comprehensively evaluated. (19, 20, 21, 22, 23)
Development of the integrated UCD-JAX PDX Resource in NSCLC
Here we describe development of a clinically and genomically-annotated PDX resource in NSCLC and initial pilot projects focusing on epidermal growth factor receptor (EGFR) pathways and EGFR-directed therapies, to directly address and overcome these limitations. This NSCLC PDX research platform integrates core components of The Jackson Laboratory (JAX) NCI-designated Basic Science Cancer Center and the JAX In Vivo Pharmacology and Clinical Lab Services together with laboratory resources and the clinical research program of the NCI-designated UCD. The platform is designed to dynamically engage external institutions, investigators, additional organizations and pharmaceutical partners as appropriate on project-specific bases. Altogether, over 25 medical centers and other partners have participated in the Primary Human Tumor Consortium. Since the launching of the program in 2009, more than 1700 tumor specimens from individual cancer patients, including over 200 lung tumors, have been xenotransplanted into immunodeficient NOD scid gamma (NSG; NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice. Information and data about the PDX models are publicly accessible from the PDX portal at the Mouse Tumor Biology database (MTB; http://tumor.informatics.jax.org/mtbwi/pdxSearch.do) (24)).
JAX NSG model for Patient Derived Xenotransplantation
The JAX NSG model is of particular interest for PDX development. Lacking functional B and T cells and natural killer (NK) cell activity, the NSG mouse is the most immunodeficient yet physiologically durable murine model available for consistent xenoengraftment of human primary tumors [24]. These attributes of the NSG mouse model facilitate engraftment of a wide variety of human cancers, with excellent correlation of histomorphological and molecular features between PDX tumors and the original human cancers as described below. Ongoing studies at JAX are focusing on optimizing the engraftment algorithm, such as use of small biopsy samples acquired through core needle biopsy (minimum of 1 mm3 of tissue) and “transportability” of viable patient tumor specimens from other institutions. For example, engraftment rates appear similar between those specimens acquired locally in Sacramento versus those express-shipped overnight from sites across the country.
Integrated Clinical and Genomic Annotation of patient tumor and corresponding PDX
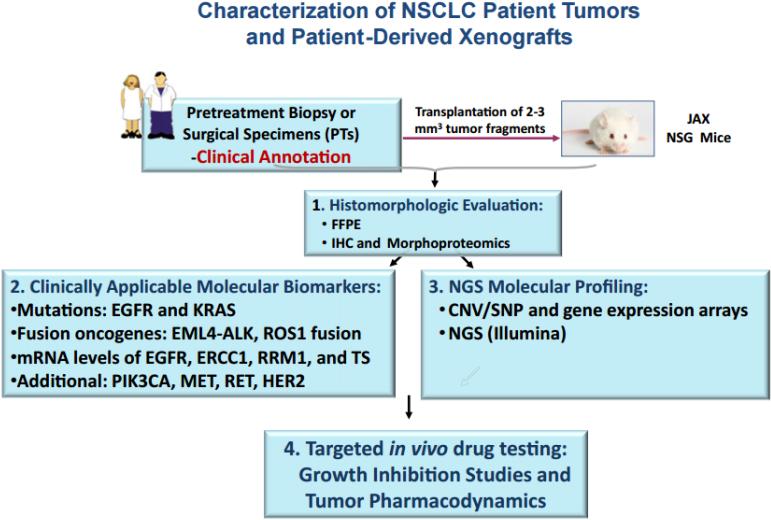
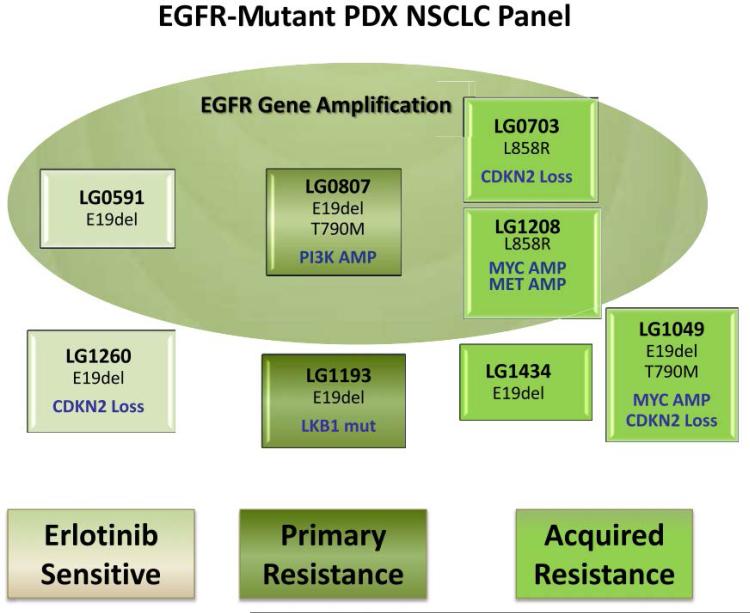
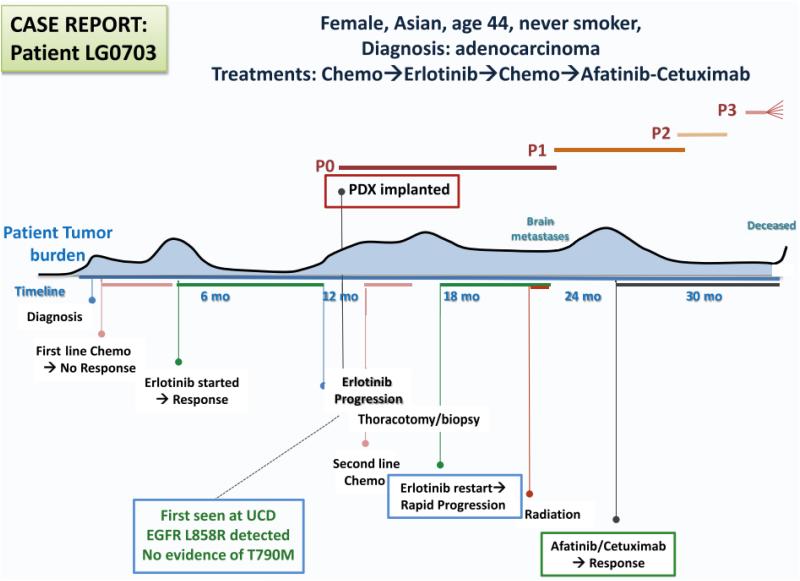
The algorithm for PDX creation and analysis following patient tumor biopsy is shown in Figure 1. In a synchronized evaluation process, both patient tumors (PTs) and subsequent PDXs undergo histomorphologic assessment by a single reference pathologist, biomarker testing through the CLIA laboratory at Response Genetics, Inc., and genomic analysis by multiple platforms including gene expression arrays and next generation sequencing. Those PDXs of interest, based on molecular subtyping (e.g. EGFR-mutated, KRAS-mutated, squamous, etc) are then grouped into panels for subsequent drug testing. Figure 2 demonstrates how a panel (e.g. EGFR-mutated) is sub-grouped by mutation type, resistance mechanism (if known) and clinical annotation (sensitive to EGFR tyrosine kinase inhibitor [TKI], de novo resistance or acquired resistance). Patient clinical annotation is tracked (case example as shown in Figure 3) to account for all pertinent patient demographics, including smoking status, treatment records and timing of PDX creation. Mutational fidelity is demonstrated in Table 1 for a representative group of PTs and PDXs characterized for EGFR and KRAS mutations and ALK gene rearrangements in a CLIA laboratory environment (Response Genetics, INC). (25, 26)
Figure 1.
Algorithm of Patient-Derived Xenograft (PDX) Creation. Candidate Patients (PTs) Were Identified and Provided Consent for Tumor Collection. After Biopsy, Pleural Effusion Fluid Collection, or Surgical Resection, Viable Portions of the Fresh Specimens Were Rapidly Transported to the Jackson Laboratories-West Facilities for Implantation Into the NSG Mouse Model. Concurrently, Remaining Portions of the PT Specimen Were Fixed and Subsequently Characterized and Molecularly Profiled. A PDX Model Was Considered “Established” After Demonstrating Growth in Passage 1, After Successful Implantation, Development and Transplantation From Passage 0. Histomorphologic Evaluation and Molecular Profiling of the PDX Model Was Conducted and Results Compared With the Contributing Human Specimen. When PDX Models Reach Passage 2, Cohorts Can Be Prepared for Growth Inhibition and Tumor Pharmacodynamic Studies. Abbreviations: CNV = copy number variation; FFPE = Formalin-fixed, paraffin-embedded; IHC = immunohistochemistry; NGS = next-generation sequencing; NSCLC = non–small-cell lung cancer; SNP = single-nucleotide polymorphism.
Figure 2.
Panel of Non–Small-Cell Lung Cancer (NSCLC) Patient-Derived Xenograft (PDX) Models That Harbor Epidermal Growth Factor Receptor (EGFR)-Activating Mutations. Models Are Organized According to Patient Clinical Status at the Time of the Originating Biopsy. “Erlotinib Sensitive-Naive” Models Were Acquired Before Treatment With Any EGFR Inhibitor; “Primary Resistance” Models Were Derived From Tumors That Showed No Clinical Benefit From Erlotinib; “Acquired Resistance” Models Were Derived From Tumors That Initially Responded to Erlotinib But Had Progressed at the Time of Biopsy. Models Were Also Subdivided According To Those With EGFR Gene Amplification (Upper Circle). Additional Information Is Provided in Each Box, Indicating Oncogenic Abnormalities of Interest. Abbreviations: AMP = amplification; mut = mutation
Figure 3.
Case Example of Clinical Annotation of Patient-Derived Xenograft (PDX) Models. A Large Amount of Anonymized, Annotated Patient Information Is Collected From Each Consenting Donor on PDX Implantation. This Information Includes Age, Sex, Race, Diagnosis, Course of Disease, and Previous Treatment, Known Mutation Status, Confounding Medical Issues, Family History of Cancer, Timing of the Establishment of PDX Relative to Treatment, and Therapeutic Outcomes. The Figure Shows Estimated Disease Burden of the Patient Who Contributed the LG0703 model. As Indicated, the PDX Was Established From a Biopsy at the Time of Disease Progression After a Successful Course of Erlotinib. The Patient Subsequently Went on to Receive the Combination of Afatinib With Cetuximab, With a Durable Response. Abbreviations: Chemo = chemotherapy; P0-3 = Passage 0-3.
Table 1.
Fidelity between PT and PDX. Abbreviations: PDX=patient-derived xenograft; PT=patient tumor.
| Mutational Status | |||
|---|---|---|---|
| EML4-ALK Fusion Transcript |
K-RAS Mutation | EGFR Mutation | |
| LG-0476 | |||
| PT | No translocation | Wild type | Wild type |
| PDX | No translocation | Wild type | Wild type |
| LG-0481 | |||
| PT | No translocation | Gly12Cys | Wild type |
| PDX | No translocation | Gly12Cys | Wild type |
| LG-0703 | |||
| PT | No translocation | Wild type | EGFR L858R |
| PDX | No translocation | Wild type | EGFR L858R |
| LG-1193 | |||
| PT | No translocation | Wild type | EGFR E19del |
| PDX | No translocation | Wild type | EGFR E19del |
| LG-0812 | |||
| PT | Fusion-positive | Wild type | Wild type |
| PDX | Fusion-positive | Wild type | Wild type |
| LG-0552 | |||
| PT | No translocation | Gly12Ala | Wild type |
| PDX | No translocation | Gly12Ala | Wild type |
| LG-0567 | |||
| PT | No translocation | Gly12Cys | Wild type |
| PDX | No translocation | Gly12Cys | Wild type |
Drug Testing Paradigm
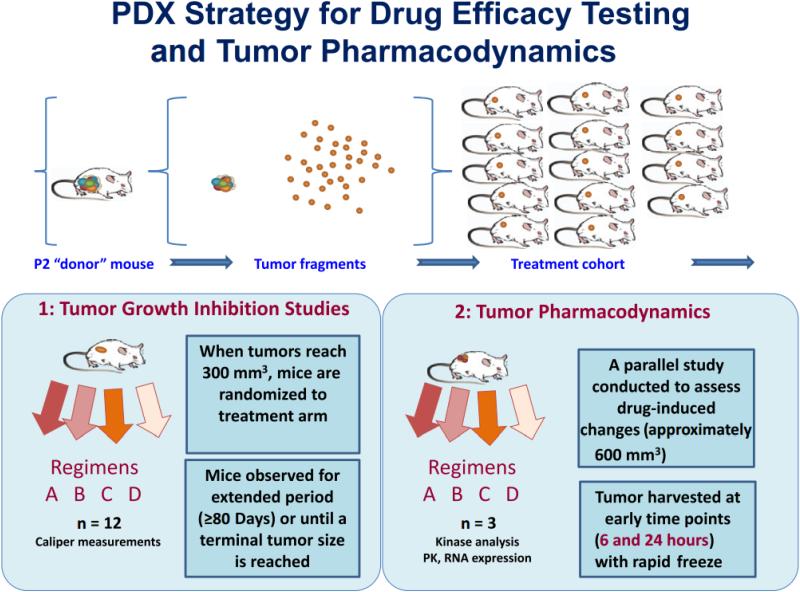
A variety of projects are ongoing testing PDX panels of interest for new drug treatment strategies, and to determine mechanisms of resistance and how to overcome them. A combination of tumor growth inhibition studies and real-time tumor pharmacodynamics are incorporated into each drug testing project. (Figure 4) An advantage of this testing paradigm in these is that degrees of responsiveness can be quantitatively measured providing a more nuanced assessment of pharmacologic effect of any drug or drug combination. Moreover, the availability of an annotated panel of xenografts for a particular genetic mutation or histology should increase the likelihood of l discovery of new genomic signatures of drug sensitivity and resistance. The use of these models in co-clinical trials is of particular interest.
Figure 4.
Diagram of Experimental Approach to Testing Drug Efficacy in Patient-Derived Xenograft (PDX) Models. Tumor Fragments From a Single Passage 2 (P2) Mouse Were Implanted Simultaneously Into a Treatment Cohort of NSG Mice. As Tumors Reach 300 mm3, They Are Randomized Into Arms and Receive Treatment for the Indicated Number of Cycles. All Treated Mice Are Further Observed for an Extended Period of Time or Until the Tumor Reached a Terminal Size and Mice Were Euthanized. In Parallel, Pharmacodynamic Studies Were Conducted, With Mice Treated for Short Periods (For Instance, 6 or 24 Hours) to Examine the Immediate Molecular Effects of Treatment Such as Degree of Target Inhibition and Effects on Signal Transduction. Tumors Were Harvested Using a Rapid Resection Protocol Designed to Minimize Tumor Ischemia. Abbreviation: PK = protein kinase.
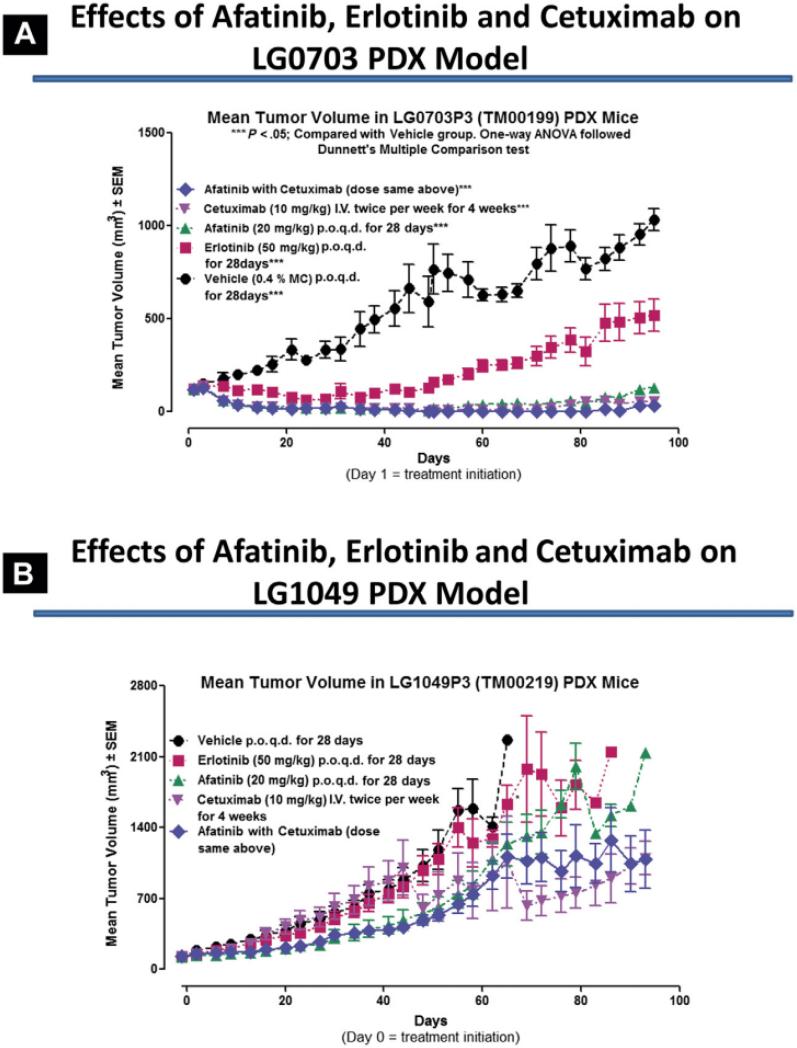
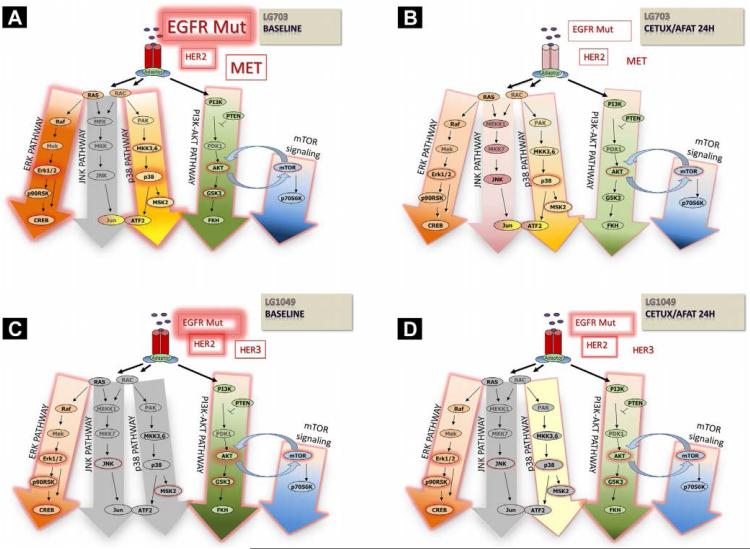
An ongoing pilot study investigating mechanisms of acquired drug resistance in EGFR-mutated lung cancer, for example, is utilizing a panel of EGFR-mutated PDXs to investigate a drug regimen of afatinib with or without cetuximab, using erlotinib and vehicle as controls. Results in two PDX models, one in which the host patient proved to subsequently be responsive to the afatinib-cetuximab combination while the other patient was unresponsive, mimicked the clinical outcomes, as shown in Figure 5. Associated tumor pharmacodynamics (Figure 6) illustrates abrogation of multiple signal transduction pathways globally mediated by maximal EGFR inhibition in the sensitive model (LG0703), in contrast to incomplete EGFR inhibition coupled with compensatory upregulation of survival pathways in the resistant model (LG1049). These data suggest that PDX models can recapitulate drug treatment outcomes seen in patients from whom the PDXs were derived, and that investigations can be designed to increase understanding of underlying biologic pathways and to devise strategies to improve effectiveness of therapy. This hypothesis will be tested in a future SWOG Phase II/III clinical trial (S1403) comparing afatinib with or without cetuximab in the 1st line therapy of patients with EGFR-mutated lung cancer, in which selected patients will undergo repeat tumor biopsy at the time of progressive disease, for genomic assessment and for PDX creation and drug testing.
Figure 5.
Activity of Epidermal Growth Factor Receptor (EGFR) Inhibitors in EGFR-Mutant Patient-Derived Xenograft (PDX) Models. The Relative Tumor Growth Inhibition Induced By Erlotinib, Afatinib, Cetuximab or the Combination of Afatinib With Cetuximab Was Tested in 2 PDX Models Derived From Erlotinib-Refractory Patients Who Subsequently (After PDX Biopsy) Were Treated With the Combination of Afatinib With Cetuximab. (A) The Patient From Whom the PDX LG0703 Was Derived (See Figure 3) Was Responsive to Subsequent Therapy With Afatinib With Cetuximab. This Combination Proved to Be Effective in The PDX Model, Recapitulating the Clinical Results. (B) In Contrast, PDX LG1049, Derived From a T790M-Positive Cancer Patient Who Did Not Respond to Subsequent Cetuximab With Afatinib Therapy, Exhibited No Significant Response to Afatinib With Cetuximab. Again, Clinical Results Were Recapitulated in the Related PDX Models. Abbreviations: ANOVA = Analysis of Variance; p.o. = orally; q.d. = once per day.
Figure 6.
Comparison of Tumor Pharmacodynamic Effects After Afatinib/Cetuximab in Two Erlotinib-Resistant Patient-Derived Xenograft (PDX) Models LG0703 (T790M-Negative) and LG1049 (T790M-Positive). (A) LG0703 Untreated, Revealing Intense Signaling Through ERK and p38. (B) LG0703 After Treatment With Afatinib/Cetuximab for 24 Hours, Showing Substantially Diminished MAPK and PI3K-AKT Signaling Concurrent With Loss of Epidermal Growth Factor Receptor (EGFR) Phosphorylation. (C) LG1049 Untreated, Showing Enhanced AKT Activity With Moderate ERK Activity at Baseline. (D) LG1049 After Treatment With Afatinib/Cetuximab Showing Minimal Diminishment of EGFR Phosphorylation and Signal Transduction, With Compensatory Upregulation of p38. Phosphorylation States of Receptors and Signaling Intermediaries Were Measured Using A Combination of immunoblot and Kinase Arrays (Data Not Shown, Publication In Preparation). Abbreviations: ERK = extracellular signal-regulated kinase; MAPK = Mitogen-activated protein kinase; mTor = mammalian target of rapamycin; Mut = mutant; PI3K = phosphatidylinositol-3-kinase.
Summary
The integrated preclinical-clinical modeling strategy described here, utilizing a large annotated NSG mouse resource of PDX models, provides a novel research tool for drug development and for understanding the genomic complexity of of lung cancer. Prospective development of PDXs from mutation-specific cohorts of patients treated in a homogeneous fashion, such as the planned PDX project associated with the S1403 clinical trial, should provide a unique resource for future study. It is anticipated that availability of such an annotated resource will assist in optimizing therapeutic strategies and lead to improved patient outcomes.
Acknowledgements
Research reported in this publication was partially supported by the National Cancer Institute award to UCDCC [P30CA034196] by The Jackson Laboratory Director’s Innovation Fund (DIF), the Addario Lung Cancer Foundation and the HOPE Foundation. The MTB database is supported by the National Institutes of Health [CA089713].
Abbreviations
- JAX
The Jackson Laboratory
- UCD
University of California Davis Comprehensive Cancer Center
- NSG
Nod SCID Gamma
- CLIA
Clinical Laboratory Improvement Amendments
References
- 1.Collins I, Workman P. New approaches to molecular cancer therapeutics. Nat Chem Biol. 2006;(12):689–700. doi: 10.1038/nchembio840. [DOI] [PubMed] [Google Scholar]
- 2.Workman P. Genomics and the second golden era of cancer drug development. Mol Biosyst. 2005;1(1):17–26. doi: 10.1039/b501751n. [DOI] [PubMed] [Google Scholar]
- 3.Daigo Y, Takano A, Teramoto K, Chung S, Nakamura Y. A systematic approach to the development of novel therapeutics for lung cancer using genomic analyses. Clin Pharmacol Ther. 2013;94(2):218–23. doi: 10.1038/clpt.2013.90. [DOI] [PubMed] [Google Scholar]
- 4.Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR, Schacht AL. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nat Rev Drug Discov. 2010;9(3):203–14. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 5.Hutchinson L, Kirk R. High drug attrition rates--where are we going wrong? Nat Rev Clin Oncol. 2011;(4):189–90. doi: 10.1038/nrclinonc.2011.34. [DOI] [PubMed] [Google Scholar]
- 6.Rubin EH, Gilliland DG. Drug development and clinical trials--the path to an approved cancer drug. Nat Rev Clin Oncol. 2012;9(4):215–22. doi: 10.1038/nrclinonc.2012.22. [DOI] [PubMed] [Google Scholar]
- 7.Li T, Kung HJ, Mack PC, Gandara DR. Genotyping and genomic profiling of non-small-cell lung cancer: implications for current and future therapies. J Clin Oncol. 2013;10;31(8):1039–49. doi: 10.1200/JCO.2012.45.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gandara DR, Li T, Lara PN, Jr, Mack PC, Kelly K, Miyamoto S, Goodwin N, Beckett L, Redman MW. Algorithm for codevelopment of new drug-predictive biomarker combinations: accounting for inter- and intrapatient tumor heterogeneity. Clin Lung Cancer. 2012;13(5):321–5. doi: 10.1016/j.cllc.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins FS. Reengineering translational science: the time is right. Sci Transl Med. 2011;3(90):90cm17. doi: 10.1126/scitranslmed.3002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosell R, Karachaliou N, Morales-Espinosa D, Costa C, Molina MA, Sansano I, Gasco A, Viteri S, Massuti B, Wei J, González Cao M, Martínez-Bueno A. Adaptive resistance to targeted therapies in cancer. Transl Lung Cancer Res. 2013;2(3):152–159. doi: 10.3978/j.issn.2218-6751.2012.12.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gandara DR, Li T, Lara PN, Kelly K, Riess JW, Redman MW, Mack PC. Acquired resistance to targeted therapies against oncogene-driven non-small-cell lung cancer: approach to subtyping progressive disease and clinical implications. Clin Lung Cancer. 2014;15(1):1–6. doi: 10.1016/j.cllc.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gower A, Wang Y, Giaccone G. Oncogenic drivers, targeted therapies, and acquired resistance in non-small-cell lung cancer. J Mol Med (Berl) 2014;92(7):697–707. doi: 10.1007/s00109-014-1165-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwasa Y, Michor F. Evolutionary dynamics of intratumor heterogeneity. PLoS One. 2011;30;6(3):e17866. doi: 10.1371/journal.pone.0017866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Politi K, Pao W. How genetically engineered mouse tumor models provide insights into human cancers. J Clin Oncol. 2011;1;29(16):2273–81. doi: 10.1200/JCO.2010.30.8304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carver BS, Pandolfi PP. Mouse modeling in oncologic preclinical and translational research. Clin Cancer Res. 2006;12(18):5305–11. doi: 10.1158/1078-0432.CCR-06-0482. [DOI] [PubMed] [Google Scholar]
- 16.Sharpless NE, Depinho RA. The mighty mouse: genetically engineered mouse models in cancer drug development. Nat Rev Drug Discov. 2006;5(9):741–54. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- 17.Regales L, Balak MN, Gong Y, Politi K, Sawai A, Le C, Koutcher JA, Solit DB, Rosen N, Zakowski MF, Pao W. Development of new mouse lung tumor models expressing EGFR T790M mutants associated with clinical resistance to kinase inhibitors. PLoS One. 2007;2(8):e810. doi: 10.1371/journal.pone.0000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weaver Z, Difilippantonio S, Carretero J, Martin PL, El Meskini R, Iacovelli AJ, Gumprecht M, Kulaga A, Guerin T, Schlomer J, Baran M, Kozlov S, McCann T, Mena S, Al-Shahrour F, Alexander D, Wong KK, Van Dyke T. Temporal molecular and biological assessment of an erlotinib-resistant lung adenocarcinoma model reveals markers of tumor progression and treatment response. Cancer Res. 2012;72(22):5921–33. doi: 10.1158/0008-5472.CAN-12-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, Arcaroli JJ, Messersmith WA, Eckhardt SG. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol. 2012;9(6):338–50. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shultz LD, Ishikawa F, Greiner DL. Humanized mice in translational biomedical research. Nat Rev Immunol. 2007;7(2):118–30. doi: 10.1038/nri2017. [DOI] [PubMed] [Google Scholar]
- 21.Olive KP, Tuveson DA. The use of targeted mouse models for preclinical testing of novel cancer therapeutics. Clin Cancer Res. 2006;12(18):5277–87. doi: 10.1158/1078-0432.CCR-06-0436. [DOI] [PubMed] [Google Scholar]
- 22.Hidalgo M, Bruckheimer E, Rajeshkumar NV, Garrido-Laguna I, De Oliveira E, Rubio-Viqueira B, Strawn S, Wick MJ, Martell J, Sidransky D. A pilot clinical study of treatment guided by personalized tumorgrafts in patients with advanced cancer. Mol Cancer Ther. 2011;10(8):1311–6. doi: 10.1158/1535-7163.MCT-11-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.John T, Kohler D, Pintilie M, Yanagawa N, Pham NA, Li M, Panchal D, Hui F, Meng F, Shepherd FA, Tsao MS. The ability to form primary tumor xenografts is predictive of increased risk of disease recurrence in early-stage non-small cell lung cancer. Clin Cancer Res. 2011;17(1):134–41. doi: 10.1158/1078-0432.CCR-10-2224. [DOI] [PubMed] [Google Scholar]
- 24.Bult CJ, Krupke DM, Begley DA, Richardson JE, Neuhauser SB, Sundberg JP, Eppig JT. Mouse Tumor Biology (MTB): a database of mouse models for human cancer. Nucleic Acids Res. 2014 Oct 20; doi: 10.1093/nar/gku987. pii: gku987. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gandara DR, Grimminger P, Mack PC, Lara PN, Jr, Li T, Danenberg PV, Danenberg KD. Association of epidermal growth factor receptor activating mutations with low ERCC1 gene expression in non-small cell lung cancer. J Thorac Oncol. 2010;5(12):1933–8. doi: 10.1097/JTO.0b013e3181fd418d. [DOI] [PubMed] [Google Scholar]
- 26.Maus MK, Grimminger PP, Mack PC, Astrow SH, Stephens C, Zeger G, Hsiang J, Brabender J, Friedrich M, Alakus H, Hölscher AH, Lara P, Danenberg KD, Lenz HJ, Gandara DR. KRAS mutations in non-small-cell lung cancer and colorectal cancer: implications for EGFR-targeted therapies. Lung Cancer. 2014;83(2):163–7. doi: 10.1016/j.lungcan.2013.11.010. [DOI] [PubMed] [Google Scholar]